Abstract
Regulatory molecules produced by stromal cells are often membrane bound until cleaved by matrix metalloproteinases (MMPs); cleavage can either activate or inactivate regulatory functions. We report here that marrow stromal cells induce the expression of MMP-9 in monocytes. Induction was contact independent and could be reproduced with recombinant MCP-1/CCL2, whereas IL-6, M-CSF, G-CSF, GM-CSF, IL-8/CXCL8, SDF-1/CXCL12, and MGSA/CXCL1 did not have this effect. Stroma-induced levels of MMP-9 in the monocyte population from healthy donors were relatively consistent, whereas induced levels varied significantly (P < .001) in the CD14+ population from 27 patients with myelodysplastic syndrome (MDS). In patients with a clonal chromosomal marker, the level of inducible MMP-9 expression in the monocyte population was inversely correlated with the percentage of marker-positive cells (n = 11, P = .01), suggesting that the ability to induce MMP-9 may be compromised in clonally derived monocytes. The inducible levels of MMP-9 were also inversely correlated with marrow cellularity observed in biopsies from MDS patients (P < .001). We conclude that monocytes can express MMP-9 in response to stromal factors and that this response may be significantly decreased in MDS-derived monocytes.
Introduction
The hematopoietic microenvironment (ME) is composed of cells of hematopoietic and nonhematopoietic origins. These cellular components function in concert to produce soluble and membrane-bound cytokines, as well as an extracellular matrix (ECM). The ECM acts, in part, to facilitate structural organization, but also serves to concentrate cytokines. Matrix metalloproteinases (MMPs) are the principal enzymes for remodeling the ECM and for facilitating the movement of cells.1,2,3,4,5,6 One proteinase, gelatinase B/MMP-9, reportedly also functions to release cytokines that are either bound to the ECM or tethered to ME-cell surfaces. Excess production of MMP-9 can disrupt the balance of available cytokines leading to tissue damage and disease.7,8,9,10
In addition to ECM remodeling, MMPs are required for bone remodeling, wound healing, angiogenesis, and stem-cell regulation.11,12 Leukocytes require MMP function to infiltrate or egress tissues, and tumor cells secrete MMPs to facilitate invasion and disseminate metastases. Two members of the MMPs, gelatinase A (also called MMP-2) and gelatinase B (also called MMP-9) have a gelatin-binding domain homologous to the fibronectin type II domain. Whereas gelatinase A/MMP-2 is constitutively produced by most cell types, gelatinase B/MMP-9 is secreted only after specific stimulation. Once secreted, MMP-9 may become bound to the ECM or tethered to cell-surface docking proteins such as integrins, CD44, or reversion-inducing cysteine-rich protein with Kazal motifs (RECK).13,14,15,16 MMP-9 is reported to degrade denatured collagen type IV and contribute to the turnover of the basement membrane.17
MMP-9 is a major secretory product of macrophages and granulocytes.18,19,20 The enzyme is also secreted by lymphocytes, keratinocytes, and fibroblasts on stimulation.21,22,23 Thus, both hematopoietic cells and stroma could be a source of MMP-9 in the marrow ME. One study, using an MMP-9 knockout model and a metalloproteinase inhibitor, reported a role for MMP-9 in the marrow ME, specifically in the mobilization of stem and progenitor cells.2 In that report, bone marrow cells were proposed as the source of MMP-9; precisely which type of marrow cell was not specified. Although subsequent studies failed to reproduce the effect of MMP-9 in knockout models,24,25,26 a role for MMP-9 in stem-cell mobilization has not been excluded since the compensatory up-regulation of other proteases may mask the effects of the MMP-9 deficiency, and subtle differences, both in genetic background and in homologous recombination, may exist in various knockout mouse lines. In addition, monoclonal antibodies against MMP-9 were shown to block the IL-8–mediated mobilization of hematopoietic progenitor cells in primates,27 suggesting a role for MMP-9 in this model.
In the present study, we investigated the induction of MMP-9 mRNA and protein expression in CD14+ peripheral-blood mononuclear cells (PBMCs) from healthy donors and from patients with myelodysplastic syndrome (MDS). We found a consistent and significant up-regulation of both mRNA and protein in normal CD14+ cells when they were cultured in stromal-cell–conditioned media. However, the levels of stroma-induced MMP-9 were highly variable in CD14+ cells from patients with MDS. Further analysis indicated that inducible levels were inversely correlated with the percentage of clonal cells as indicated by a chromosomal marker and with marrow cellularity.
Patients, materials, and methods
Patient data and samples
Bone marrow aspirates, biopsies, and peripheral blood were obtained from healthy donors, aged 25 to 70 years, and patients with MDS, aged 24 to 77 years, after written informed consent was obtained using forms approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center (FHCRC) in accordance with the Declaration of Helsinki. Patient samples were obtained prior to conditioning for transplantation and at least 3 to 5 weeks after any previous therapy. Patient data are provided in Table S1 (available on the Blood website; see the Supplemental Table link at the top of the online article). Cytogenetic analyses on all patients were provided by the Clinical Cytogenetics Laboratory of the FHCRC and Seattle Cancer Care Alliance under the direction of Dr Eileen Bryant.
PBMCs from healthy donors and MDS patients were isolated over a Ficoll-Hypaque step gradient and washed 3 times in Hanks balanced salt solution (HBSS). CD14+ cells were isolated by AutoMacs (Miltenyi Biotec, Auburn, CA) using a CD14 monoclonal antibody (clone TÜK4; Dako, Carpinteria, CA) and anti–mouse immunoglobulin G2a+b–conjugated magnetic microbeads (Miltenyi Biotec) as described before.28 The purity of isolated CD14+ cells from healthy donors and MDS patients was 98% (range, 94%-99%) and 82% (range, 60%-94%), respectively.
To prepare primary fibroblasts from bone marrow of healthy donors, mononuclear cells were isolated over a Ficoll-Hypaque step gradient and cultured for 1 day in supplemented RPMI 1640 containing 10% FCS. Adherent cells were collected by trypsinization, and residual hematopoietic cells depleted by AutoMacs using anti-CD45– and anti-CD14–conjugated magnetic beads. The remaining cells were cultured in RPMI 1640 containing FCS until primary fibroblasts reached 60% to 80% confluency. Recombinant cytokines used in this study were purchased from R&D Systems (Minneapolis, MN).
Preparation of HS-5 conditioned medium
HS-5 cells (3 × 106) were grown in supplemented RPMI 1640 medium containing 10% FCS in a T-75 flask. Conditioned medium (CM) was harvested after 5 days and clarified by centrifugation and stored at 4°C. The CM was diluted with one volume of supplemented RPMI 1640 medium before use. Cytokine levels were determined using enzyme-linked immunosorbent assays (ELISAs) performed by the Cytokine Laboratory Shared Resource at the FHCRC.
Syber Green real-time PCR analysis
Expression levels of RNA transcripts were quantitated by real-time polymerase chain reaction (PCR). Total RNA was purified using RNeasy spin column (Qiagen, Valencia, CA) and treated with RNase-free DNase (Qiagen) according to the manufacturer's protocol. Samples were then reverse-transcribed into cDNA with an oligo dT12-18 primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) by incubating for 2 hours at 42°C, followed by termination of the reaction by heat inactivation (65°C, 20 minutes). cDNAs were mixed with Syber Green (SyBr) PCR master mix (Applied Biosystems, Foster City, CA) and primers; real-time PCR was performed using ABI Prism 7900HT Sequence Detector (Applied Biosystems). PCR conditions were 15 seconds at 95°C and 2 minutes at 68°C with 40 cycles for real-time PCR. Specific primers for MMP-9 and glycerol-3-phosphate dehydrogenase (G3PDH) were designed by the Clonal Analysis Shared Resource at the FHCRC. Forward and reverse oligonucleotide primers were as follows, in 5′ to 3′ orientation: GGGCTTAGATCATTCCTCAGTGCC and GAAGATGTTCACGTTGCAGGCATC for MMP-9, and ATCGCTCTGAAATTAGCCTACTGCC and TAAGGCTGGCAAATACACCAACACC for G3PDH.
Immunoprecipitation, SDS-PAGE, and immunoblotting of MMP-9 protein
CD14+ cells isolated from PBMCs were placed in 12-well plates at 0.2 × 106 cells/well in 2 mL control medium or HS-5 conditioned medium (HS5 CM), and cultured for 5 days. The culture media were collected, and protease inhibitors (Complete Mini, Roche, Mannheim, Germany) were added. Immunoprecipitation of MMP-9 protein in the culture medium (0.6 mL) was performed using 10 μg of a monoclonal antibody against MMP-9 (MAB 13415, Chemicon, Temecula, CA). This antibody binds both 92-kDa pro- (latent) and 86-kDa active forms of MMP-9. After incubation at 4°C for 2 hours, 50 μL protein G-coupled agarose beads (Pharmacia, Uppsala, Sweden) was added, and the incubation continued for an additional 2 hours at 4°C. The agarose beads were extensively washed with phosphate-buffered saline (PBS), and the immunoprecipitated proteins were heat-denatured in 60 μL Laemmli sample buffer containing 5% β-mercaptoethanol. The denatured proteins (30 μL) and prestained marker proteins (10 μL; PageRuler, Fermentas Life Sciences, Hanover, MD) were applied to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen) with a 4% to 12% linear gradient of acrylamide and immunoblotted to a nitrocellulose membrane. The membrane was stained with Ponceau S (Sigma, St Louis, MO) for proteins to confirm an equivalent protein load, blocked in Blotto, and probed sequentially with polyclonal antibodies against pro- and activated MMP-9 (1:500 dilution; AB805, Chemicon) and IRDye 800-conjugated secondary antibodies (1:2000 dilution; Rockland, Gilbertsville, PA). The bound fluorescence was visualized using Odyssey scanner (LI-COR, Lincoln, NE) and quantitated using Odyssey Application Software v.1.2. The molecular weight of MMP-9 protein was determined using a BenchMark protein ladder (Invitrogen).
In some experiments, MMP-9 protein was purified using gelatin-conjugated agarose beads according to the methods by Goldberg et al.29 In brief, culture supernatants from CD14+ cells treated with or without HS5 CM for 5 days were harvested, and the protein inhibitors were added. The supernatants were incubated with 0.1 to 0.2 mL gelatin-agarose beads (Sigma) for 2 hours. After exhaustive washing with PBS, the bound proteins were denatured in the Laemmli sample buffer containing 5% β-mercaptoethanol. They were applied to SDS-PAGE, immunoblotted using the anti–MMP-9 antibodies, and quantitated as described.
Immune histochemistry of MMP-9 protein in bone marrow
The Experimental Histopathology Laboratory at the FHCRC performed immune cytochemistry for MMP-9 protein detection according to published methods.30 In brief, bone marrow biopsies and aspirate specimens from patients with MDS and healthy donors were immediately fixed for 2 to 3 hours in B5 fixative and decalcified in Richard-Allan Scientific Decalcifying Solution (Kalamazoo, MI) for 2.5 hours at room temperature. After the specimens were rinsed in running tap water for 5 minutes, they were processed in paraffin and embedded. Sections (5 μm) were treated with Target Retrieval (DakoCytomation, Carpinteria, CA) according to the manufacturer's protocol. After rehydration in Dako Wash Buffer, the slides were incubated with 3% H2O2 for 8 minutes to block endogenous peroxidase activity. The sections were then incubated with Dako Wash Buffer containing 15% swine serum and 5% human serum for 10 minutes; they were incubated with a monoclonal antibody against human MMP-9 (10 μg/mL; REGA-2D9) or a concentration-matched isotype control (IgG1, Chemicon) at 22°C for 30 minutes. The REGA-2D9 antibody was previously shown to work for immune cytochemistry using tissues fixed with B5 fixative.30 After washing, slides were incubated with HRP-conjugated Envision Plus (DakoCytomation) for 30 minutes. The HRP was visualized with 3,3′-diaminobenzidine (DAB; DakoCytomation) for 7 minutes, and the sections were counterstained with hematoxylin (DakoCytomation) for 2 minutes. Pictures were taken with a Nikon E802 microscope (Nikon, Tokyo, Japan) fitted with either a 40 ×/1.30 Plan Fluor or a 100 ×/1.30 Plan Fluor objective lens. Bright contrast images were acquired on a Photonomics Coolsnap cf camera (Roper Scientific, Tucson, AZ).
Marrow cellularity
Patient blood, used for CD14+-cell isolation, and marrow biopsies were obtained concurrently prior to conditioning for stem-cell transplantation and at least 3 to 4 weeks after prior treatment (Table S1). To evaluate marrow cellularity, diagnostic marrow biopsies and aspirated particle preparations were fixed, sectioned, and stained using hematoxylin and eosin as well as periodic acid-Schiff. Coded slides were analyzed by a single pathologist (R.C.H.) without knowledge of clinical or laboratory information. Marrow cellularity was expressed as the proportion of the stromal space occupied by cells.
Combined immune histochemistry and FISH
Fluorescence in situ hybridization (FISH) analysis was conducted on monocytes from patients with MDS and healthy donors. CD14+ cells were cultured in HS5 CM for 5 days, cytospun, and fixed in 10% buffered formalin. Cells were permeabilized using 1% Triton X-100, then immunostained for MMP-9 protein using REGA-2D9 monoclonal antibody or an isotype-matched control, followed by HRP-conjugated secondary antibodies and Vector SG peroxidase substrate (Vector Laboratories, Burlingame, CA) as described in “Immune histochemistry of MMP-9 protein in bone marrow.” Slides were subsequently treated with 0.2 M HCl for 20 minutes, then placed in 1 M sodium isothiocyanate at 80°C for 3 minutes, followed by enzymatic digestion with 100 μg/mL pepsin (Sigma) in 10 mM HCl at 37°C for 5 minutes. Slides were then fixed in 4% formalin for 10 minutes, subsequently dehydrated using a cold alcohol wash series, and denatured in 70% formamide/2 × SSC at 72°C for 5 minutes followed by another cold alcohol series. The chromosome 7-specific probe (Vysis CEP7 Spectrum Green) provided from FHCRC cytogenetics, was denatured by heating to 72°C for 5 minutes. The probes were then placed on the slides, sealed with coverslips, and allowed to hybridize overnight at 37°C. The following day, the coverslips were removed and the slides were washed in 0.4 × SSC/0.3% NP-40 at 72°C for 2 minutes. Slides were then rinsed in 2 × SSC/0.1% NP-40, stained with 4′6-diamidino-2-phenylindole (DAPI) at 1 μg/mL and mounted with ProLong Antifade mounting media (Molecular Probes, Eugene, OR). At least 150 cells were counted for each sample. Pictures were taken on a Nikon E800 microscope fitted with a 100 ×/1.30 Plan Fluor objective lens. Fluorescent images were acquired on a Photonomics Coolsnap HQ camera (Roper Scientific).
Statistics
Means and SDs of the gene expression were measured, and statistically significant differences in gene expression were identified by paired Student t test (P < .05). The Folded F test was used to analyze differences in variation shown in Figure 3. Pearson correlation coefficients were calculated for data in Figure 5.
Endotoxin and Mycoplasma screening
Endotoxin levels of media, cytokine, and all other reagents were tested using a Limulus Amebocyte Lysate test kit (Charles River Laboratories, Wilmington, MA) by kinetic-turbidmetric method. Mycoplasma levels of cells were tested using DAPI-based cytochemical detection method. Both assays were performed by the Biologics Production Facility at the FHCRC.
Results
HS5 CM augments MMP-9 gene expression in normal CD14+ cells isolated from blood
Levels of MMP-9 gene expression in normal CD14+ cells were determined by real-time PCR. Little or no MMP-9 gene expression was detected in CD14+ cells on day 0 (no culture). After 5 days of culture in control medium, MMP-9 gene expression was increased significantly; culturing cells in HS5 CM resulted in an additional 3-fold increase in expression (Figure 1)
Up-regulation of MMP-9 gene expression in CD14+ cells by conditioned media from HS-5 stromal cells. Total RNA was isolated from CD14+ cells from 10 healthy donors prior to cultures (No culture) or after culturing for 5 days in control medium or in conditioned medium from HS-5 (HS5CM). MMP-9 gene expression was determined by the real-time PCR assay and normalized with G3PDH gene expression. P values were calculated by paired Student t test. Data are shown as mean ± STD.
Up-regulation of MMP-9 gene expression in CD14+ cells by conditioned media from HS-5 stromal cells. Total RNA was isolated from CD14+ cells from 10 healthy donors prior to cultures (No culture) or after culturing for 5 days in control medium or in conditioned medium from HS-5 (HS5CM). MMP-9 gene expression was determined by the real-time PCR assay and normalized with G3PDH gene expression. P values were calculated by paired Student t test. Data are shown as mean ± STD.
MMP-9 is known to be expressed in monocytes and granulocytes. The CD14+ cells used in this study were monocytes isolated from the mononuclear fraction of peripheral blood followed by purification using magnetic microbeads. The purity of the CD14+ cells from healthy donors was more than 95% by flow cytometric analysis. Given the low level of potential granulocyte contamination and the fact that granulocytes only survive a few hours, we conclude that the MMP-9 gene expression detected after 5 days of culture could be attributed to the CD14+ monocytes. This was confirmed by immune cytochemistry using an antibody against MMP-9 (data not shown).
Primary stromal fibroblasts from bone marrow of healthy donors were also tested for the ability to augment MMP-9 gene expression in monocytes. CD14+ cells were cocultured with the marrow fibroblasts or with HS-5 stromal cells for 5 days, and MMP-9 gene expression levels were determined in nonadherent CD14+ cells. Comparable levels of MMP-9 gene expression were induced in CD14+ cells by normal marrow fibroblasts and HS-5 cells (normalized MMP-9 gene expression, 11.19 ± 0.87 and 12.21 ± 1.77, respectively).
All media, culture supernatants, and cells used in these experiments were tested for endotoxin and Mycoplasma and found to be negative, thereby excluding the possibility of endotoxin- and Mycoplasma-induced MMP-9 gene expression.
HS-5 CM stimulates MMP-9 protein secretion in normal CD14+ cells
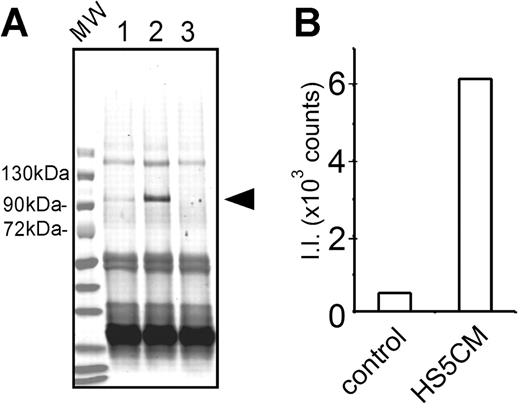
Next, we used immune precipitation to determine if MMP-9 protein expression correlated with gene expression in CD14+ cells. MMP-9 protein was detected after culture in control medium, but was increased 12-fold after culture in HS5 CM (Figure 2) The molecular weight of MMP-9 protein produced by CD14+ cells was determined to be 92 kDa, which is the expected molecular weight for the intact form of human MMP-9. The protease was not detected in fresh medium used as a negative control (Figure 2A lane 3) nor in conditioned medium from HS-5 cells (data not shown). MMP-9 secretion was confirmed by gelatin-agarose affinity chromatography, followed by SDS-PAGE and Western blot analysis (data not shown).
Secretion of MMP-9 protein. (A) CD14+ cells (0.1 × 106 cells) were cultured for 5 days in control medium or conditioned medium from HS-5 (lanes 1 or 2, respectively). Culture media were harvested. The media were immunoprecipitated, proteins were denatured, run on SDS-PAGE, and transferred onto a nitrocellulose membrane. The membrane was incubated with polyclonal anti–MMP-9 antibodies, followed by fluorescent-labeled secondary antibodies. As a control, fresh medium was used as a mock immunoprecipitation (lane 3). The conditioned medium from HS-5 cells was also negative (data not shown). An arrowhead indicates the position of MMP-9 protein. Immunoglobulin heavy and light chains (∼60 kDa and ∼25 kDa) are stained in all 3 lanes. This is a representative blot of 3 similar experiments. MW indicates a lane for prestained molecular weight markers. (B) Fluorescence intensity of MMP-9 protein bands in lanes 1 and 2 was determined by Odyssey scanner (Control and HS5CM, respectively). II indicates integrated intensity.
Secretion of MMP-9 protein. (A) CD14+ cells (0.1 × 106 cells) were cultured for 5 days in control medium or conditioned medium from HS-5 (lanes 1 or 2, respectively). Culture media were harvested. The media were immunoprecipitated, proteins were denatured, run on SDS-PAGE, and transferred onto a nitrocellulose membrane. The membrane was incubated with polyclonal anti–MMP-9 antibodies, followed by fluorescent-labeled secondary antibodies. As a control, fresh medium was used as a mock immunoprecipitation (lane 3). The conditioned medium from HS-5 cells was also negative (data not shown). An arrowhead indicates the position of MMP-9 protein. Immunoglobulin heavy and light chains (∼60 kDa and ∼25 kDa) are stained in all 3 lanes. This is a representative blot of 3 similar experiments. MW indicates a lane for prestained molecular weight markers. (B) Fluorescence intensity of MMP-9 protein bands in lanes 1 and 2 was determined by Odyssey scanner (Control and HS5CM, respectively). II indicates integrated intensity.
Recombinant MCP-1/CCL2 and IL-1β induce MMP-9 gene expression and protein secretion in normal CD14+ cells
We previously reported that HS-5 cells robustly express and secrete many cytokines and chemokines.31,32 These data prompted us to test the ability of those HS-5 products to stimulate MMP-9 gene expression in CD14+ cells.
MMP-9 gene expression was analyzed after normal CD14+ cells were cultured for 5 days in the presence or absence of 9 different recombinant factors (Table 1) Factors to be tested were also chosen on the basis of known receptor expression by monocytes. Change in MMP-9 gene expression by each cytokine was compared to that obtained with control medium. Conditioned medium from HS-5 cells was used as a positive control and induced a 5-fold increase compared to the control medium. The MMP-9 gene expression increased 4.1-fold with 0.5 ng/mL MCP-1 (CCL2), which is the concentration of MCP-1 in HS5 CM. Higher and lower concentrations of MCP-1 were less effective. IL-1 present in HS5 CM at 0.25 ng/mL increased gene expression 3-fold at a concentration of 0.05 to about 0.5 ng/mL. IL-6 and M-CSF had only marginal effects (∼2-fold). Other cytokines known to be secreted by HS-5 cells, including IL-8/CXCL8, G-CSF, or GM-CSF, did not alter MMP-9 gene expression in CD14+ cells at the concentrations tested. All media and cytokines used in these experiments were tested for endotoxin and found to be negative, thereby excluding the possibility of endotoxin-induced MMP-9 gene expression.
Effect of recombinant cytokines and HS5 CM on MMP-9 gene expression and protein secretion in CD14+ cells
| Concentration, ng/mL . | MMP-9 gene expression . | MMP-9 protein secretion . |
|---|---|---|
| HS5 CM* | 500 | 8756 |
| MCP-1/CCL2 | ||
| 0.05 | 236 | 153 |
| 0.5 | 409 | 1089 |
| 5 | 263 | 136 |
| IL-1β | ||
| 0.005 | 211 | 95 |
| 0.05 | 312 | 155 |
| 0.5 | 356 | 576 |
| IL-6 | ||
| 0.5 | 146 | 119 |
| 5 | 244 | 124 |
| 50 | 206 | 121 |
| 500 | 161 | 205 |
| M-CSF | ||
| 0.05 | 180 | 163 |
| 0.5 | 152 | 287 |
| G-CSF, 10 | 193 | ND |
| GM-CSF, 100 | 59 | ND |
| MGSA/CXCL1, 100 | 111 | ND |
| IL-8/CXCL8, 10 | 125 | ND |
| SDF-1a/CXCL12, 100 | 125 | ND |
| Concentration, ng/mL . | MMP-9 gene expression . | MMP-9 protein secretion . |
|---|---|---|
| HS5 CM* | 500 | 8756 |
| MCP-1/CCL2 | ||
| 0.05 | 236 | 153 |
| 0.5 | 409 | 1089 |
| 5 | 263 | 136 |
| IL-1β | ||
| 0.005 | 211 | 95 |
| 0.05 | 312 | 155 |
| 0.5 | 356 | 576 |
| IL-6 | ||
| 0.5 | 146 | 119 |
| 5 | 244 | 124 |
| 50 | 206 | 121 |
| 500 | 161 | 205 |
| M-CSF | ||
| 0.05 | 180 | 163 |
| 0.5 | 152 | 287 |
| G-CSF, 10 | 193 | ND |
| GM-CSF, 100 | 59 | ND |
| MGSA/CXCL1, 100 | 111 | ND |
| IL-8/CXCL8, 10 | 125 | ND |
| SDF-1a/CXCL12, 100 | 125 | ND |
Data are represented as percent control. Control values were derived from CD14 cells cultured for 5 days in control media consisting of 10% FCS in RPMI 1640 medium.
ND indicates not determined.
HS5 CM (conditioned medium) was diluted with an equal volume of fresh medium for use in culturing CD14+ cells. Concentrations of the cytokines in HS5 CM were: IL-1β, 0.25 ng/mL; MCP-1, 0.5 ng/mL; M-CSF, 0.25 ng/mL; and IL-6, 250 ng/mL. CD14 cells were harvested for RNA and supernatants collected for protein detection after 5 days of culture.
MMP-9 gene expression in CD14+ cells from patients with MDS
Peripheral blood was obtained from 27 patients with primary MDS. Patient characteristics are provided in Table S1. CD14 cells were isolated as described, and MMP-9 gene expression was determined before and after cells were cultured with HS5 CM or control media. Before culture and after 5 days of culture in control medium, there was no significant difference in MMP-9 gene expression detected between CD14+ cells collected from MDS patients and healthy donors (Figure 3) In contrast, after 5 days of culture in HS5 CM, the gene expression of MMP-9 in patient monocytes was highly variable, a result that differed significantly from that observed with normal cells (P < .001 by the Folded F test; Figure 3A-B). A separate analysis of inducible levels in monocytes isolated from only the 11 patients with refractory anemia (RA) indicated that the variability of their levels also differed significantly from controls (P < .01). Monocytes from the 10 patients with refractory anemia with excess blasts (RAEB) showed greater variability and differed from controls (P < .001).
MMP-9 gene expression in CD14+ cells from healthy donors and patients with MDS. Total RNA was isolated from CD14+ cells from 21 healthy donors and 27 MDS patients either immediately (No culture) or after culturing for 5 days in control media or HS5 CM. The inducible levels of MMP-9 gene expression were determined by real-time PCR and normalized with G3PDH gene expression. The induced levels were significantly more variable in monocytes from MDS patients compared to those from healthy donors (P < .001 by Folded F test). The 11 MDS patients with RA are shown in red; the remaining patients are in black. RA patient monocytes analyzed alone also differed significantly from controls (P < .01); all non-RA patient monocytes (including 10 RAEB patients) analyzed as a group differed from controls at P < .001.
MMP-9 gene expression in CD14+ cells from healthy donors and patients with MDS. Total RNA was isolated from CD14+ cells from 21 healthy donors and 27 MDS patients either immediately (No culture) or after culturing for 5 days in control media or HS5 CM. The inducible levels of MMP-9 gene expression were determined by real-time PCR and normalized with G3PDH gene expression. The induced levels were significantly more variable in monocytes from MDS patients compared to those from healthy donors (P < .001 by Folded F test). The 11 MDS patients with RA are shown in red; the remaining patients are in black. RA patient monocytes analyzed alone also differed significantly from controls (P < .01); all non-RA patient monocytes (including 10 RAEB patients) analyzed as a group differed from controls at P < .001.
The purity of CD14+ cells within the mononuclear-cell fraction differed significantly between healthy donors and MDS patients (P < .001). To determine if this difference influenced the amount of MMP-9 detected, we tested whether there was a correlation between gene expression and CD14-cell purity. We found no significant correlation between purity of CD14+ cells and gene expression of MMP-9 at time 0 or after 5 days in culture in the presence or absence of HS5 CM, suggesting that the difference in CD14-cell purity between patients with MDS and healthy donors did not contribute to the difference in inducible MMP-9 levels.
We next considered that the variability in inducible levels of MMP-9 among MDS patient samples might be related to differences in the proportion of CD14+ cells in the samples that were derived from the MDS clone. To approach this issue, cytospins of isolated CD14+ cells were prepared and analyzed by FISH to determine the percentage of cells containing the cytogenetic marker specific for the MDS clone (Table 2) Among 3 informative patients, one patient had only 7% cytogenetically marked monocytes, and this patient had normal levels of inducible MMP-9 gene expression. In the other 2 patients, the majority of CD14 cells were cytogenetically marked, and both cell preparations showed low levels of inducible MMP-9. In addition, using clinical cytogenetic data available for 11 of the 27 patients, we found a significant negative correlation between the percentage of cytogenetically marked cells in the marrow and inducible levels of MMP-9 (P = .01). These data are summarized in Table 2.
MMP-9 gene expression and percent cytogenetically marked cells in patients with MDS
| Patient no. . | Chromosomal abnormality . | Percent marked CD14+ cells* . | Percent marked marrow† . | Percent marrow cellularity . | Induced expression of MMP-9 . |
|---|---|---|---|---|---|
| 3 | del 20q | NA | 55 | 70 | 5.18 |
| 4 | t(2:11), del 5p, del X | NA | 100 | 90 | 6.84 |
| 5 | del 20q | 64 | 75 | 100 | 7.23 |
| 6 | inv 16 | NA | 45 | 98 | 9.25 |
| 7 | t(1:7), del 7p | NA | 65 | 90 | 11.66 |
| 8 | t(1:7), del 7 | NA | 17 | 20 | 14.10 |
| 10 | tri 15 | 7 | 18 | 40 | 15.61 |
| 12 | del 12 | NA | 85 | 95 | 3.15 |
| 13 | tri 8 | NA | 95 | 98 | 7.02 |
| 17 | del 20q, t(13:14) | NA | 85 | 60 | 11.41 |
| 25 | del 20q | 84 | 100 | 65 | 6.45 |
| Patient no. . | Chromosomal abnormality . | Percent marked CD14+ cells* . | Percent marked marrow† . | Percent marrow cellularity . | Induced expression of MMP-9 . |
|---|---|---|---|---|---|
| 3 | del 20q | NA | 55 | 70 | 5.18 |
| 4 | t(2:11), del 5p, del X | NA | 100 | 90 | 6.84 |
| 5 | del 20q | 64 | 75 | 100 | 7.23 |
| 6 | inv 16 | NA | 45 | 98 | 9.25 |
| 7 | t(1:7), del 7p | NA | 65 | 90 | 11.66 |
| 8 | t(1:7), del 7 | NA | 17 | 20 | 14.10 |
| 10 | tri 15 | 7 | 18 | 40 | 15.61 |
| 12 | del 12 | NA | 85 | 95 | 3.15 |
| 13 | tri 8 | NA | 95 | 98 | 7.02 |
| 17 | del 20q, t(13:14) | NA | 85 | 60 | 11.41 |
| 25 | del 20q | 84 | 100 | 65 | 6.45 |
Del indicates deletion; NA not available; inv, inversion; tri 15, trisomy 15.
Determined by FISH analysis of blood-derived CD14+ cells by FHCRC Cytogenetics Laboratory.
Determined by karyotype analysis in cultured marrow cells conducted by FHCRC Cytogenetics Laboratory.
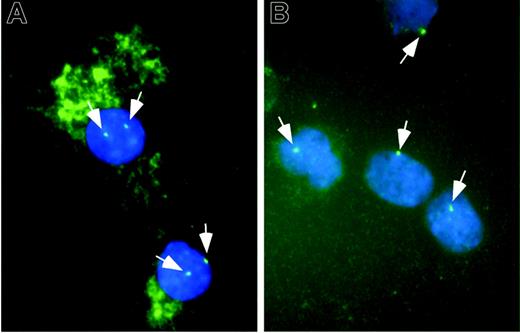
Taken together, these data led to the hypothesis that the ability to induce MMP-9 was decreased in MDS-derived monocytes. To test this directly, CD14 cells from a control subject and a patient with monosomy 7 were sorted, cultured in HS5 CM for 5 days, stained for MMP-9 protein, and probed for chromosome 7. The results (Figure 4) indicated that the majority of cells with monosomy 7 did not express MMP-9. Although this represents data from only one patient, this observation, together with the presented data, suggests that monocytes derived from the MDS clone have a reduced ability to up-regulate the expression of MMP-9 in response to stromal signals.
Immune cytochemical and FISH analyses of monocytes. CD14+ cells from a healthy donor (A) and a patient with a deletion of chromosome 7 in the MDS clone (B) were cultured in the presence of HS5 CM for 5 days then spun onto glass slides and labeled with REGA-2D9 monoclonal antibody against MMP-9 revealed with an HRP-conjugated secondary antibody and Vector SG peroxidase substrate. The cells were then probed for chromosome 7 by FISH. Green dots in the nuclei represent the FISH signal of chromosome 7 (indicated by white arrows). Positive staining of MMP-9 is detected as green fluorescence in the cytoplasm as a result of autofluorescence of the peroxidase substrate. Nuclei are counterstained with DAPI (blue). The images were captured and flattened prior to leveling contrast and brightness. Original objective × 100.
Immune cytochemical and FISH analyses of monocytes. CD14+ cells from a healthy donor (A) and a patient with a deletion of chromosome 7 in the MDS clone (B) were cultured in the presence of HS5 CM for 5 days then spun onto glass slides and labeled with REGA-2D9 monoclonal antibody against MMP-9 revealed with an HRP-conjugated secondary antibody and Vector SG peroxidase substrate. The cells were then probed for chromosome 7 by FISH. Green dots in the nuclei represent the FISH signal of chromosome 7 (indicated by white arrows). Positive staining of MMP-9 is detected as green fluorescence in the cytoplasm as a result of autofluorescence of the peroxidase substrate. Nuclei are counterstained with DAPI (blue). The images were captured and flattened prior to leveling contrast and brightness. Original objective × 100.
Correlation of inducible MMP-9 expression with marrow cellularity
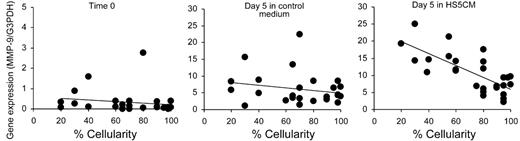
Given the putative roles for monocytes/macrophages in the marrow ME, we hypothesized that the proportion of abnormal monocytes, either measured directly or inferred by levels of inducible MMP-9, may contribute to some aspects of MDS pathogenesis. To test this hypothesis, we measured the correlation between MMP-9 expression and marrow cellularity, blast counts, white blood cell counts, platelet counts, peripheral-blood monocyte counts, hematocrit, patient age, sex, disease category, cytogenetic abnormality, and International Prognostic Symptom Score (IPSS), all recorded prior to study (Table S1). After a preliminary analysis showed a significant correlation between marrow cellularity and MMP-9 levels using the hospital database, the marrow cellularity was reanalyzed by a single pathologist (R.C.H.) who was blinded as to clinical and gene expression data. The results again indicated that there was no significant correlation between MMP-9 expression and the parameters studied except marrow cellularity. Figure 5 shows the significant negative correlation between HS5 CM-inducible MMP-9 gene expression and percentage of marrow cellularity (P < .001, r = −0.62, n = 27). There was no correlation with gene expression at time 0 or after 5 days of culture in control medium.
Correlation between MMP-9 gene expression and the percentage of marrow cellularity in MDS patients. Gene expression data obtained from CD14+ cells at time 0 (top panel), and after 5 days of culture in control medium (middle panel) or HS5 CM (bottom panel) were analyzed with respect to marrow cellularity in 27 patients with MDS. MMP-9 gene expression was determined by the real-time PCR assay and normalized with G3PDH gene expression (y-axis). The x-axis shows the percentage of marrow cellularity. Regression analysis indicates a significant negative correlation with cellularity for HS5 CM inducible MMP-9 expression in the bottom panel (R = −0.62; P < .001), but not for time 0 expression (R = 0.15; P = .42) or control medium expression (R = −0.21; P = .30) in the top and middle panels, respectively.
Correlation between MMP-9 gene expression and the percentage of marrow cellularity in MDS patients. Gene expression data obtained from CD14+ cells at time 0 (top panel), and after 5 days of culture in control medium (middle panel) or HS5 CM (bottom panel) were analyzed with respect to marrow cellularity in 27 patients with MDS. MMP-9 gene expression was determined by the real-time PCR assay and normalized with G3PDH gene expression (y-axis). The x-axis shows the percentage of marrow cellularity. Regression analysis indicates a significant negative correlation with cellularity for HS5 CM inducible MMP-9 expression in the bottom panel (R = −0.62; P < .001), but not for time 0 expression (R = 0.15; P = .42) or control medium expression (R = −0.21; P = .30) in the top and middle panels, respectively.
Immune cytochemical localization of MMP-9 protein in bone marrow biopsies
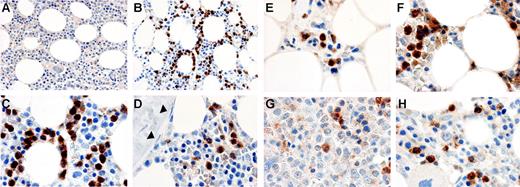
Figure 6A-D shows immune cytochemical analysis of MMP-9 protein in bone marrow biopsies from a healthy donor and 4 patients with MDS. Comparison of the isotype control and anti–MMP-9 antibody staining at low magnification showed that some hematopoietic cells were strongly stained with the anti–MMP-9 antibody. At high magnification, mature myeloid cells including granulocytes and monocytes showed strong staining for MMP-9 protein, whereas stromal cells, fat cells, megakaryocytes, immature myeloid cells, blasts, and cells of the erythroid lineage were negative. Although the types of cells that were positive for MMP-9 were the same in all samples, the frequency of positive cells was generally more variable in MDS samples than that observed in normal marrows. No attempt was made to quantitate these observations.
Immune cytochemical detection of MMP-9 protein in bone marrow. Bone marrow biopsies from a healthy donor (A-D) or MDS patients (E-H) were fixed in B5 fixative, antigen-retrieved, and incubated with a monoclonal antibody against MMP-9 (clone REGA 2D9; B-H) or an isotype-matched control antibody (A). The bound antibody was detected with HRP-conjugated secondary antibodies and Vector ABC stain kit. Positive staining of MMP-9 is brown; nuclei counterstained with hematoxylin are blue. Mature myeloid cells are MMP-9+ in both the healthy donor and MDS patients. Fibroblastic cells (stroma) were negative (nuclei of the fibroblasts are indicated by arrowheads in panel D). Megakaryocytes, evident in panel H, are also negative, as are endothelial cells and blood cells in sinusoids shown in panel E. Specific patient samples shown include: E, patient no. 9; F, patient no. 20; G, patient no. 12; H, patient no. 22 (Table S1 provides patient data). The images were captured and flattened prior to leveling contrast and brightness. Original objective × 40 for panels A-B; × 100 for panels C-H.
Immune cytochemical detection of MMP-9 protein in bone marrow. Bone marrow biopsies from a healthy donor (A-D) or MDS patients (E-H) were fixed in B5 fixative, antigen-retrieved, and incubated with a monoclonal antibody against MMP-9 (clone REGA 2D9; B-H) or an isotype-matched control antibody (A). The bound antibody was detected with HRP-conjugated secondary antibodies and Vector ABC stain kit. Positive staining of MMP-9 is brown; nuclei counterstained with hematoxylin are blue. Mature myeloid cells are MMP-9+ in both the healthy donor and MDS patients. Fibroblastic cells (stroma) were negative (nuclei of the fibroblasts are indicated by arrowheads in panel D). Megakaryocytes, evident in panel H, are also negative, as are endothelial cells and blood cells in sinusoids shown in panel E. Specific patient samples shown include: E, patient no. 9; F, patient no. 20; G, patient no. 12; H, patient no. 22 (Table S1 provides patient data). The images were captured and flattened prior to leveling contrast and brightness. Original objective × 40 for panels A-B; × 100 for panels C-H.
Discussion
Growing evidence suggests that monocytes/macrophages within the marrow ME contribute to hematopoietic regulation by providing signals that can act directly on progenitor cells and indirectly by modifying stromal-cell function within a circumscribed ME.33,34,35,36,37 In a previous study, we investigated the interactions between monocytes and stroma in an in vitro coculture system using cloned human marrow stromal cell lines and normal peripheral-blood monocytes.28,38 Transcriptome analysis identified MMP-9 as one of several genes strongly up-regulated in cocultures of HS-5 and CD14+ cells (M.I. and B.T.-S., unpublished data, 2004). This observation, together with the proposed functions attributed to MMP-9,1,2,15 prompted our interest in this protease. In the current report, we show that the increased MMP-9 gene expression seen in cocultures occurs in monocytes because these cells increased both gene and protein expression when cultured in HS5 CM.
Additional studies showed that the MMP-9–inducing activity could be attributed in part to MCP-1/CCL2 and IL-1β, because both are secreted by HS-5 and their recombinant forms induced MMP-9 gene expression in isolated CD14 cells. These data are in keeping with previous studies that showed IL-1β and TNF-α could induce MMP-9 expression in monocytes, macrophages, and lymphocytes through IL-1/toll-like and TNF receptors.15,21,39,40 Also, in addition to MCP-1 (CCL2),41 up-regulation of MMP-9 has been attributed to other β-chemokines, such as MIP-1α (CCL3), and RANTES (CCL5), which signal through the CCR1, CCR2, and CCR5 receptors on monocytes.42 Interestingly, although levels of MMP-9 gene expression comparable to that induced with HS5 CM could be induced in CD14+ monocytes with recombinant IL-1β and MCP-1, the amount of MMP-9 protein secreted by cells cultured in HS5 CM was significantly higher (> 8-fold) than that secreted by cells cultured with IL-1β or MCP-1. This suggests that there are additional activities in the conditioned medium that contribute to MMP-9 protein secretion by monocytes. Such multilevel regulation may provide for localized control of MMP-9 secretion by monocytes entering the ME.
Our data also indicate that as a group, populations of CD14+ cells isolated from patients with MDS expressed lower levels of inducible MMP-9 compared to normal CD14+ cells (P = .05 with unequal variance). However, of greater significance was the observation that the inducible levels in patient cells were variable to a significant extent when compared to the relatively consistent levels induced in normal cells (P < .001 by the Folded F test). This variability appeared to be associated with the proportion of cells within the patient CD14+-cell population that was estimated to be from the MDS clone; the higher the proportion of cytogenetically marked cells, the lower the amount of inducible MMP-9. This, in turn, suggested that the MDS-derived monocytes may be defective in terms of inducible MMP-9. In one patient we combined immune cytochemistry for MMP-9 with FISH analysis for chromosome 7 and showed that the majority of CD14+ cells that contained only one chromosome 7 did not express MMP-9.
Staining marrow biopsies with an antibody specific for MMP-9 showed, in general, a more consistent pattern of positive cells per field in normal marrows compared to patient marrows. However, in both normal and MDS marrows, MMP-9 was restricted to monocytic cells and granulocytes. In granulocytes the pro-MMP-9 peptide is known to be stored in intracellular granules and released quickly in response to agonists such as IL-8 without the need for transcription.15 In contrast, monocyte-derived MMP-9 is induced by cytokine exposure resulting in transcription, translation, and eventually secretion. It appears that these 2 sources of MMP-9 are under different controls and may have different functions.
A previously published study43 showed that there was no correlation between MDS patient characteristics and MMP-9 expression in freshly isolated marrow mononuclear cells. In agreement with this, our data also showed that MMP-9 levels in freshly isolated blood monocytes did not correlate with any clinical parameters. However, we did find a significant negative correlation between the level of inducible MMP-9 gene expression in circulating monocytes and marrow cellularity in MDS patients (r = −0.15, P < .001, n = 27). Specifically, lower levels of inducible MMP-9 were associated with higher marrow cellularity.
One speculative interpretation for our observations is that monocyte-derived MMP-9 is induced locally by stoma to facilitate egress of cells from the marrow. The substrates for MMP-9 in bone marrow include ECM components (eg, denatured collagens), cytokines (eg, SCF), and CXC chemokines including IL-8/CXCL8 and SDF-1/CXCL12.2,15,44,45,46,47,48 SDF-1/CXCL12 is an important chemokine that serves as a ligand for the CXCR4 receptor expressed on hematopoietic cells. Blocking SDF-1/CXCR4 interactions in vivo with the administration of AMD3100, an antagonist of CXCR4, results in a rapid mobilization of CXCR4 expressing cells into the periphery.49 Given this observation, it is reasonable to speculate that inactivation of SDF-1 by MMP-9 might also contribute to the release of hematopoietic cells, and failure to do so results in areas of hypercellularity. An alternative interpretation would be that inducible MMP-9 levels are not causally related to marrow cellularity, but rather provide an estimate of the proportion of otherwise abnormal cells within an MDS patient's mononuclear-cell population.
The presence of an abnormal monocyte population makes it reasonable to speculate that the altered ME function observed with MDS marrows50,51,52 may be the consequence of inappropriate modulation of stromal function by MDS-derived monocytes. This speculation is supported by the observation that stromal cells harvested from MDS marrow are not derived from the malignant clone53 and are presumably intrinsically normal. If true, this speculation has relevance for stem-cell transplantation protocols that use nonmyeloablative conditioning. In such protocols the monocyte stromal interactions that fail to support normal endogenous stem-cell function would remain in place after conditioning and likewise fail to support donor stem-cell function. A better understanding of the functional repertoire of the monocyte in the ME is needed to more completely address this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: M.I. designed and performed experiments, analyzed data, and wrote the manuscript; M.P. performed cocultures and prepared RNA/cDNA from some MDS patients and obtained patients' data; A.R. performed immune cytochemistry and FISH analysis of monocytes and provided patient data; R.C.H. analyzed bone marrow biopsies from MDS patients; H.J.D.wrote the clinical protocol, supplied MDS patients' samples, and obtained biopsies of healthy donors; G.O. supplied REGA-2D9 anti–MMP-9 antibody for immune cytochemistry and assisted with editing; B.T.-S. designed and supported research, provided oversight, and edited the paper.
This work was supported by National Institutes of Health grants HL62923, DK56465, and CA87948. M.P. was supported by T32CA09515 and by the Ladies Auxiliary of the Veterans of Foreign Wars. A.R. was supported by 2T32CA009515-21. G.O. was supported by Federation Against Cancer, Belgium.
The authors thank Drs David Myerson and Julie Randolf-Habecker at the Experimental Histopathology Laboratory at FHCRC for preparing and evaluating immunohistologic sections, Dr David Madtes at FHCRC for fruitful discussion and advice, Dr Barry Storer for statistical evaluation, Dr Eileen Bryant for FISH analysis, Ludmila Golubev and Gretchen Johnson for maintaining tissue culture, and Bonnie Larson and Helen Crawford for preparing the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal