Abstract
Alteration of lineage-specific transcriptional programs for hematopoiesis causes differentiation block and promotes leukemia development. Here, we show that AML1/ETO, the most common translocation fusion product in acute myeloid leukemia (AML), counteracts the activity of retinoic acid (RA), a transcriptional regulator of myelopoiesis. AML1/ETO participates in a protein complex with the RA receptor alpha (RARα) at RA regulatory regions on RARβ2, which is a key RA target gene mediating RA activity/resistance in cells. At these sites, AML1/ETO recruits histone deacetylase, DNA methyltransferase, and DNA-methyl-CpG binding activities that promote a repressed chromatin conformation. The link among AML1/ETO, heterochromatic RARβ2 repression, RA resistance, and myeloid differentiation block is indicated by the ability of either siRNA-AML1/ETO or the DNA methylation inhibitor 5-azacytidine to revert these epigenetic alterations and to restore RA differentiation response in AML1/ETO blasts. Finally, RARβ2 is commonly silenced by hypermethylation in primary AML blasts but not in normal hematopoietic precursors, thus suggesting a role for the epigenetic repression of the RA signaling pathway in myeloid leukemogenesis.
Introduction
The postgenomic era has shown that correct gene expression is regulated by epigenetic mechanisms, such as DNA methylation, posttranslational modifications of histone proteins, remodeling of nucleosomes, and expression of small regulatory RNAs. These events are essential during development and for the maintenance of tissue- and cell-type–specific functions.1–3 The contribution of epigenetic mechanisms for a correct cell function is highlighted by the effects of their deregulation that in cooperation with genetic alterations lead to the establishment and progression of tumors.4–9
In vivo and in vitro models of hematopoiesis indicate a physiological role for retinoic acid (RA), a natural derivative of vitamin A, in regulating myelopoiesis through its binding to the RA receptor alpha (RARα).10–13 RARα is a member of the RA nuclear receptor family (also including RARβ and RARγ) that acts as a ligand-inducible transcription factor by binding to specific response elements (RAREs) in the regulatory regions of target genes.14 Here, the heterodimer RARα–retinoid X receptor (RXR) recruits a corepressor complex containing histone deacetylase (HDAC) activities that result in local chromatin condensation and transcriptional repression. Physiological concentrations of RA (1 to 10 nM) cause the exchange of the corepressor complex from the RARα-RXR, with transcriptional adapters and coactivators with intrinsic histone acetyltransferase activities. This causes histone hyperacetylation at RARE sites, chromatin remodeling, and transcriptional activation of RARα target genes.14 Among the RARα target genes, RARβ2, the RA-inducible splicing isoform of the RARβ gene, is a biochemically and biologically relevant model. It contains in its promoter the strongest natural RA response element (βRARE).14,15 Moreover, RARβ2 is an RA-regulated tumor suppressor gene silenced by aberrant DNA methylation in a variety of human malignancies and in acute promyelocytic leukemia (APL), the acute myeloid leukemia (AML)–M3 subtype by the French-American-British (FAB) classification.8,16–19
In APL, the aberrant DNA methylation of RARβ2 and consequent gene silencing is caused by the leukemogenetic PML/RARα fusion protein generated by the t(15;17) translocation involving the PML and the RARα genes. By forming oligomeric structures, PML/RARα stably recruits a corepressor complex containing HDAC, histone methyltransferase (HMT), DNA methyltransferase (DNMT), and methyl-CpG binding activities at the βRARE site on the RARβ2 gene. Therefore, PML/RARα acts as a constitutive transcriptional repressor of RARα target genes. Pharmacologic doses of RA, however, release this repressor complex from PML/RARα and provoke histone hyperacetylation and DNA demethylation at these RARβ2 promoter sites, restoring APL blast differentiation in vitro and in vivo.6,18–25 To date, APL represents a paradigm for differentiation therapy of cancer.
The t(8;21) generating the AML1/ETO fusion product is one of the most common genetic events associated with AML. This chromosomal translocation is present in up to 40% in AML FAB M2 cases and, at a lower frequency, in AML-M4 and other subsets.26–28 The AML1/ETO product shares several features with PML/RARα, therefore suggesting unifying pathogenic mechanistic links in AML. Similarly to PML/RARα, the oncogenic effect of AML1/ETO is mainly linked to its ability to form oligomeric complexes with an increased affinity for HDACs and DNMT1, which render AML1/ETO a potent transcriptional repressor of AML1-target genes.29–35 However, besides the direct targets of AML1, other transcriptional regulators may be aberrantly modulated in AML1/ETO-AMLs.26,27
Non-APL AMLs are resistant to the differentiating action of RA.36 A dysfunctional control of RA activity in myelopoiesis might represent per se an oncogenic hit relevant to the development and progression of leukemia. In agreement with this hypothesis, we have shown that the RA signaling pathway is silenced in AMLs regardless of their underlying genetic lesion.37,38
In this study, we identify the epigenetic silencing of the RA signaling pathway as an additional genomic alteration caused by the AML1/ETO fusion product in myeloid cells. Here, we show AML1/ETO and RARα as “in vivo” components of a protein complex recruiting HDAC1/DNMT/MeCP2 activities on regulatory sites of the RARβ2 gene, resulting in its heterochromatic transcriptional silencing. Our findings also link the hypermethylation of specific CpG dinucleotides and repressive chromatin status at these sites of RARβ2 to RA resistance and differentiation block in AMLs.
Materials and methods
Reagents
All-trans-retinoic acid (RA) and 5-azacytidine were from Sigma-Aldrich (Milan, Italy) and both used at a concentration of 1μM.
Clinical samples
Bone marrow (BM) and/or peripheral blood (PB) samples were obtained from 20 informed, newly diagnosed AML patients showing a percentage of BM blasts of at least 80%. AML cases were classified as AML-M2 (9 cases), AML-M3 (1 case), and AML-M4 (10 cases) according to the FAB classification.16 Blasts isolation and molecular analysis to evaluate the presence of the AML-associated fusion genes were performed as described.39 CD34+ cells were isolated from the BM of informed healthy donors as reported.40
Cell lines and cell cultures
The U937–A/E-HA clone was obtained by electroporation into U937 wild-type (WT) cells of a hemagglutinin (HA)–tagged AML1/ETO cDNA subcloned into a vector carrying the Zn2+-inducible mouse MT1 promoter as described.29,37 Mock cells were U937 WT cells transfected with an empty MT1 promoter vector. The leakiness of the MT1 promoter was used to select the U937 MT-MHA-AE clone 9, which in the absence of Zn2+ expressed an amount of AML1/ETO fusion product at levels comparable to those expressed by the t(8;21) SKNO-1 cell line,41 as detected by Western blot analysis using an α-AML1/RHD (PC285) antibody (Oncogene Science, Boston MA). Similarly to SKNO-1, the U937 MT-MHA-AE clone 9 was viable and tolerated the expression of the fusion product without relevant apoptosis. Apoptosis was quantified by propidium iodide staining (50 μg/mL) of permeabilized cells using a Epics XL Cytometer (Beckman Coulter, Hialeah, FL) as described.42 The relative percentages of apoptotic cells measured in these cells were as follows: U937-MT, 0.8%; U937 MT-MHA-AE, 0.5%; SKNO-1, 0.7%. Cell lines were maintained in RPMI 1640 medium supplemented with 10% FCS at 37°C.
Immunophenotypic analysis
Cell differentiation was evaluated by direct immunofluorescence staining of cells using an allophycocyanin (APC)–conjugated mouse anti–human CD11b antibody and a peridinin chlorophyll protein (PerCP)– conjugated mouse anti–human CD14 antibody (Becton Dickinson, San Jose, CA). A minimum of 50 000 events was recorded for each sample by a FACScan flow cytometer (Becton Dickinson) using CellFit software (Becton Dickinson) for data acquisition and analysis.
Assay for RA binding activity
Nuclear extracts were prepared from 2 × 106 cells, incubated for 18 hours at 4°C with 10 nM [3H]-RA, and fractionated at 4°C by high-performance liquid chromatography (HPLC) using a Superpose 6 HR 10/30 column (Pharmacia, Milan, Italy) at a flow rate of 0.4 mL/min as described.43
Plasmid constructs and siRNA assay
The sequence of the siRNA oligonucleotide siAGF144 was used to derive a short hairpin that, upon cloning into the plasmid psiUx,45 allowed the stable expression of siRNAs against the AML1/ETO mRNA fusion (siA/E-RNAs). This psiU-derived expression cassette was subcloned into the long terminal repeat (LTR) of the lentiviral vector pRRLcPPT.hPGK.EGFP.WPRE. Cells were infected with empty control vector (mock) or vector encoding the siAGF1 oligonucleotide (siA/E) and purified by fluorescence-activated cell sorting (FACS) as reported.46
Transient cotransfection and transactivation assays
Human embryonic kidney 293T cells were transiently cotransfected by the Ca3(PO4)2 method with 20 ng or 40 ng of the pcDNA3 vector containing the HA-tagged AML1/ETO cDNA29 and 200 ng of the −5kb+155bp RARβ2pr-LUC reporter15,37 carrying (RAREmut) or not (RAREwt) the 5′-GGTTCAC-3′ direct motif of the βRARE site substituted with the 5′-aacTCAC-3′ sequence. A cotransfected plasmid encoding β-galactosidase (pCMV-βgal) was used as a control for normalization of the reactions. Twelve hours after transfection, cells were treated with 1 μM RA for 24 hours, lysed, and assayed using the Luciferase Assay Kit (Promega, Milan, Italy).
RNA extraction and analysis
Total RNA was extracted from cells using the TRIzol RNA isolation system (Invitrogen, Milan, Italy). Northern blot analysis to evaluate the siRNA oligo (siAGF1) expression was performed using 5 μg total RNA electrophoresed in a 10% polyacrylamide-7M urea gel and transferred by electroblotting onto a HybondN+ membrane (Amersham Biosciences, Milan, Italy). Hybridization was performed with terminally 32 P-labeled DNA oligonucleotides as described.44,46 U2 snRNA expression levels were measured to normalize for RNA content among samples.46
The relative quantity of RARβ2 mRNA was measured on 1 μg total RNA by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) in the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Monza, Italy), and it was determined by the comparative CT method using GAPDH mRNA levels for normalization as reported.19 AML1/ETO mRNA levels were normalized using c-abl mRNA quantity values and quantified by the absolute standard curve method.39 Semiquantitative RT-PCR was performed to evaluate the RARβ2 gene expression in AML blasts.19,37
Western blot and coimmunoprecipitation assays
Western blot analyses were performed on total cell lysates (50 μg) using the following rabbit polyclonal IgG antibodies: anti-RARα, anti-RXR (Santa Cruz Biotechnology, Santa Cruz, CA), and anti–AML1/RHD-PC285 (Oncogene Science). The anti–β-tubulin mouse monoclonal IgG (Sigma-Aldrich) was used to normalize the amount of the samples analyzed. The immunoreactivity was determined by the enhanced chemiluminescence (ECL) method (Amersham Biosciences). Coimmunoprecipitation experiments were performed as described.47 Rabbit polyclonal IgG (Santa Cruz Biotechnology) was used as a nonspecific antibody. The secondary antibody employed for these detections was the antirabbit polyclonal IgG provided by the TrueBlot Set (eBioscience, San Diego, CA) that preferentially detects the nonreduced form of rabbit IgG over the reduced SDS-denaturated forms of IgG.
Chromatin immunoprecipitation assay
Crosslinking of proteins to DNA was obtained by the addition to cultured cells (2 × 106) of formaldehyde at 1% final concentration for 10 minutes at 37°C. After sonication, the chromatin was immunoprecipitated overnight with 5 μL of the following antibodies recognizing RARα, RXRα, HDAC1 (Santa Cruz Biotechnology), HA monoclonal (Babco, Richmond, CA), AML1/RHD-PC285, ETO (Ab-1) (Oncogene Science), DNMT1 (New England BioLabs, Ipswich, MA), DNMT3a and DNMT3b (Abcam, Cambridge, United Kingdom), MeCP2, acetyl-histone-4, acetyl-histone-3, acetyl-histone-3-Lys9, methyl-histone-3-Lys9 (Upstate Biotechnology, Lake Placid, NY). PCR for RARβ2, p14ARF, p16INK4a, interleukin-3 (IL-3) promoter, IL-3 exon 7, and GAPDH was performed using the conditions and primers already described.19,33,35 Chromatin immunoprecipitation (ChIP) using the cytosine(5-methyl) (Abcam) antibody was performed on naked and sonicated DNA extracted from the same cell samples. Genomic regions of about 200 bp of a RARβ distal gene region (exon 7) were amplified with the primer sequences designed by Primer Express software (Applied Biosystems): (fwd) 5′-AATTCCAGTGCTGACCATCG-3′ and (rev) 5′-GCCTTCAGCAGGGTAATTTG-3′.
Bisulfite sequencing and methylation-specific PCR assays
We digested 5 μg of DNA extracted from SKNO-1, si-A/E, U937, A/E-HA cells, and human BM samples with 5 units of EcoRV. Sodium bisulfite treatment was performed as described.48 For bisulfite sequencing assay, a fragment of 608 bp (nucleotides [nt] −370 to +238) of the RARβ2 promoter/exon 1 region was amplified using specific primer pairs as described.18,19,49 Single-band PCR products were gel purified and cloned into the TOPO TA Cloning/pCR2.1 TOPO kit (Invitrogen). We subjected individual bacterial colonies to PCR using vector-specific primers (sequences available upon request), and the products were sequenced for the analyses of DNA methylation. Methylation-specific PCR (MSP) assay was performed on bisulfite-treated genomic DNA as described.18,19
Results
AML1/ETO knockdown restores the transcriptional and differentiation response of t(8;21) AMLs to RA
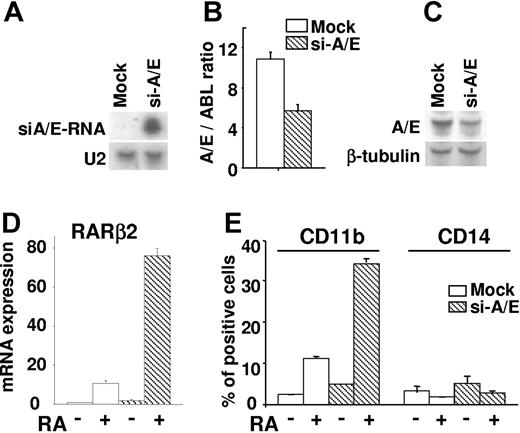
We investigated the consequences of the AML1/ETO knockdown on the response of the SKNO-1 cell line to RA. SKNO-1 is a human AML cell line that is resistant to the differentiation effect of RA and harbors the chromosomal translocation t(8;21) and the AML1/ETO fusion product.41,47 AML1/ETO mRNA interference was obtained by the infection of SKNO-1 cells with a GFP lentiviral vector expressing siRNA against the AML1/ETO mRNA (siA/E-RNA).44,46,50 Northern blot analysis revealed that the siA/E-RNA transcripts were specifically expressed in SKNO-1 cells (si-A/E) (Figure 1A), while qRT-PCR and Western blot analyses assessed a diminution of about 50% in the AML1/ETO mRNA and fusion protein levels in si-A/E cells as compared with mock-infected cells (Figure 1B-C). Notably, this reduction of the AML1/ETO product was sufficient to restore the transcriptional action of RA as indicated by the potent induction of the endogenous RARβ2 mRNA transcripts measurable by qRT-PCR in RA-treated si-A/E cells (Figure 1D). The AML1/ETO knockdown also restored the differentiation response of SKNO-1 cells to RA as shown by the specific induction of the expression levels of CD11b in RA-treated si-A/E cells, while the levels of CD14 remained unchanged (Figure 1E). These data suggested that threshold levels of AML1/ETO are required for the silencing of the RA signaling pathway and for inducing the differentiation block, which occurs toward the granulocytic lineage. In agreement with evidence showing the epigenetic silencing of RARβ2 as the basis of RA resistance in cells,51 our findings pointed at the RA-induced maturation of SKNO-1 cells as the consequence of a combined effect on RARβ2 gene promoter: transcriptional derepression caused by the reduced AML1/ETO levels and transcriptional activation induced by RA treatment.
AML1/ETO siRNAs support differentiation response to RA of human t(8;21)-positive cells (SKNO-1). SKNO-1 cells were infected (si-A/E) or not (mock) with a lentiviral vector containing a construct expressing siRNAs against the region of fusion of the AML1/ETO mRNA (siA/E-RNA). (A) Expression levels of siRNAs as evaluated by Northern blot analysis on total RNA (5 μg) from mock and si-A/E samples. U2 snRNA was used as a loading control. (B) AML1/ETO mRNA amounts were normalized using c-abl mRNA quantity values and quantified in mock and si-A/E cells by qRT-PCR. The results represent the average of 3 independent evaluations ± SE. (C) Western blot analysis was performed on total cell lysates (50 μg) from mock and si-A/E cells using an anti-AML1 antibody. The immunodetection of the anti–β-tubulin antibody was used as loading control. (D) RARβ2 mRNA expression from mock and si-A/E–infected cells treated or not with RA (1 μM) for 48 hours was measured by qRT-PCR. The results represent the average of 3 independent evaluations ± SE. (E) Effect of 72-hour treatment with RA (1 μM) on the percentage of cells positively stained for CD11b and CD14 surface markers as measured by FACS analysis in mock and si-A/E cells. The results represent the average of 3 independent evaluations ± SE.
AML1/ETO siRNAs support differentiation response to RA of human t(8;21)-positive cells (SKNO-1). SKNO-1 cells were infected (si-A/E) or not (mock) with a lentiviral vector containing a construct expressing siRNAs against the region of fusion of the AML1/ETO mRNA (siA/E-RNA). (A) Expression levels of siRNAs as evaluated by Northern blot analysis on total RNA (5 μg) from mock and si-A/E samples. U2 snRNA was used as a loading control. (B) AML1/ETO mRNA amounts were normalized using c-abl mRNA quantity values and quantified in mock and si-A/E cells by qRT-PCR. The results represent the average of 3 independent evaluations ± SE. (C) Western blot analysis was performed on total cell lysates (50 μg) from mock and si-A/E cells using an anti-AML1 antibody. The immunodetection of the anti–β-tubulin antibody was used as loading control. (D) RARβ2 mRNA expression from mock and si-A/E–infected cells treated or not with RA (1 μM) for 48 hours was measured by qRT-PCR. The results represent the average of 3 independent evaluations ± SE. (E) Effect of 72-hour treatment with RA (1 μM) on the percentage of cells positively stained for CD11b and CD14 surface markers as measured by FACS analysis in mock and si-A/E cells. The results represent the average of 3 independent evaluations ± SE.
AML1/ETO expression modulates the RARβ2 gene promoter activity through the βRARE site
The transcriptional regulatory functions of AML1/ETO on RARβ2 gene were investigated by cotransfecting the human 293T cell line with an AML1/ETO expression vector along with the RARβ2Pr-LUC, a reporter vector containing the entire 5 Kb promoter region of RARβ2 gene cloned upstream of the luciferase coding region.15 Increasing concentrations of ectopically expressed AML1/ETO down-regulated the RARβ2 promoter activity in a dose-dependent manner in untreated and RA-treated 293T cells. Both the transcriptional repression exerted by AML1/ETO product on RARβ2pr-LUC basal activity and the RA-dependent activation of RARβ2 promoter were abrogated by mutating 3 bases of the 5′-GGTTCAC direct motif of the βRARE binding site of the RARβ2Pr-LUC vector (RAREmut) (Figure 2A-B). These results pointed at the βRARE site on the RARβ2 promoter as a target of the repressive effect exerted by AML1/ETO expression on the RA signaling pathway.
AML1/ETO modulates RARβ2 promoter activity through the βRARE binding site due to its interaction with RARα. (A) Schematic representation of the RARβ2 reporter constructs. The sequences of the region of the −5kb+155bp RARβ2pr-LUC reporter vector15 containing the βRARE and the TATA box (from −59 bp to −23 bp) carrying the wild-type 5′-GGTTCAC-3′ direct motif of the βRARE site (RAREwt)15 or a 5′-aacTCAC-3′ selective mutation at this site (RAREmut) are indicated. (B) Human 293T cells were transiently cotransfected with increasing amounts of the pCDNA3-AML1/ETO expression vector (20 ng and 40 ng) and 1 μg of RARβpr-LUC carrying the RAREwt or the RAREmut site. A cotransfected vector encoding the β-galactosidase (pSV-βgal) was used as an internal control for normalization of the reactions. After transfections the cells were treated (⊡) or not (□) with 1 μM RA for 24 hours. The results represent the average of 3 independent evaluations ± SD. (C) Coimmunoprecipitation and Western blot experiments performed in human U937 WT, U937 stably transfected with an HA-tagged AML1/ETO cDNA (A/E-HA), or with an empty vector (mock), and in human SKNO-1 cells lines. Ip- indicates the antibody used for the coimmunoprecipitation; IgG, rabbit serum used as nonspecific antibody. Coimmunoprecipitates were analyzed by Western blot (WB) with antibodies detecting the expression levels of AML1/ETO (A/E), AML1, and RARα proteins in different samples and in whole cell lysates (input). The difference in size of the AML1/ETO fusion protein between the A/E-HA and SKNO-1 is due to the HA-tagged domain, which increased the molecular weight of the AML1/ETO product in A/E-HA cells. (D) Nuclear extracts were prepared from U937 mock and A/E-HA cells and analyzed (50 μg) by Western blot for the expression of the RARα and AML1/ETO products by using α-RARα and α-HA antibodies, respectively. The expression of β-tubulin confirmed protein loading. RA binding activity was measured in (•) mock and (○) A/E-HA nuclear extracts labeled with 10 nM [3H]-RA in the absence or in the presence of 200-fold molar excess of unlabeled RA (▴, mock; △, A/E-HA) to determine nonspecific binding by HPLC using a Superose 6 HR 10/30 size exclusion column.
AML1/ETO modulates RARβ2 promoter activity through the βRARE binding site due to its interaction with RARα. (A) Schematic representation of the RARβ2 reporter constructs. The sequences of the region of the −5kb+155bp RARβ2pr-LUC reporter vector15 containing the βRARE and the TATA box (from −59 bp to −23 bp) carrying the wild-type 5′-GGTTCAC-3′ direct motif of the βRARE site (RAREwt)15 or a 5′-aacTCAC-3′ selective mutation at this site (RAREmut) are indicated. (B) Human 293T cells were transiently cotransfected with increasing amounts of the pCDNA3-AML1/ETO expression vector (20 ng and 40 ng) and 1 μg of RARβpr-LUC carrying the RAREwt or the RAREmut site. A cotransfected vector encoding the β-galactosidase (pSV-βgal) was used as an internal control for normalization of the reactions. After transfections the cells were treated (⊡) or not (□) with 1 μM RA for 24 hours. The results represent the average of 3 independent evaluations ± SD. (C) Coimmunoprecipitation and Western blot experiments performed in human U937 WT, U937 stably transfected with an HA-tagged AML1/ETO cDNA (A/E-HA), or with an empty vector (mock), and in human SKNO-1 cells lines. Ip- indicates the antibody used for the coimmunoprecipitation; IgG, rabbit serum used as nonspecific antibody. Coimmunoprecipitates were analyzed by Western blot (WB) with antibodies detecting the expression levels of AML1/ETO (A/E), AML1, and RARα proteins in different samples and in whole cell lysates (input). The difference in size of the AML1/ETO fusion protein between the A/E-HA and SKNO-1 is due to the HA-tagged domain, which increased the molecular weight of the AML1/ETO product in A/E-HA cells. (D) Nuclear extracts were prepared from U937 mock and A/E-HA cells and analyzed (50 μg) by Western blot for the expression of the RARα and AML1/ETO products by using α-RARα and α-HA antibodies, respectively. The expression of β-tubulin confirmed protein loading. RA binding activity was measured in (•) mock and (○) A/E-HA nuclear extracts labeled with 10 nM [3H]-RA in the absence or in the presence of 200-fold molar excess of unlabeled RA (▴, mock; △, A/E-HA) to determine nonspecific binding by HPLC using a Superose 6 HR 10/30 size exclusion column.
AML1/ETO interacts with RARα-RXR at specific sites on AML1 and RA target gene promoters
To clarify the molecular events by which AML1/ETO silences the RA signaling pathway, initially we investigated the possibility of an in vivo interaction of AML1/ETO fusion protein with the RARα receptor. This hypothesis was tested in human SKNO-1 cells constitutively expressing the AML1/ETO and in the myeloid cell line U937 stably transfected with an empty vector (mock) or with an HA-tagged AML1/ETO (A/E-HA) expression vector. Immunoblot analysis revealed that AML1 and RARα proteins are constitutively expressed in these cell lines and that similar levels of the AML1/ETO fusion protein are detectable in SKNO-1 and A/E-HA cells (Figure 2C, input lanes). By coimmunoprecipitation experiments and reciprocal immunoblotting, we found that α-RARα antibodies coimmunoprecipitated the AML1/ETO fusion protein but not the AML1 product present in A/E-HA and SKNO-1 lysates (Figure 2C, IP-αRAR). Moreover, α-AML1 antibodies immunoprecipitated the RARα proteins exclusively from lysates of AML1/ETO-expressing cells (Figure 2C, Ip-αAML1). While, similar “in vivo” interactions were detected when α-ETO and α-RXR antibodies were used in coimmunoprecipitation assays, pulldown experiments performed using in vitro–translated AML1/ETO and the GST-RARα or GST-RXR did not show a direct interaction between these proteins (not shown). This suggested that the RARα-RXR interacts with other components of the aberrant repressive protein complex generated by AML1/ETO.
We tested whether the in vivo interaction with the AML1/ETO fusion complex affects the RA binding properties of RARα receptor. Nuclear extracts were prepared from U937-MT and U937–A/E-HA cells. Immunoblot analysis confirmed the presence of AML1/ETO in U937–A/E-HA nuclear extracts and the similar expression levels of the endogenous RARα receptors in these cell lines (Figure 2D). The extracts were labeled with [3H]-RA, and the RA binding was measured by HPLC analysis.43 The RA binding affinity and the morphology of the elution profile of U937-A/E cell nuclear extracts did not differ from that of mock cells (Figure 2D). These cells indeed displayed a major peak of specific RA binding activity, eluting at retention times corresponding to an apparent molecular weight of 50 kDa (about 44 minutes), which represents binding by wild-type RARα.43 This indicated that the interaction with AML1/ETO does not affect the RA binding properties of RARα.
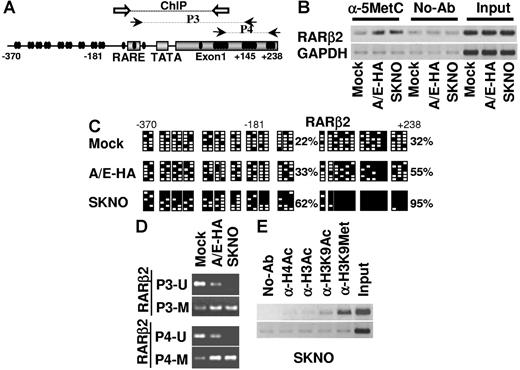
Thus, we investigated whether in vivo the AML1/ETO fusion protein is aberrantly present on βRARE binding sites. ChIP experiments were performed in mock, A/E-HA, and SKNO-1 cells using anti-RARα, -RXR, -AML1, and -ETO antibodies. Previously described PCR primers were used to amplify the βRARE site on RARβ2 promoter and the AML1 sites on p14arf gene promoter.19,35 The p14arf gene is known to be specifically repressed by the AML1/ETO fusion protein.35 As expected, in these cells, the RARα-RXR heterodimer and AML1 were present at their target promoters, RARβ2 and p14arf, respectively (Figure 3A). Remarkably, DNA sequences containing regions including the βRARE of the RARβ2 promoter were immunoprecipitated using α-AML1 and α-ETO antibodies in A/E-HA and SKNO-1 cells but not in mock cells (Figure 3A-B). ChIP analysis performed with the α-HA antibody enabled the AML1/ETO-HA fusion protein to be distinguished from the endogenous AML1 or ETO proteins expressed by A/E-HA cells and confirmed the presence of AML1/ETO at this RARα-RXR target site. Furthermore, RARα-RXR immunocomplexes were aberrantly present at the AML1 sites on the p14arf promoter in AML1/ETO-expressing cells (Figure 3A). As a control for the specificity of binding at these chromatin sites, distal sequences in the RARβ2 gene (exon 7) or sequences of the p16 promoter (p16INK4a), which is not regulated by AML1/ETO and lacks the AML1 binding site,35 were amplified in the same samples and showed no signal (Figure 3A-B). These results identify both AML1 and RARα-RXR gene promoters as molecular targets for the AML1/ETO fusion protein.
AML1/ETO recruits DNMTs, HDAC1, and MeCP2 to the RARβ2 promoter. (A) ChIPs were performed in mock, A/E-HA, and SKNO-1 cells using the indicated antibodies or in absence of antibody (no-Ab) or the anti-ETO antibody (B) and analyzed by PCR with primer for the βRARE binding site on RARβ2 promoter or for the AML1 binding site on p14ARF promoter. A region including the p16INK4a transcriptional start site and RARβ2 exon 7 (EX7), each lacking the AML1 and the RARE binding sites, respectively, were amplified to evaluate the specificity of AML1/ETO binding. (C-D) ChIPs were performed on mock, A/E-HA, and SKNO-1 cell lines using antibodies specific for MeCP2, DNMT1, DNMT3a, DNMT3b, and HDAC1 or with no antibody (no-Ab) as a negative control. Immunoprecipitated chromatins were analyzed by PCR using primer pairs specific for the amplification of the promoter region containing the βRARE and the 5-UTR exon 1 sequences on RARβ2 gene (RARβ2pr) or regions surrounding the AML1 site of the interleukin-3 promoter (IL-3pr). SKNO-1 cells were treated or not with 1 μM RA for 24 hours. Samples representing 0.02% of sonicated chromatin (input) were included in the PCR analysis. The amplification of a GAPDH coding region was used as a control of nonspecific precipitated sequences.
AML1/ETO recruits DNMTs, HDAC1, and MeCP2 to the RARβ2 promoter. (A) ChIPs were performed in mock, A/E-HA, and SKNO-1 cells using the indicated antibodies or in absence of antibody (no-Ab) or the anti-ETO antibody (B) and analyzed by PCR with primer for the βRARE binding site on RARβ2 promoter or for the AML1 binding site on p14ARF promoter. A region including the p16INK4a transcriptional start site and RARβ2 exon 7 (EX7), each lacking the AML1 and the RARE binding sites, respectively, were amplified to evaluate the specificity of AML1/ETO binding. (C-D) ChIPs were performed on mock, A/E-HA, and SKNO-1 cell lines using antibodies specific for MeCP2, DNMT1, DNMT3a, DNMT3b, and HDAC1 or with no antibody (no-Ab) as a negative control. Immunoprecipitated chromatins were analyzed by PCR using primer pairs specific for the amplification of the promoter region containing the βRARE and the 5-UTR exon 1 sequences on RARβ2 gene (RARβ2pr) or regions surrounding the AML1 site of the interleukin-3 promoter (IL-3pr). SKNO-1 cells were treated or not with 1 μM RA for 24 hours. Samples representing 0.02% of sonicated chromatin (input) were included in the PCR analysis. The amplification of a GAPDH coding region was used as a control of nonspecific precipitated sequences.
AML1/ETO recruits chromatin remodeling enzymes on target promoters
We investigated whether specific active multifactor complexes, including chromatin remodeling activities such as DNMTs, HDAC1, and the methyl binding protein MeCP2, are aberrantly recruited by AML1/ETO at promoter sites on RARβ2 as demonstrated for the oncoprotein PML/RARα in APL blasts.9,18,19 DNMTs are responsible for cytosine methylation within CpG dinucleotides (CpGs), which are often gathered in clusters (“CpG islands”) and are mainly present in promoter regions.3,4,52 Methylated CpGs are docking sites for proteins with high affinity for methylated DNA sequences (MeCPs and MBDs). Often, DNMTs, MeCPs, and MBDs are associated with corepressor complexes (mSin3A, NCoR, SWI/SNF, NuRD) and chromatin remodeling factors such as HDACs, HMTs, and heterochromatin protein-1.3,4,52 ChIP analysis revealed the presence of HDAC1, DNMT3a, DNMT3b, and MeCP2 on a chromatin region including the βRARE site at the RARβ2 promoter in both A/E-HA and SKNO-1 cells, whereas HDAC1 was faintly present in mock cells (Figure 3C-D). DNMT1, which was absent at the βRARE site on RARβ2 in A/E-HA and SKNO-1 cells, was detectable when the immunoprecipitated DNA was amplified with primers located in regions surrounding the AML1 sites of IL-3 promoter (Figure 3C-D) as previously shown in a t(8;21) cell line.33 Thus, it appears that transcriptional silencing of myeloid target genes can be induced by AML1/ETO through the aberrant recruitment of specific chromatin remodeling enzymes at defined sites on AML1 and RARα target gene promoters. Interestingly, ChIP assay performed in SKNO-1 cells treated with or without 1 μM RA for 24 hours showed that both the AML1/ETO and its associated corepressor complex are present at the βRARE site on the RARβ2 promoter when RARα receptor binds RA (Figure 3A-D). This indicated that the AML1/ETO-induced chromatin remodeling events cannot be reversed by RA alone, further explaining the RA resistance of the SKNO-1 cells.
Epigenetic modifications induced by AML1/ETO at RARβ2 promoter and exon 1 regions
We addressed whether the aberrant recruitment of chromatin-modifying activities by AML1/ETO on the RARβ2 promoter is linked to its transcriptional silencing by investigating in mock, A/E-HA, and SKNO-1 cell lines the modifications of DNA and chromatin structure at the promoter regions including the βRARE site and the 5′–untranslated region (5′-UTR) exon 1 region of this gene (Figure 4A). ChIP experiments performed with an α-5′-methylcytosine antibody to immunoprecipitate-sonicated naked DNA revealed an increase of methylated cytosines in regulatory regions on RARβ2 gene in both A/E-HA and SKNO-1 cells when compared with the levels measured in mock cells or in the absence of antibody (Figure 4B). These results are consistent with the abnormal presence of de novo DNMT activities (DNMT3a and DNMT3b) at this site in AML1/ETO-expressing cells (Figure 3C-D). Genomic bisulfite sequencing was performed to detect the methylation status of each CpG within the RARβ2 promoter and the 5′-UTR exon 1 region of RARβ2 gene. As seen in Figure 4C, the percentage of 5-methylcytosine residues in these 2 respective regions was constitutively higher in A/E-HA cells (33% and 55%) and in SKNO-1 cells (62% and 99%) than in mock cells (22% and 32%), therefore confirming the basal hypermethylated status of RARβ2 gene in AML1/ETO-expressing cells. A comparable high percentage of 5-methylcytosine was detected in these samples by methylation-specific PCR (MSP). MSP was performed using previously described methylation-sensitive primers designed to amplify regions of the promoter/βRARE (P3 primers) and the exon 1 (P4 primers) of RARβ2 (Figure 4A).19 The annealing of the DNA-methylated selective primers (P3-M and P4-M), resulting in a progressive accumulation of PCR products, increased in A/E-HA and SKNO-1 cells as compared with mock cells, in which PCR products corresponding to unmethylated DNA are mainly detectable (Figure 4D). Moreover, in SKNO-1 cells only PCR products corresponding to methylated genomic regions on RARβ2 gene were detectable, therefore confirming that the CpGs at these sites of RARβ2 gene are heavily methylated in this cell line (Figure 4D).
AML1/ETO induces epigenetic modification of the RARβ2 gene in t(8;21)-positive cells. (A) Schematic representation of the distribution of the CpGs (black circles) along the promoter and exon 1 (nt 370 to +238) of the RARβ2 gene. The black arrows indicate the DNA sequences amplified by different sets of primers sensitive to the methylation status of CpGs (P3 and P4). White arrows indicate the regions of annealing of the PCR primers used in ChIP assays. (B) ChIP analysis was performed in mock, A/E-HA, and SKNO-1 cell lines using an antibody specific for 5-methylcytosine (α-5MetC) or with no antibody (no-Ab) as negative control. Immunoprecipitated chromatins were analyzed by PCR primers specific for the amplification of a genomic region containing the βRARE binding site and the 5′-UTR exon 1 of RARβ2 gene. Samples representing 0.02% of sonicated chromatin (input) were included in the PCR analysis. The amplification of a GAPDH coding region was performed as a control of nonspecific precipitated sequences. (C) Bisulfite sequencing assay was performed to detect the methylation status of each CpG along the RARβ2 promoter/exon 1 sequence on genomic DNA isolated from mock, A/E-HA, and SKNO-1 cells. Black (▪) and empty (□) squares represent methylated and unmethylated CpGs, respectively. For each sample, the percentages of global methylation level in the promoter and in the 5′-UTR exon 1 regions are indicated. (D) MSP PCR assay was performed on bisulfite-treated DNA isolated from mock, A/E-HA, and SKNO-1 cell lines to analyze the methylation status of RARβ2 promoter/exon 1 regions. U and M indicate the unmethylated and methylated forms amplified by MSP primers, respectively. (E) ChIP assay was performed on the SKNO-1 cell line using antibodies specific for the acetyl-H4, acetyl-H3, acetyl-H3-Lys9, and methyl-H3-Lys9 forms or without antibody (no-Ab) as a negative control.
AML1/ETO induces epigenetic modification of the RARβ2 gene in t(8;21)-positive cells. (A) Schematic representation of the distribution of the CpGs (black circles) along the promoter and exon 1 (nt 370 to +238) of the RARβ2 gene. The black arrows indicate the DNA sequences amplified by different sets of primers sensitive to the methylation status of CpGs (P3 and P4). White arrows indicate the regions of annealing of the PCR primers used in ChIP assays. (B) ChIP analysis was performed in mock, A/E-HA, and SKNO-1 cell lines using an antibody specific for 5-methylcytosine (α-5MetC) or with no antibody (no-Ab) as negative control. Immunoprecipitated chromatins were analyzed by PCR primers specific for the amplification of a genomic region containing the βRARE binding site and the 5′-UTR exon 1 of RARβ2 gene. Samples representing 0.02% of sonicated chromatin (input) were included in the PCR analysis. The amplification of a GAPDH coding region was performed as a control of nonspecific precipitated sequences. (C) Bisulfite sequencing assay was performed to detect the methylation status of each CpG along the RARβ2 promoter/exon 1 sequence on genomic DNA isolated from mock, A/E-HA, and SKNO-1 cells. Black (▪) and empty (□) squares represent methylated and unmethylated CpGs, respectively. For each sample, the percentages of global methylation level in the promoter and in the 5′-UTR exon 1 regions are indicated. (D) MSP PCR assay was performed on bisulfite-treated DNA isolated from mock, A/E-HA, and SKNO-1 cell lines to analyze the methylation status of RARβ2 promoter/exon 1 regions. U and M indicate the unmethylated and methylated forms amplified by MSP primers, respectively. (E) ChIP assay was performed on the SKNO-1 cell line using antibodies specific for the acetyl-H4, acetyl-H3, acetyl-H3-Lys9, and methyl-H3-Lys9 forms or without antibody (no-Ab) as a negative control.
Finally, we investigated whether posttranslational modifications of histone tails in AML1/ETO-expressing cells correlated with DNA hypermethylation and with the aberrant recruitment of epigenetic modifiers at these sites on the RARβ2 gene. Deacetylated histones within regions of DNA methylation and/or methylation of specific histone residues (ie, lysine 9 of histone 3) are a hallmark of repressive chromatin status.3 ChIP analysis was therefore performed on SKNO-1 cells using antibodies against the acetylated forms of histone H3 and H4 and the acetylated (H3-K9Ac) or methylated (H3-K9Met) lysine 9 on histone H3. By PCR we amplified the RARβ2 region encompassing the βRARE, the TATA box, the transcription start site, and the CpGs present in exon 1 (Figure 4A, arrows and “ChIP”). Figure 4E shows that in SKNO-1 cells histones H3 and H4 were fully deacetylated and low levels of acetylation were measurable on histone H3 at lysine 9 (H3-K9), which indeed was highly methylated at these regulatory regions on the RARβ2 gene. Thus, the aberrant presence of chromatin remodeling activities causing a repressive chromatin status of the RARβ2 gene appears functionally related to the RA resistance of AML1/ETO-positive blasts.
5-azacytidine relieves AML1/ETO transcriptional silencing of the RA signaling pathway and supports RA-induced differentiation
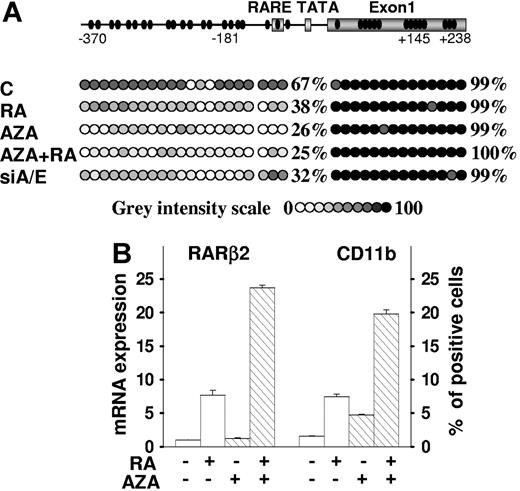
Next we investigated the respective roles of DNA hypermethylation of the RARβ2 gene and of AML1/ETO expression in the silencing of the RA signaling pathway and in the differentiation block of SKNO-1 cells. Initially, we tested the ability of the DNA methylation inhibitor 5-azacytidine to restore cell sensitivity to RA in terms of reactivation of the endogenous RARβ2 gene through demethylation of its promoter region and of restoration of RA differentiation activity in this RA-resistant AML cell line.47 In untreated SKNO-1 cells the basal percentage of 5-methylcytosine residues on the RARβ2 gene in the promoter/βRARE and in the exon 1 regions was 67% and 99%, respectively (Figures 4C and 5A). Following treatment with 5-azacytidine, the percentage of 5-methylcytosine residues diminished to 26% in the promoter/βRARE region of RARβ2 gene, and a similar percentage was reached when given simultaneously with RA, while RA alone decreased the methylation of this region to 38%. Notably, the knockdown of AML1/ETO in SKNO-1-siA/E–infected cells also resulted in a reduction of the percentage of 5-methylcytosine residues at that promoter/βRARE site to 32%. Although we did not observe demethylation of the exon 1 region in SKNO-1–treated or siA/E-infected cells, the demethylation of the promoter/βRARE region following the 5-azacytidine treatment or AML1/ETO knockdown was sufficient to restore RA-dependent induction of RARβ2 expression in these cells (Figures 1D and 5B). These results are in agreement with the regulatory function of the βRARE region and with its RA inducibility that can be accomplished in the presence of a reduced CpGs methylation status at this site, as shown by methylation data. Remarkably, in SKNO-1 cells the expression level of the myeloid-specific cell-surface antigen CD11b is poorly modified by treatment with RA or 5-azacytidine when used as single agents, while it is strongly induced by the combined treatment of RA/5-azacytidine, thus suggesting the ability of 5-azacytidine to restore the differentiation responsiveness of SKNO-1 cells to RA (Figure 5B). These findings also established a correlation between AML1/ETO expression levels, RARβ2 demethylation, RARβ2 reexpression, and induction of differentiation of RA-treated SKNO-1 cells.
5-Azacytidine relieves AML1/ETO transcriptional silencing of the RA signaling pathway and supports RA-induced differentiation. (A) The bisulfite sequencing assay was performed on genomic DNA isolated from SKNO-1 cells and SKNO-1 –infected (siA/E) cells. Cells were treated or not with 5-azacytidine (1 μM) for 48 hours, and then RA (1 μM) was added for an additional 48 hours in the indicated samples. The methylation status of each CpG from nt −370 to +238 of the RARβ2 promoter/exon 1 DNA sequence was measured and represented by a circle depicted by increasing gray intensities. The increasing gray-intensity scale indicates a 10% rise in methylation status of the CpG of interest and summarizes the results of the analysis of 6 different clones for each sample. For each sample, the percentages of global methylation level in the promoter and in the 5′-UTR exon 1 regions are indicated. (B) SKNO-1 cells were treated or not with 5-azacytidine (1 μM), RA (1 μM), or in combination for 48 hours as described. The qRT-PCR results of RARβ2 mRNA expression and the percentage of CD11b+ SKNO-1 cells measured by FACS analysis are shown. The results represent an average of 3 independent evaluations ± SD.
5-Azacytidine relieves AML1/ETO transcriptional silencing of the RA signaling pathway and supports RA-induced differentiation. (A) The bisulfite sequencing assay was performed on genomic DNA isolated from SKNO-1 cells and SKNO-1 –infected (siA/E) cells. Cells were treated or not with 5-azacytidine (1 μM) for 48 hours, and then RA (1 μM) was added for an additional 48 hours in the indicated samples. The methylation status of each CpG from nt −370 to +238 of the RARβ2 promoter/exon 1 DNA sequence was measured and represented by a circle depicted by increasing gray intensities. The increasing gray-intensity scale indicates a 10% rise in methylation status of the CpG of interest and summarizes the results of the analysis of 6 different clones for each sample. For each sample, the percentages of global methylation level in the promoter and in the 5′-UTR exon 1 regions are indicated. (B) SKNO-1 cells were treated or not with 5-azacytidine (1 μM), RA (1 μM), or in combination for 48 hours as described. The qRT-PCR results of RARβ2 mRNA expression and the percentage of CD11b+ SKNO-1 cells measured by FACS analysis are shown. The results represent an average of 3 independent evaluations ± SD.
The RARβ2 promoter/exon 1 region is methylated in non-APL AML blasts
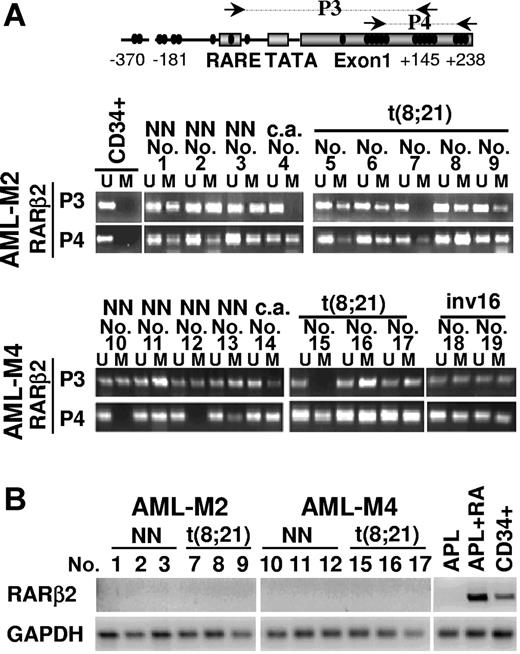
To determine the significance of our findings in human leukemia, we investigated the methylation status of RARβ2 promoter/exon 1 and RARβ2 mRNA expression levels in primary blasts from AML patients containing or not specific genetic lesions (Figure 6). We used MSP as a sensitive and reliable approach for the identification of CpG sites that differ in their methylation status in DNA samples from a series of AML patients (Figure 6A). Remarkably, by using primers for the promoter/exon 1 regions of RARβ2 (P3 primers), we found that the region containing the βRARE binding site is methylated in 6 of 8 AML1/ETO-positive samples, and all of the samples (8 of 8) presented hypermethylation of a region located in the 5′-UTR exon 1 of RARβ2 as detected by using P4 primers. Neither of these 2 regions on the RARβ2 gene was found methylated in CD34+ normal hematopoietic precursors (Figure 6A).
Methylation status of CpGs along the RARβ2 promoter/exon 1 regions in human primary AML blasts. (A) Methylation-sensitive PCR (MSP) assay performed to detect the methylation status of CpGs along the RARβ2 promoter/exon 1 regions on bisulfite-treated genomic DNAs from 19 primary AML blasts at diagnosis (9 cases of AML-M2, 10 cases of AML-M4 according to FAB classification) and CD34+ hematopoietic progenitors obtained from healthy donors. Black arrows in the diagram of RARβ2 promoter/exon 1 region indicate the fragments amplified by the different methylation-sensitive sets of primer pairs (P3 and P4). U and M indicate the unmethylated and methylated forms amplified by methylation-sensitive primer pairs, respectively. The presence of chromosomal translocations is indicated as follows: NN, normal karyotype; c.a., complex aberration. (B) RT-PCR was performed to evaluate the expression levels of RARβ2 mRNA in samples from human AML blasts, normal CD34+ hematopoietic progenitors, and an APL-representative patient treated or not “in vitro” for 48 hours with RA (1 μM). GAPDH expression level was used as internal control to evaluate the amount and the integrity of RNA samples.
Methylation status of CpGs along the RARβ2 promoter/exon 1 regions in human primary AML blasts. (A) Methylation-sensitive PCR (MSP) assay performed to detect the methylation status of CpGs along the RARβ2 promoter/exon 1 regions on bisulfite-treated genomic DNAs from 19 primary AML blasts at diagnosis (9 cases of AML-M2, 10 cases of AML-M4 according to FAB classification) and CD34+ hematopoietic progenitors obtained from healthy donors. Black arrows in the diagram of RARβ2 promoter/exon 1 region indicate the fragments amplified by the different methylation-sensitive sets of primer pairs (P3 and P4). U and M indicate the unmethylated and methylated forms amplified by methylation-sensitive primer pairs, respectively. The presence of chromosomal translocations is indicated as follows: NN, normal karyotype; c.a., complex aberration. (B) RT-PCR was performed to evaluate the expression levels of RARβ2 mRNA in samples from human AML blasts, normal CD34+ hematopoietic progenitors, and an APL-representative patient treated or not “in vitro” for 48 hours with RA (1 μM). GAPDH expression level was used as internal control to evaluate the amount and the integrity of RNA samples.
To verify whether RARβ2 promoter/exon 1 methylation could represent a common lesion in myeloid leukemia, we next analyzed AML-M2 and AML-M4 blasts with apparently normal karyotype or presenting recurrent AML genetic alterations. Notably, the RARβ2 region containing the βRARE binding site was methylated in 7 of 9 AML-M2 samples (P3 primers) and in 9 of 10 AML-M4 samples, whereas the 5′-UTR exon 1 region of RARβ2 (P4 primers) was methylated in samples from 9 of 9 AML-M2 and 8 of 10 AML-M4.
In agreement with previous results obtained in BM cells from 15 APL (AML-M3) patients,18,19 RARβ mRNA was not expressed in samples isolated from the BM of 1 APL or 12 non-APL AML patients (AML-M2–AML-M4) (Figure 6B). However, RARβ gene expression was fully detectable in CD34+ hematopoietic progenitors isolated from the BM of healthy donors and, as expected, its expression was restored by RA treatment in primary APL blasts (Figure 6B).
Overall, these results indicate the magnitude of the heterochromatic silencing of the RA signaling pathway in myeloid leukemias and support previous in vitro and in vivo evidence indicating that epigenetic deregulation of the developmental program of normal myelopoiesis such as that of RA is shared by different AML subtypes irrespective of the presence of specific genetic lesions.37,38,42
Discussion
Aberrant DNA methylation seems to be a dominant factor in epigenetic gene silencing. Indeed, disruption of DNA methylation patterns is frequently present in aberrant development and neoplastic transformation.4,5 Here, we show that the expression of AML1/ETO, the most common AML-associated fusion protein, increases the methylation status at genomic loci physiologically regulated by RA, a regulator of myelopoiesis. Our findings identify RARα-RXR heterodimer as a component of the macromolecular complex formed by AML1/ETO. As a consequence, AML1/ETO recruits HDAC1/DNMT/MeCP2 activities on specific DNA binding domains of RARα and AML1 genes and methylates CpG-rich sequences and deacetylates/methylates specific lysine residues on nucleosomal histones, thereby promoting the formation of nonpermissive chromatin at these sites. Therefore, the association of the “gain of function” of the AML1/ETO oncoprotein and the “loss of function” of RARα-RXR heterodimer results in the heterochromatic silencing of a key transcriptional pathway of myelopoiesis, contributing to the differentiation block of myeloid progenitors. This hypothesis is supported by findings indicating that the differentiation response to RA can be restored in AML1/ETO blasts by (1) impairing the interaction between the AML1/ETO and the transcriptional corepressor complex using protein fragments representative of their interaction surfaces47 and (2) changing the methylation status at regulatory sites on the RA target gene RARβ2 by AML1/ETO knockdown by RNAi or pharmacologic treatment with the demethylating agent 5-azacytidine.
While this paper was in preparation, Tabe et al addressed the methylation status of the RARβ2 gene and the ability of demethylating agents to reverse RA resistance in AML blasts and in PML/RARα- or AML1/ETO-expressing cell lines.53 We found some discrepancies between the results of this study and our findings18,19 that are probably due to the different cell lines, MSP primers, RARβ2 region analyzed, drug dosage, and treatment time used. For example, by limiting their MSP analysis to the exon 1 region of RARβ2 gene, they found this region methylated in the AML1/ETO-positive Kasumi-1 cell line and in primary AML blasts but not in 3 cases carrying the t(8;21) translocation.53 Considering that MSP is not a quantitative assay for methylation, it should be noted that all the primary AML samples tested in this study, including those carrying the AML1/ETO fusion gene, showed a uniform lack of RARβ2 mRNA expression as measured by qRT-PCR analysis,53 which is consistent with our findings.
Transgenic mouse models and bone marrow transplantation approaches in mice have shown, however, that AML1/ETO expression predisposes myeloid precursors to transformation but is not sufficient to induce leukemia, which appears to require further molecular damages.27,54 In this context, a crucial question is the role of genetic and epigenetic alterations in the initiation or maintenance of AML1/ETO-positive leukemia. RNAi experiments indicated that a titrated AML1/ETO production is necessary for its activity as a transcriptional repressor of RARβ2 and for the block of myeloid differentiation. Interestingly, RARβ2 is the RA-regulated tumor suppressor gene silenced in a variety of human malignancies.8,16–19 Of note, hypermethylation of RARβ2 is commonly detectable in AML blasts independently from the presence of specific genetic lesions.
The relevance of this finding in AMLs is underlined by the fact that aberrant heterochromatic gene silencing can represent an alternative mechanism to gene mutation or deletion for the transcriptional repression of tumor suppressor genes.4,5,48 In AML subtypes lacking the PML/RARα or the AML1/ETO translocations, other mechanistic factors involved, which are presently unknown, might implicate chromatin regulators initiating the gene silencing. This raises the important question of whether RARβ2 epigenetic silencing “addicts” cancer cells to altered signal transduction pathways for tumor initiation. This “addiction” might in turn confer cell-survival benefit and, by allowing the accumulation of additional genetic and/or epigenetic events, promote tumor progression.
In conclusion, our study provides new insight into molecular pathways deregulated in myeloid leukemogenesis and further supports the reversion of transformed phenotype by targeting of gene silencing as a promising and powerful therapeutic strategy for AMLs.
Authorship
Contribution: F.F. and G.Z. designed and performed experiments and cowrote the manuscript; V.G., L.T., and A.C. designed and performed experiments; L.D.C., A.R., I.B., and F.G. generated essential new reagents and contributed to experimental design; F.L.-C. provided primary leukemia samples fully characterized at the molecular level and contributed to experimental design; P.G.P. contributed to experimental design; and C.N. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Clara Nervi, University La Sapienza Rome & San Raffaele Bio-medical Park Foundation, Via di Castel Romano 100, Rome, 00128, Italy; e-mail: clara.nervi@uniroma1.it.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Italian Association for Cancer Research (AIRC and AIRC–Rome Oncogenomic Center [ROC]), Ministero dell'Istruzione, dell'Università e della Ricerca, University of Rome “La Sapienza,” Ministero della Salute, Sixth Research Framework Programme of the European Union, Project RIGHT (LSHB-CT-2004 005276), and a fellowship from University La Sapienza Rome (L.T.). We are grateful to Silvia Di Cesare and Fabrizio Padula for FACS analysis and to Linda Starnes for critical reading of the manuscript.

![Figure 2. AML1/ETO modulates RARβ2 promoter activity through the βRARE binding site due to its interaction with RARα. (A) Schematic representation of the RARβ2 reporter constructs. The sequences of the region of the −5kb+155bp RARβ2pr-LUC reporter vector15 containing the βRARE and the TATA box (from −59 bp to −23 bp) carrying the wild-type 5′-GGTTCAC-3′ direct motif of the βRARE site (RAREwt)15 or a 5′-aacTCAC-3′ selective mutation at this site (RAREmut) are indicated. (B) Human 293T cells were transiently cotransfected with increasing amounts of the pCDNA3-AML1/ETO expression vector (20 ng and 40 ng) and 1 μg of RARβpr-LUC carrying the RAREwt or the RAREmut site. A cotransfected vector encoding the β-galactosidase (pSV-βgal) was used as an internal control for normalization of the reactions. After transfections the cells were treated (⊡) or not (□) with 1 μM RA for 24 hours. The results represent the average of 3 independent evaluations ± SD. (C) Coimmunoprecipitation and Western blot experiments performed in human U937 WT, U937 stably transfected with an HA-tagged AML1/ETO cDNA (A/E-HA), or with an empty vector (mock), and in human SKNO-1 cells lines. Ip- indicates the antibody used for the coimmunoprecipitation; IgG, rabbit serum used as nonspecific antibody. Coimmunoprecipitates were analyzed by Western blot (WB) with antibodies detecting the expression levels of AML1/ETO (A/E), AML1, and RARα proteins in different samples and in whole cell lysates (input). The difference in size of the AML1/ETO fusion protein between the A/E-HA and SKNO-1 is due to the HA-tagged domain, which increased the molecular weight of the AML1/ETO product in A/E-HA cells. (D) Nuclear extracts were prepared from U937 mock and A/E-HA cells and analyzed (50 μg) by Western blot for the expression of the RARα and AML1/ETO products by using α-RARα and α-HA antibodies, respectively. The expression of β-tubulin confirmed protein loading. RA binding activity was measured in (•) mock and (○) A/E-HA nuclear extracts labeled with 10 nM [3H]-RA in the absence or in the presence of 200-fold molar excess of unlabeled RA (▴, mock; △, A/E-HA) to determine nonspecific binding by HPLC using a Superose 6 HR 10/30 size exclusion column.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/10/10.1182_blood-2006-09-045781/4/m_zh80100700950002.jpeg?Expires=1765891425&Signature=0eeMob7V6C3QihjlHXxU6EDehc92Yc6rhhrG~Lnr6C-ZC~jKVRc60A2esxIzNVU6zLMKfgYl~1WIyQn5BdUMJJN86S~d~X4Q29XvO8BFNMut1zuWQOjkz4656X1Gyplhl5YXKM5Pk7ZbpQYc~00c1ujX13S~AeWmTG7htB-sccly1I~T5P-vbuZPi8GjingjuoH-B6c1UHGKLgarvHiWo1RFiZnyvGJVlM6Qy5RmIl~nYPJ0q6C0tylnEjUDfQsQnA~teJ4VqCwlLVTXF7IGVO0oRTBrWEJEAKtIwFu4fn8XYfgsGvrJMQYpBiEHUoV~31hBVzebh8LKfTOyOgYRbw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal