Abstract
Stats (signal transducers and activators of transcription) regulate multiple aspects of T-cell fate. T regulatory (Treg) cells are a critical subset that limits immune responses, but the relative importance of Stat5a/b versus Stat3 for Treg cell development has been contentious. We observed that peripheral CD25+CD4+ T cells were reduced in Stat5ΔN mice; however, the levels of Foxp3, a transcription factor that is critical for Treg cells, were normal in splenic CD4+ T cells even though they were reduced in the thymus. In contrast, complete deletion of Stat5a/b (Stat5−/−) resulted in dramatic reduction in CD25- or Foxp3-expressing CD4+ T cells. An intrinsic requirement was demonstrated by reduction of Stat5a/b in CD4-expressing cells and by stem cell transplantation using Stat5−/− fetal liver cells. Stat5a/b were also required for optimal induction of Foxp3 in vitro and bound directly to the Foxp3 gene. Reduction of Stat3 in T cells did not reduce the numbers of Treg cells in the thymus or spleen; however, Stat3 was required for IL-6–dependent down-regulation of Foxp3. Therefore, we conclude that Stat5a/b have an essential, nonredundant role in regulating Treg cells, and that Stat3 and Stat5a/b appear to have opposing roles in the regulation of Foxp3.
Introduction
The development and differentiation of immune cells is carefully orchestrated by an array of cytokines. Signal transducers and activators of transcription (Stats) represent a small but critical family of transcription factors that play important roles in transmitting cytokine signals. Consequently, Stats are critical for immunoregulation and the development of immune cells.1,2 Stat5a and Stat5b are two closely related proteins that have overlapping functions with respect to lymphoid development and differentiation.3,4 Gene targeting of Stat5a and Stat5b (collectively referred to as Stat5), results in impairment in the development of T, B, and natural killer (NK) cells.5–7 In mice in which the amino termini of Stat5a and Stat5b are deleted (denoted as Stat5ΔN mice), major disruption of various immune cell parameters was noted.8,9 However, residual Stat5 function permits T cell development, albeit suboptimally.10 This contrasts with the complete absence of Stat5a/b, which results in dramatic reduction in thymocyte numbers, in part due to effects on lymphoid stem cell function.5
T regulatory (Treg) cells comprise a population of cells enriched in CD4+CD25+ T cells that suppresses T-cell proliferation and function and attenuates immune responses against self- or nonself-antigens.11–13 Naturally arising Treg cells are produced in the thymus as a functionally distinct T-cell subpopulation, whereas adaptive Treg cells are induced from naive T cells after antigen exposure in the periphery.14–17 In classic studies, mice develop organ-specific autoimmune disease following neonatal thymectomy, which is corrected by reconstitution with CD4+CD25+ T cells.13 The essential role of Treg cells in maintaining tolerance has been confirmed by findings that defective function of this subset is a feature of many models of autoimmunity.18
More recently, it was discovered independently by several groups that a subset of CD4+CD25+ T cells expresses the transcription factor Foxp3, which is necessary and sufficient for Treg cell development and function.19–22 Foxp3 is highly conserved in mice and humans. Mutation of Foxp3 in mice (scurfy) results in early autoimmune disease,23 whereas mutations of human Foxp3 are associated with a disorder known as immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX).24 In mice, Foxp3 is a reliable marker for the Treg lineage.
Multiple lines of evidence have indicated that IL-2 is an important growth factor for Treg development and maintenance. Mice lacking IL-2 or its receptor subunits, IL-2Rα (CD25) and IL-2Rβ (CD122), have deficits in CD4+CD25+ Treg cells and develop autoimmune disease similar to Foxp3−/− mice.25–27 However, IL-2 is dispensable for Treg cell development, as some Foxp3-expressing cells are present in Il2−/− and Il2ra−/− mice, suggesting the involvement of other cytokines.28 In vitro culture of CD4+ T cells with transforming growth factor-β1 (TGF-β1) can promote the generation of Foxp3+ Treg cells from naive CD4+ T cells. In contrast, in vitro culture of CD4+ T cells with TGF-β1 and IL-6 promotes the differentiation of inflammatory T helper 17 (Th17) cells and suppresses Treg cells.29
A first step in IL-2 signaling is the activation of the Janus kinase, Jak3, which associates with the IL-2Rγ chain (CD132), also known as the common gamma chain (γc).30 Jak3 and γc are essential for Treg cell development and maintenance, as Jak3−/− and Il2rg−/− mice lack CD25 and Foxp3 expression in the thymus and spleen.28,31 Activation of Jaks results in phosphorylation of Stats, and Stat5a/b are the most prominent Stats activated by IL-2.32 Stat5ΔN mice, which express N-terminally–truncated Stat5 proteins, have reduced numbers of CD25+CD4+ cells in the periphery and autoimmunity, although assessment of these mice has led to conflicting conclusions regarding the importance of Stat5 in Treg cell development.33–35 Interpretation of these studies is further complicated by the finding that Stat5ΔN mice expressed truncated Stat5 proteins that are partially functional; as such, these actually represent hypomorphic Stat5 alleles.5,6 As noted, the residual function in Stat5ΔN mice was illustrated by comparing immune defects in these mice with those of a different model of Stat5a/b deletion in which both Stat5a and Stat5b were completely deleted.5 However, the role of Stat5 in regulating Foxp3 was not assessed, and other studies have argued for a role of Stat3 in regulating Foxp3.36,37 This suggests that Stat3 and Stat5 may play redundant roles in regulating this transcription factor. These findings prompted reassessment of the roles of Stat5 and Stat3 in Treg cell development and maintenance with focus on their effects on Foxp3 expression.
In this report, we compared and contrasted Treg cell development in Stat5ΔN mice and Stat5a/b−/− (called Stat5−/−) mice. Stat5 was demonstrated to be critical for both Treg cell development and maintenance and critical for Foxp3 expression. The intrinsic requirement for Stat5a/b in Treg cells was evident through the use of tissue-specific Stat5 deletion and stem cell transplantation experiments. Moreover, Stat5 directly binds the Foxp3 gene. In contrast, reduction of Stat3 in T cells did not reduce the numbers of Treg cells in the thymus or spleen. However, the ability of IL-6 to down-regulate Foxp3 expression was greatly attenuated in Stat3-deleted T cells. Thus, Stat5a/b have an essential and direct positive role in regulating Foxp3 and Treg cells. Although Stat3 is not essential for the development or maintenance of Treg cells, it does appear to have an important role in mediating IL-6 signals to attenuate Foxp3 expression.
Materials and methods
Mice
Stat5a/b−/− (Stat5−/−), Stat5a/bfl/−, CD4cre (Stat5fl/−, CD4cre), and Stat5ΔN mice were described previously5–7 and housed at NIH under approved protocols. Stat5fl/−, CD4cre, Yfp mice were generated by crossing Stat5fl/−, CD4cre with Yfp indicator mice (ROSA26–stop-floxed–YFP reporter mice; Jackson Laboratory, Bar Harbor, ME). Cre-mediated deletion was monitored by yellow fluorescent protein (YFP) expression from a “ROSA26–stop-floxed–YFP” reporter. Stat3fl/fl mice were bred with mice expressing Cre under the control of the MMTV (MMTV-Cre) to produce Stat3fl/fl, MMTV-Cre mice.38 CD45.1 congenic Rag2−/−, Jak3−/−, and Il2rg−/−(Y) mice were obtained from Taconic Farms (Hudson, NY). Animals were handled and housed in accordance with the guidelines of the NIH Animal Care and Use Committee.
Antibodies
Biotin-, FITC-, PE-, Per-CP–, PE-Cy5.5–, and APC-conjugated antibodies to mouse CD4, CD25, CD8, CD62L, CD44, and CD122 were purchased from BD Biosciences (San Jose, CA). The mouse Foxp3 staining kit was purchased from eBioscience (San Diego, CA).
Cell preparation, DNA, RNA, and protein expression analysis
CD4+ single-positive (SP) thymocytes and splenocytes from Stat5fl/fl mice and YFP+CD4+ SP thymocytes and splenocytes from Stat5fl/−, CD4cre, Yfp mice were sorted using a Moflo cell sorter (Dako Cytomation, Glostrup, Denmark). The purity of the sorted cells was greater than 99%. These sorted cells were subjected to DNA, RNA, and protein analysis essentially as described before.5 In brief, DNA was purified using the DNA easy kit from Qiagen (Valencia, CA). RNA was prepared with Trizol reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). For real-time polymerase chain reaction (PCR), cDNA was generated using a first-strand cDNA synthesis kit (Applied Biosystems, Foster City, CA). The primers and probes for real-time PCR were purchased from Applied Biosystems (Applied Biosystems). Real-time (RT)–PCR was performed on ABI PRISM 7700 (Applied Biosystems, Foster City, CA).
Cell culture
CD25−CD4+ splenic T cells from Stat5fl/fl mice, YFP+CD25−CD4+ splenic T cells from Stat5fl/−, CD4cre, Yfp mice or Stat3fl/fl mice and Stat3fl/fl, MMTVcre mice were isolated by the Moflo cell sorter and cultured for 3 days with plate-bound anti-CD3 (5 μg/mL) and anti-CD28 (5 μg/mL) (BD PharMingen, San Diego, CA) plus TGF-β1 (5 ng/mL, PeproTech, Rocky Hill, NJ), hIL-2 (100 U/mL; provided by National Cancer Institute [NCI]–Frederick, MD) and IL-6 (10 ng/mL; PeproTech) as indicated, with or without anti–murine IL-2 antibody (20 μg/mL; R&D Systems, Minneapolis, MN) in complete RPMI 1640 medium containing 10% fetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin and 2 mM β-mercaptoethanol.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as previously described.39 CD4+CD25+ and CD4+CD25− cells were sorted from thymi and spleens and stimulated with IL-2 (100 U/mL) for 1 hour. Formaldehyde (final concentration, 1%) was then added to cross-link proteins and DNA. The cell lysates were sonicated and immunoprecipitated with normal rabbit serum (Upstate Biotechnology, Charlottesville, VA), α-Stat5 (R&D Systems), and α-Stat3 (Santa Cruz Biotechnology, Santa Cruz, CA). The immunopreciptated DNA was eluted and amplified by real-time PCR using an ABI 7700 (Applied Biosystems). Values were normalized to corresponding input control and are expressed as fold enrichment relative to normal rabbit serum for each experiment. The sequences specific primers and probes used for amplification of the Foxp3 gene surrounding putative Stat binding sites were as follows: site in I, 5′-CCTCCTGGAAACCTGTGTCAC-3′, 5′-AACTTGGTCAGAGAGGTGGCA-3′, and 5′-6FAM-TACCCCTCATTTACTTATC-3′; site in II, 5′-CTTCTGGGAGCCAGCCATT-3′, 5′-GCTGTACTCCCCCCACAAATT-3′, and 5′-6FAM-TGAGACTCTCTGATTCTGT-3′; sites in III, 5′-ACAACAGGGCCCAGATGTAGA-3′, 5′-GGAGGTTGTTTCTGGGACATAGA-3′, and 5′-6FAM-CCCGATAGGAAAACA-3′. The primers and probe used for irrelevant IV are 5′-CACCAAAGGCTGGAAGCCT-3′, 5′-CAGACGAGCCTCCACAGAGTT-3′, and 5′-6FAM-CCGTGCCTTGTCAGG-3′.
Stem cell transplants
Single-cell suspensions were generated from E14.5 Stat5+/+ or Stat5−/− fetal livers, and cells (2 × 106) were injected into tail veins of lethally irradiated (9 Gy [900 rad]) Rag2−/− CD45.1 congenic recipient mice housed under pathogen-free conditions with acidified water as previously described.5 At 7 to 8 weeks later, tissues were harvested. Thymi and spleens were analyzed by flow cytometry for donor-derived CD45.2+ cells.
Results
Stat5a/b are critical for thymic development of Treg cells
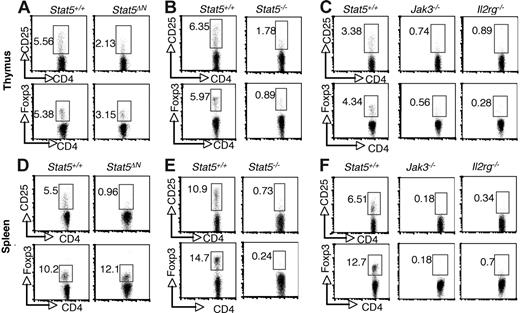
Previous studies using Stat5ΔN mice have reached conflicting conclusions regarding requirement for Stat5a/b in thymic Treg cell development, but because of the aforementioned limitations with this animal model, we sought to revisit this issue.34,35 As shown in Figure 1, the proportion of CD4+ CD25+ cells in thymi from Stat5ΔN mice was substantially reduced compared with wild-type (WT) littermates (Figure 1A). This is consistent with the well-documented effect of Stat5 on CD25 expression.40 In addition, the proportion of CD4+ Foxp3+ thymocytes in Stat5ΔN mice was reduced (Figure 1A; bottom panels). Furthermore, the level Foxp3 expression, as assessed by mean fluorescence intensity (MFI), was also reduced in Stat5ΔN by approximately 20% compared with that of controls. The specificity of Foxp3 staining was documented by comparison with isotype control (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). As the cellularity of the thymus in adult Stat5ΔN mice is roughly comparable with that of WT mice, the absolute numbers of Foxp3+ CD4 SP and CD25+ CD4 SP thymocytes were also reduced.8,10 In interpreting data using Stat5ΔN mice, one needs to bear in mind that truncated Stat5a/b proteins are expressed. However, Stat5−/− mice in which the entire Stat5a and Stat5b loci were deleted die perinatally, but a small numbers of mice (approximately 2%) survive for 6 to 8 weeks after birth.5–7 Stat5−/− viable mice exhibited marked reductions in the proportions and absolute numbers of CD25- and Foxp3-expressing CD4 SP thymocytes (Figure 1B). In fact, the reductions in CD25- and Foxp3-expressing cells from Stat5−/− mice were comparable to that observed in Jak3−/− and Il2rg−/−(Y) mice (Figure 1C). CD4 SP thymocytes were generated (albeit in substantially lower numbers) in these mice; nonetheless, there was profound reduction in Foxp3-expressing cells.28,31 The presence of Treg cells in Stat5ΔN mice is likely due to the residual activity of the truncated Stat5 protein rather than Stat5-independent development.
Stat5a/b are critical for the generation of thymic Foxp3+ CD4+ T cells and maintenance of peripheral Foxp3+ CD4 T+ cells. CD25 and Foxp3 expression were analyzed by flow cytometry in CD4 SP thymocytes (A-C) and splenocytes (D-F) from 4- to 6-week-old Stat5+/+ and Stat5ΔN mice (A,D), Stat5−/− mice (B,E), and Jak3−/− and Il2rg−/− mice (C,F). Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
Stat5a/b are critical for the generation of thymic Foxp3+ CD4+ T cells and maintenance of peripheral Foxp3+ CD4 T+ cells. CD25 and Foxp3 expression were analyzed by flow cytometry in CD4 SP thymocytes (A-C) and splenocytes (D-F) from 4- to 6-week-old Stat5+/+ and Stat5ΔN mice (A,D), Stat5−/− mice (B,E), and Jak3−/− and Il2rg−/− mice (C,F). Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
Stat5a/b are critical for peripheral Foxp3 expression and Treg cell maintenance
Previous studies using Stat5ΔN mice noted a substantial reduction of CD25+CD4+ T cells in the periphery; however, expression of Foxp3 was not examined.33,34 As shown in Figure 1D, the proportion of Foxp3-expressing CD4+ T cells was not reduced in 4- to 5-week-old Stat5ΔN mice, despite the dramatic reduction in CD25-expressing cells. Since the cellularity of spleens from Stat5ΔN mice is comparable with that of WT mice at this young age,8 the absolute number of Foxp3+ Treg cells was normal in Stat5ΔN mice, which might lead one to conclude that peripheral Foxp3 expression does not require Stat5a/b. In contrast, though, the proportions of Foxp3-expressing and CD25-expressing splenic CD4 T cells were both markedly reduced in Stat5−/− mice (Figure 1E). This is consistent with the notion that Stat5a/b are essential for Treg cell maintenance, and the expression of Foxp3 in Stat5ΔN mice is the result of residual Stat5 expression. The loss of peripheral Foxp3 expression was again comparable with that seen in spleens from Jak3−/− and Il2rg−/− mice (Figure 1F).
Intrinsic requirement for Stat5a/b but not Stat3 for Treg cells
While these data indicate a requirement for Stat5a/b in Foxp3 expression in the thymus and periphery, Stat5a/b are lacking in all tissues from Stat5−/− viable mice (data not shown), and an intrinsic requirement for Stat5a/b in T cells cannot be inferred. We therefore approached this problem in two ways: (1) stem cell transplantation using Stat5−/− precursors; and (2) tissue-specific deletion of Stat5 using transgenic expression of Cre.
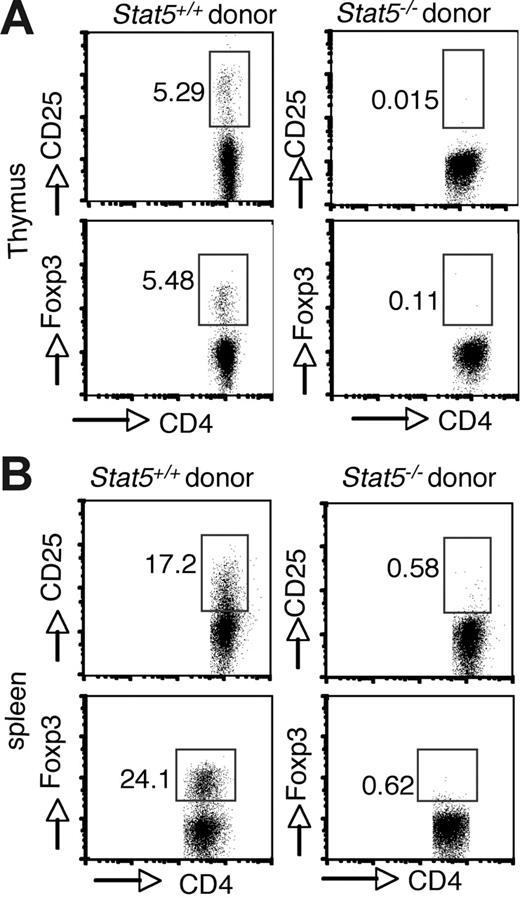
Because Stat5 deficiency is usually lethal (approximately 98%) and affects multiple cell lineages and pathways, we first reconstituted irradiated Rag2−/− recipient mice with Stat5−/− fetal liver cells to analyze Treg cell development. As noted, reconstitution with Stat5−/− precursors is inefficient due to defective hematopoietic/lymphoid stem cell functions.5,41 Nonetheless, T-cell development with production of SP thymocytes does occur.5 However, no CD25+ or Foxp3+ cells were detected in the CD4+ population of thymocytes from mice that received transplants (Figure 2A; right panel). In contrast, these cells were readily detected when normal stem cells were transplanted into Rag2−/− recipients (Figure 2A; left panel). CD25+ or Foxp3+ CD4+ T cells were also absent in spleens of Rag2−/− recipient mice that received transplants of Stat5−/− precursors (Figure 2B), consistent with what we observed in viable Stat5−/− mice.
Intrinsic requirement of Stat5a/b for generation of Foxp3+ CD4 T cells. Irradiated C57BL/6 Rag2−/− CD45.1+ congenic mice were reconstituted with 2 × 106 total fetal liver cells from CD45.2+ WT (left panels) or Stat5−/− (right panels) mice. After reconstitution (8 weeks), cell populations in the thymus (A) and spleen (B) were analyzed. Donor-derived CD4 SP T cells were analyzed for CD25 and Foxp3 expression. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
Intrinsic requirement of Stat5a/b for generation of Foxp3+ CD4 T cells. Irradiated C57BL/6 Rag2−/− CD45.1+ congenic mice were reconstituted with 2 × 106 total fetal liver cells from CD45.2+ WT (left panels) or Stat5−/− (right panels) mice. After reconstitution (8 weeks), cell populations in the thymus (A) and spleen (B) were analyzed. Donor-derived CD4 SP T cells were analyzed for CD25 and Foxp3 expression. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
We also approached the issue of an intrinsic requirement for Stat5 by selectively reducing Stat5a/b levels in T cells by breeding Stat5fl/− mice with CD4 cre mice.5 This approach had advantages, but also had some significant limitations in that Stat5 levels were reduced but not totally absent. To monitor Cre-mediated deletion, we also introduced YFP into the mice by breeding Stat5fl/−, CD4cre mice with indicator mice in which the gene encoding YFP was inserted into the Rosa locus (ROSA26–stop-floxed–YFP reporter mice). Flow cytometric analysis of thymocytes from 4- to 5-week-old Stat5fl/−, CD4cre, Yfp+ mice showed normal proportions of CD4 and CD8 thymocytes.5 Cre-mediated gene deletion as assessed by YFP expression was apparent in 90% of CD4+CD8+ double-positive (DP) thymocytes (data not shown), and in 98% of CD4+ SP thymocytes (Figure S2A). In sorted YFP+ CD4 SP thymocytes, genomic deletion of the Stat5a/b loci was observed as previously published.5 In YFP+ CD4+ cells, the levels of Stat5a/b mRNA were reduced to approximately 10%, and the levels of Stat5 protein were reduced to 20% to 30% (Figure S2B-C); not surprisingly, in YFP− cells, the levels of Stat5 were even greater. Thus, the presence of residual Stat5 protein is a caveat that needs to be considered in interpreting experiments using these cells.
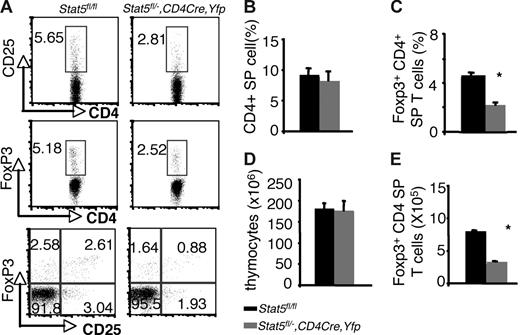
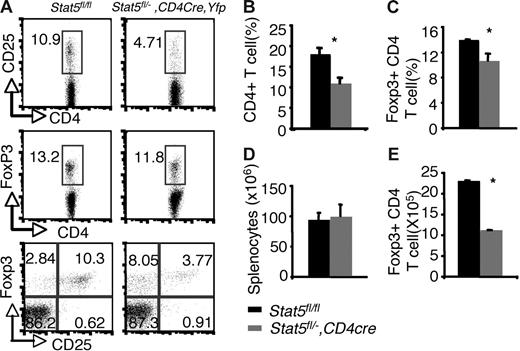
Examination of thymi from these mice revealed that the percentages of Foxp3- and CD25-expressing CD4 SP thymocytes were reduced by about 50% compared with that of Stat5fl/fl or Stat5fl/− littermates (5.18% and 5.65% versus 2.52% and 2.81% for Foxp3+CD4+ and CD25+CD4+ cells respectively; Figure 3A,C). The absolute numbers of Foxp3+CD4 SP thymocytes were also reduced by 50% compared with that of WT (Figure 3E), since the numbers of total thymocytes and proportions of CD4 SP T cells were normal in Stat5fl/−, CD4cre, Yfp mice (Figure 3B,D). In the spleen, 97% of the T cells were YFP+, and Stat5 protein levels were reduced by approximately 70% (Figure S2A,C). Flow cytometric analysis of the splenic CD4+ T cells showed that the percentage of CD25+ CD4+ T cells was reduced (Figure 4A), and the proportion of Foxp3-expressing cells from Stat5fl/−, CD4cre, Yfp mice was modestly and consistently reduced compared with that in Stat5fl/fl or Stat5fl/− mice (Figure 4A,C). The reduction in CD25-expressing cells was more dramatic than that of Foxp3-expressing cells, and in fact, most of the Foxp3-expressing CD4+ T cells from Stat5fl/−, CD4cre, Yfp mice were CD25dim (Figure 4A; bottom panel). The total numbers of splenic CD4+ T cells in Stat5fl/−, CD4cre, Yfp mice were roughly half of that seen in control mice (Figure 4B,D); as a result, the absolute number of Foxp3+CD4+ T cells in Stat5fl/−, CD4cre, Yfp mice was reduced by approximately 60% (Figure 4E). These results contrast with the near absence of Treg cells found in Stat5−/− mice. While this could be interpreted to suggest that extrinsic Stat5 expression contributes to the loss of Treg cells, in view of the stem cell transplantation experiments, we would argue that the modest reductions of Treg cells in Stat5fl/−, CD4cre, Yfp mice are more likely due to persistence of Stat5 expression in this model of tissue-specific deletion. Thus, these results further demonstrate the importance of Stat5 in both Treg cell development and maintenance. Consistent with the reduction in Foxp3+CD4+ T cells, we noted that systemic autoimmune disease was evident in Stat5fl/−, CD4cre, Yfp mice (Figure S3), similar to what has been observed in Stat5ΔN mice.33
Reduction of thymic Foxp3+ CD4+ T cells with tissue-specific diminution of Stat5a/b levels. (A) CD25 and Foxp3 expression were assessed on sorted CD4 SP thymocytes from Stat5fl/fl mice and YFP+CD4 SP thymocytes from Stat5fl/−, CD4Cre, Yfp mice. (B) Average proportion of CD4 SP thymocytes. (C) Mean percentage of Foxp3+ CD4+ T cells. (D) Average total numbers of thymocytes. (E) Absolute numbers of Foxp3+ CD4+ T cells. Means ± SE (n = 6) are shown; *P < .01 as determined by Student t test. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
Reduction of thymic Foxp3+ CD4+ T cells with tissue-specific diminution of Stat5a/b levels. (A) CD25 and Foxp3 expression were assessed on sorted CD4 SP thymocytes from Stat5fl/fl mice and YFP+CD4 SP thymocytes from Stat5fl/−, CD4Cre, Yfp mice. (B) Average proportion of CD4 SP thymocytes. (C) Mean percentage of Foxp3+ CD4+ T cells. (D) Average total numbers of thymocytes. (E) Absolute numbers of Foxp3+ CD4+ T cells. Means ± SE (n = 6) are shown; *P < .01 as determined by Student t test. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles.
Reduction of peripheral Foxp3+ CD4+ T cells with tissue-specific decrease of Stat5a/b levels. (A) CD25 and Foxp3 expression were analyzed in sorted CD4+ SP splenocytes from Stat5fl/fl mice and YFP+CD4+ SP splenocytes from Stat5fl/−, CD4Cre, Yfp mice. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles. (B) Average proportion of CD4+ SP splenocytes. (C) Mean percentage of Foxp3+ CD4+ T cells. (D) Average total numbers of splenocytes. (E) Absolute numbers of Foxp3+ CD4 T cells. Means ± SE (n = 6) are shown (*P < .01).
Reduction of peripheral Foxp3+ CD4+ T cells with tissue-specific decrease of Stat5a/b levels. (A) CD25 and Foxp3 expression were analyzed in sorted CD4+ SP splenocytes from Stat5fl/fl mice and YFP+CD4+ SP splenocytes from Stat5fl/−, CD4Cre, Yfp mice. Numbers indicate the percentage of CD25+ or FoxP3+ cells delineated by the rectangles. (B) Average proportion of CD4+ SP splenocytes. (C) Mean percentage of Foxp3+ CD4+ T cells. (D) Average total numbers of splenocytes. (E) Absolute numbers of Foxp3+ CD4 T cells. Means ± SE (n = 6) are shown (*P < .01).
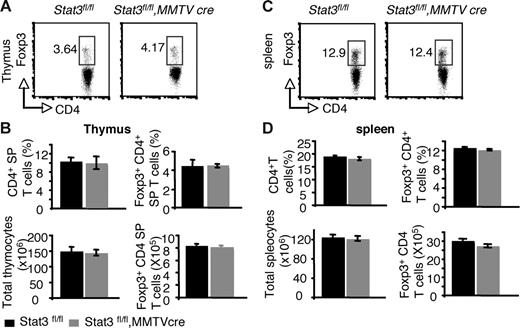
Recent studies using transient transfection have suggested that Stat3 and Stat5a/b may both positively regulate Foxp3.36 The reduction in Foxp3+ CD4+ T cells in the thymus and periphery of Stat5−/− mice argues against an essential role for Stat3 in regulating Foxp3 under normal circumstances. Nonetheless, it was important to formally document whether Stat3 was a significant contributor to the regulation of Foxp3. In Stat3fl/fl, MMTVCre mice, Stat3 mRNA levels were reduced to approximately 16% of normal levels in CD4+ T cells (Figure S4A). Examination of thymi and spleens from these mice revealed that the proportion and absolute numbers of Foxp3+ CD4+ T cells were normal (Figure 5).
Reduction of Stat3 levels in CD4 T cells has no effect on Foxp3 expression from both thymi and spleens. (A) Foxp3 expression was analyzed in CD4 SP thymocytes by flow cytometry. (B) The mean proportion of CD4 SP thymocytes, the mean percentage of Foxp3+ CD4+ T cells, the average total numbers of thymocytes, and the absolute numbers of Foxp3+ CD4 T cells in thymocytes are shown. (C) Foxp3 expression was analyzed in CD4+ splenocytes by flow cytometry. (D) The average proportion of CD4+ SP splenocytes, the mean percentage of Foxp3+ CD4+ T cells, the average total numbers of splenocytes, and the absolute numbers of Foxp3+ CD4 T cells are shown. Means ± SE (n = 5) are shown. In panels A and C, numbers indicate the percentage of FoxP3+ cells delineated by the rectangles.
Reduction of Stat3 levels in CD4 T cells has no effect on Foxp3 expression from both thymi and spleens. (A) Foxp3 expression was analyzed in CD4 SP thymocytes by flow cytometry. (B) The mean proportion of CD4 SP thymocytes, the mean percentage of Foxp3+ CD4+ T cells, the average total numbers of thymocytes, and the absolute numbers of Foxp3+ CD4 T cells in thymocytes are shown. (C) Foxp3 expression was analyzed in CD4+ splenocytes by flow cytometry. (D) The average proportion of CD4+ SP splenocytes, the mean percentage of Foxp3+ CD4+ T cells, the average total numbers of splenocytes, and the absolute numbers of Foxp3+ CD4 T cells are shown. Means ± SE (n = 5) are shown. In panels A and C, numbers indicate the percentage of FoxP3+ cells delineated by the rectangles.
Stat5 and Stat3 have opposing effects on cytokine-dependent FoxP3 regulation
Despite the persistence of residual Stat5 protein expression in Stat5fl/−, CD4cre, Yfp mice, this system was more amenable to analyzing T cells with reduced Stat5 levels. Previous studies have shown that Foxp3 can be induced in vitro by addition of exogenous TGF-β1.29,42,43 We therefore assessed the in vitro induction of Foxp3 in YFP+ CD25−CD4+ T cells from Stat5fl/−, CD4cre, Yfp mice. As shown in Figure 6A, CD25−CD4+ T cells expressed little Foxp3 prior to stimulation. Stimulation with anti-CD3, anti-CD28, and TGF-β1 was a potent inducer of Foxp3 in WT cells (Figure 6A). When endogenous IL-2 production was neutralized with anti–mIL-2 antibody, the percentage of Foxp3+ CD4+ T cells induced by TGF-β1 was markedly reduced. Conversely, addition of exogenous hIL-2 enhanced the generation of Foxp3+ CD4+ T cells. However, under all conditions, the proportion of Foxp3+ cells was markedly reduced when cells from Stat5fl/−, CD4cre, Yfp mice were used, and few Foxp3-expressing cells were induced even with addition of exogenous IL-2. Moreover, the level of induction of Foxp3 indicated by MFI was also lower, only 35% of that in wild-type cells, even under optimal conditions. Thus, despite the limitations of residual, low-level Stat5 expression in T cells from Stat5fl/−, CD4cre, Yfp mice, this system clearly supports the importance of Stat5a/b in Foxp3 regulation.
Stat5a/b are important for the in vitro induction of Foxp3. (A) Sorted splenic CD25−CD4+ T cells from Stat5fl/fl and YFP+ CD25−CD4+ T cells from Stat5fl/−, CD4Cre, Yfp+ mice were cultured with plate-bound anti-CD3 and anti-CD28 combined with or without TGF-β1, IL-2 (100 U/mL), and/or anti–IL-2 antibody as indicated. Representative of 3 experiments. Numbers in quadrants indicate FoxP3+ or FoxP3− cells. (B) Sorted splenic CD25−CD4+ T cells from Stat3fl/fl and Stat3fl/fl, MMTVcre mice were cultured with plate-bound anti-CD3 and anti-CD28 combined with or without TGF-β1 and IL-6 as indicated. Numbers indicate the percentage of FoxP3+ cells delineated by the rectangles.
Stat5a/b are important for the in vitro induction of Foxp3. (A) Sorted splenic CD25−CD4+ T cells from Stat5fl/fl and YFP+ CD25−CD4+ T cells from Stat5fl/−, CD4Cre, Yfp+ mice were cultured with plate-bound anti-CD3 and anti-CD28 combined with or without TGF-β1, IL-2 (100 U/mL), and/or anti–IL-2 antibody as indicated. Representative of 3 experiments. Numbers in quadrants indicate FoxP3+ or FoxP3− cells. (B) Sorted splenic CD25−CD4+ T cells from Stat3fl/fl and Stat3fl/fl, MMTVcre mice were cultured with plate-bound anti-CD3 and anti-CD28 combined with or without TGF-β1 and IL-6 as indicated. Numbers indicate the percentage of FoxP3+ cells delineated by the rectangles.
TGF-β1 in combination with IL-2 promotes in vitro differentiation of Treg cells and enhances Foxp3 expression, whereas IL-6 inhibits Foxp3 expression.29,44 To determine the role of Stat3 in mediating this effect of IL-6, we next analyzed Treg cell differentiation in CD25−CD4+ cells from Stat3fl/fl, MMTVcre mice. As shown in Figure 6B, Foxp3 levels were induced to comparable levels in CD25−CD4+ T cells from Stat3fl/fl, MMTVcre mice and WT mice. However, when IL-6 was added to cultures of WT T cells, the induction of Foxp3 was blocked, consistent with previously published data.29 In contrast, addition of IL-6 to cultures of T cells from Stat3fl/fl, MMTVcre mice failed to block the up-regulation of Foxp3 (Figures 6B, S4). We interpret these data to indicate that Stat5a/b are critical in vivo and in vitro for enhancing Foxp3 expression. In contrast, Stat3 is not required for induction of Foxp3; however, it does appear to be critical for mediating IL-6–dependent negative regulation of this key transcription factor.
Foxp3 is a direct target of Stat5
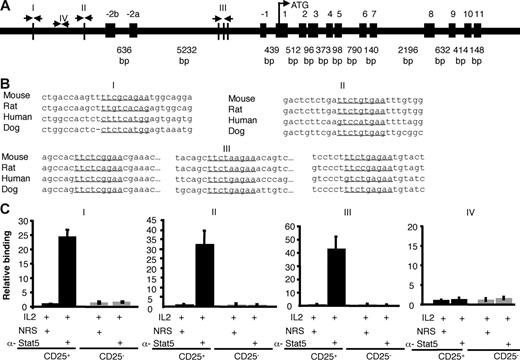
The present data suggest that Stat5a/b are critical for promoting Foxp3 expression in vivo and in vitro, likely through their role in mediating IL-2 signals, but it was possible that actions of Stat5a/b may not be direct. However, multiple consensus Stat-binding sites are present in the mouse Foxp3 gene promoter region (I, II) and the first intron (III). The most highly conserved sites are in the first intron (Figure 7). Stat5 binding to the native Foxp3 gene in murine primary cells was assessed using sorted CD25+CD4+ and CD25−CD4+ SP thymocytes and chromatin immunoprecipitation with anti-Stat5 antibodies. Subsequent real-time PCR amplification of the Foxp3 gene surrounding 3 putative Stat binding sites showed significant IL-2–inducible Stat5 binding. As a control, we also assessed a segment that does not contain consensus Stat binding sites, but found that this segment was not amplified with anti-Stat5 immunoprecipitation (Figure 7C). Stat5 binding to the Foxp3 gene was also not detected in CD25−CD4+ SP cells, but was demonstrable in CD25+CD4+ splenocytes (data not shown). These data suggest that Stat5 may play a direct role in regulating Foxp3 transcription.
Stat5 binds the Foxp3 gene. (A) Schematic of the mouse Foxp3 gene. Vertical lines depict potential Stat binding sites in the first intron and the putative promoter (I, II, III). Site IV does not contain a Stat-binding site and was used as a control. (B) Stat-binding sites in the mouse Foxp3 gene are underlined and aligned with sequences from other species. Site I is located between 006010267 and 006010275 in the mouse genome (http://genome.ucsc.edu/cgi-bin/hgGateway?hgsid = 83 436 504&clade = vertebrate&org = Mouse&Db = mm7). Site II is located between 006012112 and 006012120. Intronic sites designated III are located between 006017209 and 006017217, 006017406 and 006017414, and 006017523 and 006017531. All sequences are from the sense strand; note that the previous Stat-binding site sequences identified by Zorn et al36 were from the antisense strand, although the same sites were interrogated in our analysis. (C) Sorted thymic CD25+CD4 SP (▪) and CD25−CD4 SP (⊡) T cells were treated with IL-2 for 1 hour. Proteins and DNA were cross-linked with formaldehyde, cells were lysed, and DNA was sheared. Chromatin immunoprecipitation was performed using either normal rabbit serum or anti-Stat5 antibody. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers and probes for sites I, II, III, and the irrelevant site IV. Values were normalized to corresponding input control and are expressed as fold enrichment relative to normal rabbit serum for each experiment. Means ± SE are shown.
Stat5 binds the Foxp3 gene. (A) Schematic of the mouse Foxp3 gene. Vertical lines depict potential Stat binding sites in the first intron and the putative promoter (I, II, III). Site IV does not contain a Stat-binding site and was used as a control. (B) Stat-binding sites in the mouse Foxp3 gene are underlined and aligned with sequences from other species. Site I is located between 006010267 and 006010275 in the mouse genome (http://genome.ucsc.edu/cgi-bin/hgGateway?hgsid = 83 436 504&clade = vertebrate&org = Mouse&Db = mm7). Site II is located between 006012112 and 006012120. Intronic sites designated III are located between 006017209 and 006017217, 006017406 and 006017414, and 006017523 and 006017531. All sequences are from the sense strand; note that the previous Stat-binding site sequences identified by Zorn et al36 were from the antisense strand, although the same sites were interrogated in our analysis. (C) Sorted thymic CD25+CD4 SP (▪) and CD25−CD4 SP (⊡) T cells were treated with IL-2 for 1 hour. Proteins and DNA were cross-linked with formaldehyde, cells were lysed, and DNA was sheared. Chromatin immunoprecipitation was performed using either normal rabbit serum or anti-Stat5 antibody. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers and probes for sites I, II, III, and the irrelevant site IV. Values were normalized to corresponding input control and are expressed as fold enrichment relative to normal rabbit serum for each experiment. Means ± SE are shown.
Because IL-6–mediated inhibition of Foxp3 expression was Stat3 dependent, we also assessed whether we could also detect direct binding of Stat3 to the Foxp3 locus. However, IL-6–dependent binding of Stat3 to the regions of Foxp3 gene to which Stat5 binds was marginal (Figure S4C). That is, in contrast to Stat5, which enriched the binding regions by more than 20-fold, the same regions were enriched by less than 4-fold with anti-Stat3 antibody compared with normal rabbit serum. This is of interest because the binding sites for Stat3 and Stat5 binding are thought to be quite similar.45 It is notable therefore that Stat5 evidently binds the Foxp3 gene well, whereas Stat3 binds this locus poorly despite the clear Stat3-dependent functional effects on Foxp3 in T cells.
Discussion
In this study, we explored the role of Stat5a/b in regulating Foxp3 expression and Treg cells. Our data indicate that Stat5a/b are critical for both the development and maintenance of Treg cells. This appears to be an intrinsic requirement for Stat5a/b in Treg cells and is likely mediated through direct effects of Stat5a/b on the transcription of the Foxp3 gene, which has multiple Stat-binding sites.
Stat5a/b have long been recognized to mediate IL-2 signals, and the importance of IL-2 in Treg cell development and maintenance has been established by a number of approaches. IL-2 signaling not only leads to the activation of Jaks and Stats, but also Ras-MAPK and PI3K-AKT pathways. Jak3 and γc are essential for Treg cells, but previous studies using CD25 as a marker in Stat5ΔN mice led to the conclusion that Stat5 is required for peripheral Treg cell maintenance but not for Treg cell development.34 This would suggest that Stat5 might not be the key factor regulating Foxp3 expression, or that it was redundant with other factors. However, it is now clear that Stat5ΔN mice have residual Stat5 function that may support Foxp3+ CD4+ T-cell development. Indeed, analysis of mice completely deficient in Stat5 showed that Foxp3+ cells were severely reduced (Figures 1,3) documenting the criticality of this transcription factor for Foxp3 expression. Of note, we found reduced but not absent Treg cells in both thymi and spleens of Stat5fl/−, CD4cre, Yfp mice, but, like cells from Stat5ΔN mice, some residual Stat5 protein is also present in T cells from Stat5fl/−, CD4cre, Yfp mice. We interpret these data to suggest that low levels of Stat5 in Stat5ΔN and Stat5fl/−, CD4cre mice permit Foxp3 expression, even in circumstances where CD25 levels are dramatically reduced. We have found other circumstances where selective pressure for Stat5 expression allows for escape of cells in which the Stat5 genes are not deleted. We believe that this is simply a limitation of this system and needs to be carefully considered in assessing phenotypes using Cre-mediated deletion.
Because of its role in binding IL-2, CD25 is a critical factor in regulating Foxp3. Thus it was possible that the importance of Stat5 for Treg cells was primarily due to its requirement in regulating CD25 expression rather than directly regulating Foxp3. However, we found that in vitro induction of Foxp3 was poor in Stat5-deficient cells, despite levels of IL-2 that would obviate the need for CD25. While regulation of CD25 may contribute to the poor expression of Foxp3, we believe our data also argue for a more direct role independent of effects on CD25 expression. In addition, analysis of Il2−/− and Il2ra−/− (CD25−/−) mice showed that the impairment of Foxp3 expression is less severe than what was observed in mice lacking Jak3, Il2rg, or Stat5.28 This also suggests that other γc cytokines, which signal predominantly through Stat5, may also play a role in promoting Foxp3 expression and Treg cell development.
Previous studies have noted two consensus Stat-binding elements located in intron 1 of the human FOXP3 gene.36 Gain-of-function Stat5 and Stat3 alleles were also found to trans-activate a FOXP3 reporter construct in transient transfection assays.36 This led to the conclusion that IL-2–induced Foxp3 expression is mediated through both Stat5 and Stat3, and both of the Stats might play positive roles in regulating Foxp3 expression. Similarly, constitutive Stat3 activation was observed in malignant T cells that expressed the NPM/ALK fusion protein. These T cells express IL-10, TGF-β, and Foxp3, and have immunosuppressive properties reminiscent of Treg cells.37 These authors attributed this phenotype to constitutive activation of Stat3,37 but Stat5 is also constitutively activated in this setting.46 However, we found that reduction of Stat3 levels in T cells did not have a major effect on Foxp3 expression in Stat3fl/fl, MMTVcre mice, indicating that Stat3 and Stat5 do not have redundant roles in Foxp3 regulation in normal mice. Indeed, IL-6–dependent Stat3 activation was suggested to inhibit Treg development during allergic airway inflammation.44 Recently, it was shown that in vitro stimulation of CD4+ T cells with IL-6 and TGF-β1 down-regulated Foxp3 expression.29 Our data demonstrate that the inhibitory effect of IL-6 on Foxp3 expression is dependent on Stat3 by using Stat3-deficient T cells.
In addition, our data clearly indicate that Stat5a/b bind to the murine Foxp3 gene, implying a direct role of Stat5 in regulating Foxp3 transcription. In contrast, Stat3 binding to these sites in the Foxp3 locus was marginal. Given that IL-6 inhibits Foxp3 expression in Stat3-dependent manner, Stat3 must either bind to the Foxp3 locus at sites distinct from the Stat5 binding sites, or it must act indirectly. Regardless, it is clear that Stat5a/b and Stat3 function distinctly in regulating Foxp3, and it will be important to elucidate precisely how they mediate cytokine effects. It will be of great interest to define how they interact with other transcription factors and coactivators and to determine their ability to influence epigenetic modifications of the Foxp3 locus.
Authorship
Contribution: Z.Y. was the prinicipal participant designing and performing research, analyzing data, and writing the manuscript; Y.K., G.S., M.K., and L.D. performed research and analyzed data; Y.K. and W.T.W. performed research and prepared the manuscript; A.L., G.W.R., and L.H. provided materials; E.M.S. and R.M. analyzed data and participated in the preparation of the manuscript; and J.J.O'S. and C.W. were responsible for the overall study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John J. O'Shea, Bldg 10, Rm 9N262, 10 Center Drive, MSC-1820, NIH, Bethesda, MD 20892-1820; e-mail: osheajo@mail.nih.gov; or Changyou Wu, Department of Immunology, Zhongshan Medical School, Sun Yat-sen University, China; e-mail: changyou_wu@yahoo.com.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank the NIAMS flow cytometry core facility for cell sorting. We also thank Dr Richard Siegel and Yasmine Belkaid for critically reading this manuscript.
This work was supported by the Intramural Research Programs of NIAMS, NIAID, and NIDDK at NIH. R.M. was supported by the Austrian Science Fund (FWF) grant SFB F28.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal