Abstract
The JAK2 V617F mutation has recently been described as an essential oncogenic event associated with polycythemia vera (PV), idiopathic myelofibrosis (IMF), and essential thrombocythemia. This mutation has been detected in all myeloid lineages but has not yet been detected in lymphoid cells. This raises the question whether this molecular event occurs in a true lymphomyeloid progenitor cell. In this work, we studied the presence of the mutation in peripheral blood cells and sorted B, T, and natural killer (NK) cells from PV and IMF. We detected the JAK2 V617F mutation in B and NK cells in approximately half the patients with IMF and a minority of those with PV. Moreover, in a few cases patients with IMF had mutated peripheral T cells. The mutation (homozygous or heterozygous) could be subsequently detected in B/NK/myeloid progenitors from PV and IMF, with a much higher frequency in clones derived from IMF. Using the fetal thymus organ culture (FTOC) assay, the mutation was also detected in all T-cell fractions derived from IMF and PV CD34+ cells. These results demonstrate that myeloproliferative disorders take their origin in a true myeloid/lymphoid progenitor cell but that their phenotype is related to a downstream selective proliferative advantage of the myeloid lineages.
Introduction
Myeloproliferative disorders (MPDs) are hematologic malignancies characterized by a clonal proliferation of one or several myeloid lineages.1 Polycythemia vera (PV), essential thrombocythemia (ET), idiopathic myelofibrosis (IMF), and chronic myeloid leukemia (CML), defined as the classical MPDs, are considered to arise from the transformation of a multipotent hematopoietic stem cell (HSC).2,3,4,5 However, this stem origin of the malignant clone has only been demonstrated in CML by the detection of either the characteristic t(9;22) translocation or the BCR/ABL transcript in all hematopoietic lineages by fluorescent in situ hybridization (FISH) and reverse transcription-polymerase chain reaction (RT-PCR) techniques.6 Previous studies reported the presence of the bcr/abl translocation in B- and T- lineage progenitors (including CD34+CD7+, CD34+CD7+CD5+, and CD34+CD19+ cells) but were unable to detect the translocation in mature T cells. Thus, there is an obvious discrepancy between immature and mature cells from the T lineage in this disorder. As an explanation for this lack of Ph1 in mature T cells, a long-standing hypothesis is that the majority of T cells are long-lived and are born before the occurrence of the clonal oncogenic event.6 Alternatively thymocytes with the Ph1 chromosome may have no proliferative advantages or even fail to differentiate in mature T cells as a consequence of BCR-ABL signaling.
In the Ph1− classical myeloproliferative disorders (PV, ET, and IMF), an acquired activating mutation of the protein kinase JAK2 (JAK2 V617F) has been recently reported by our group and others.7,8,9,10,11 All these disorders are considered as hematologic malignancies arising from an HSC, based on studies with glucose-6-phophaste dehydrogenase (G6PD) isoenzymes, X-chromosome inactivation pattern (XCIP) analysis, and loss of heterozygosity of the short arm of chromosome 9 in lymphoid cells.3,5,12,13,14
The JAK2 mutation is found in almost all patients with PV, in 35% to 70% of patients with ET, and 50% of those with IMF.7,8,9,10,11 This raises the question of how a single mutation can give rise to diseases with different phenotypes. Overexpression of the murine mutated JAK2 in an animal model recapitulates PV in mice and its classical evolution to myelofibrosis called the “spent phase,” but none of these animals developed a thrombocytosis or a myelofibrosis in the absence of a polycythemia.15,16 Thus, the JAK2 mutation can be reported as the main oncogenic event responsible for PV development, but its precise role in ET and IMF development remains questionable. This may suggest the requirement of other genetic events to induce ET and IMF. Another explanation for the heterogeneity in the diseases induced by JAK2 V617F could be related to the type of leukemic stem cell that drives the disease, that is, a lymphomyeloid progenitor versus a common myeloid progenitor or even a committed progenitor. Indeed, it has been shown that the JAK2 mutation is present in erythroid and granulomonocytic progenitor cells7 as well as in CD34+ cells17,18 and their erythroblastic progeny.8,18 These observations demonstrate that the mutation occurs either at the level of the common myeloid progenitor or upstream, at the level of a lymphomyeloid multipotent progenitor cell or an HSC.
The aim of this study was thus to determine whether the JAK2 mutation involves lymphoid and myeloid lineages or was only restricted to the myeloid lineages by studying its presence in mature peripheral lymphoid and myeloid cells from patients with PV and IMF and in the cell progenitors.
Patients, materials, and methods
Patients and samples
The study was approved by the Local Research Ethics Committee from the Hôtel-Dieu and the Henri Mondor hospitals and informed consent was obtained from each patient in accordance with the Declaration of Helsinki. Baseline characteristics and diagnostic information were collected prospectively, and clinical diagnosis was defined according to standard criteria. Patient diagnoses were defined according to the modified Polycythemia Vera Study Group (PVSG) criteria for PV and the Italian criteria for IMF.19 Only patients presenting the JAK2 V617F mutation detected in the granulocytic compartment were enrolled in this study. Three normal bone marrow samples were obtained from patients undergoing hip surgery after informed consent was provided.
Cell purification
Platelets were purified from blood samples using the platelet-rich plasma (PRP) technique. Granulocytes were obtained using a dextran (Sigma, St Louis, MO) basic method followed by a Ficoll separation and red blood cell lysis. Monocytes, natural killer (NK), B, and T cells were purified from peripheral blood mononuclear cells (PBMCs) by means of the Miltenyi Biotec (Paris, France) immunomagnetic bead technique according to the manufacturer's protocol. To obtain highly purified cell populations, immunomagnetic bead-isolated cells were labeled with anti–CD3-FITC, anti–CD19-PE, anti–CD14-PE-Cy7 (Becton Dickinson, Le Pont de Claix, France) and anti–CD56-APC (Beckman Coulter, Roissy, France) antibodies and sorted using a FACS-DIVA cell sorter (Becton Dickinson). Isotype-matched antibodies were used as controls. Marrow or peripheral blood CD34+ cells were separated after Ficoll-metrizoate gradient (Lymphoprep, Nycomed Pharma, Oslo, Norway) by the same immunomagnetic procedure (Miltenyi Biotec). The CD34+CD38− cell population was subsequently isolated after staining of immunomagnetic purified CD34+ cells with CD34-PeCy5, CD38-FITC antibodies (Immunotech, Marseille-Lumigny, France) by flow cytometric cell sorting.
Assessment of simultaneous B, NK, and granulomonocytic differentiation
After the enrichment step described (see “Cell purification”), the low concentration CD34+CD38− cell suspension was sorted slowly at one cell per well in 96-well plates using the FACS-DIVA cell sorter (Becton Dickinson). According to the settings of the cell sorter, this procedure discarded all doublets and cell aggregates. The absence of wells containing more than 1 cell was assessed by a careful microscopic examination of each individual well 2 hours after sorting. These single CD34+CD38− cells were incubated on a confluent layer of MS-5 cells in RPMI medium supplemented with 10% human serum, 5% fetal calf serum (FCS), and a combination of 7 cytokines: 10 ng/mL IL-3 (generous gift from Novartis, Basel, Switzerland), 50 ng/mL SCF, 50 ng/mL FLT3-L (generous gifts from Immunex, Seattle WA), 10 ng/mL TPO (generous gift from Kyrin Laboratories, Tokyo, Japan), 20 ng/mL IL-7, 10 ng/mL IL-15 (PeproTech, London, United Kingdom) and 5 ng/mL IL-2 (generous gift from Chiron Laboratories, Emeryville, CA). Wells with significant cell proliferation were collected after 4 to 6 weeks, and cell phenotype was determined by flow cytometry in half the cell suspension using anti–CD15-FITC, anti–CD19-PE (Becton Dickinson) and anti–CD56-APC (Beckman Coulter) antibodies, the remaining cells being pelleted for genotyping analysis.
Assessment of T-cell potential in fetal thymus organ culture
Isolation of murine embryonic C57/Bl6 thymic lobes, irradiation, incubation with human CD34+ cells by means of the hanging drop procedure, and organotypic cultures were performed following a standard procedure, initially described to analyze mouse T-lymphoid differentiation and adapted to the identification of human T-cell potential.20,21 Cells recovered from the thymic lobes after about 21 days were labeled by anti–CD45-FITC, anti–CD4-PE, and anti–CD8-APC antibodies (Becton Dickinson) and were cell sorted in 4 different subpopulations: CD45+CD4−CD8−, CD45+CD4+CD8+ (double positive), CD45+CD4+CD8−, and CD45+CD4−CD8+ as described (see “Cell purification”). Genotyping was performed on the sorted populations.
Nucleic acid extraction
DNA and RNA were extracted from the different cell fractions using, respectively, the RNA+ reagent (Quantum; Appligene, Illkirch, France) and the Qiagen DNA extraction kit (Qiagen, Hilden, Germany).
Genotyping
Direct sequencing was performed on the different cell fractions on DNA or RNA. For RNA analysis, a reverse transcription was first performed. cDNA (1-6 μL) served as template. Samples were subjected to 40 cycles of amplification under the following conditions: denaturing at 94°C for 30 seconds, annealing at 40°C for 2 minutes, extension at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. The resulting PCR products were purified on a Qiagen column and subjected to DNA sequencing as previously reported on an ABI PRISM 3100 Genetic Analyser (Applied Biosystems, Foster City, CA).8 Alternatively, the mutational status was determined by a real-time PCR single nucleotide polymorphism (SNP) detection system with fluorescent competitive probes using an ABI 7500 analyzer (Applied Biosystems).22 To ensure the semiquantitative estimation of the JAK2 V617F–total JAK2 ratio, the DNA from a homozygous mutated sample (100% of mutated JAK2) from a PV patient with a normal karyotype was diluted in various proportions in normal control DNA. Each dilution was tested by sequencing and real-time PCR methods (Figure S1, available at the Blood website; see the Supplemental Figures link at the top of the online article). Sequencing semiquantitative results were obtained using peak height analysis. Quantitative results were obtained by the real-time PCR technique using the ΔCt method as described.23 Semiquantitative results were expressed as follows: +++ for samples with more than 50% of mutated allele, ++ for samples with mutated allele ranging from 25% to 50%, + for samples with 5% to 25% of mutated allele, and +low for positive samples wherein the mutated allele was at the threshold of detection (about 5% for the sequencing technique and 2% for the real-time PCR assay; Figure S1).
Results
Detection of the JAK2 V617F mutation in peripheral blood lymphocytes from patients with IMF and PV
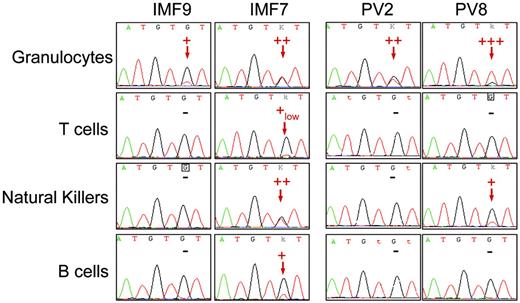
Sequence analysis detected the JAK2 G1849T (JAK2 V617F) mutation in the peripheral blood myeloid lineages, including granulocytes, platelets, and monocytes, from 10 patients with IMF and 10 with PV. Among PV patients, 7 displayed a high level of JAK2 V617F (> 50% of mutated allele as assessed by the real-time PCR technique) either in granulocytes or in bone marrow mononuclear cells, suggesting the presence of cells harboring a biallelic (homozygous) mutation (Figure 1; Table 1) All remaining PV and IMF patients had an equal or lower level of mutated JAK2 than the wild-type JAK2 (Figure 1; Table 1). To study the presence of the JAK2 V617F mutation in peripheral blood lymphocytes, T, B, and NK cells were purified by a combined immunomagnetic and flow cytometric methods on the basis of CD3, CD19, and CD56 expression, respectively. This purification procedure gave a purity greater than 98% as assessed by flow cytometry analysis. Sequence analysis revealed the presence of the mutation in B cells for 6 of 10 patients and in NK cells for 5 of 8 patients with IMF. Surprisingly, the mutation was also found in T cells for 2 of 8 patients with IMF at a very low level when compared to the JAK2 V617F level in myeloid, B, or NK cells, demonstrating that a low number of mature T cells belong to the clone (Figure 1; Table 1). In contrast, only 2 of the 10 patients with PV harbored the mutation in peripheral lymphocytes, one in B cells and the other in NK cells (Table 1). These 2 patients had a homozygous mutation detected in granulocytes (Figure 1; Table 1).
Detection of the JAK2 G1849T mutation in peripheral lymphoid cells from IMF and PV patients. Peripheral granulocytes and lymphocytes from JAK2-mutated IMF and PV patients were isolated using standard density, immunomagnetic, and flow cytometric methods for further DNA extraction and sequence analysis. The JAK2 G1849T mutation was detected in some lymphoid cells from a majority of IMF patients and a minority of PV patients. The sequence traces of granulocytes and T, NK, and B cells from 2 IMF (patients IMF9 and IMF7) and 2 PV patients (patients PV2 and PV8) are shown; – indicates that the mutation was not detected. Red arrows indicate the presence of a mutant peak. Semiquantitative estimate of the percentage of mutated allele is presented on each sequence trace.
Detection of the JAK2 G1849T mutation in peripheral lymphoid cells from IMF and PV patients. Peripheral granulocytes and lymphocytes from JAK2-mutated IMF and PV patients were isolated using standard density, immunomagnetic, and flow cytometric methods for further DNA extraction and sequence analysis. The JAK2 G1849T mutation was detected in some lymphoid cells from a majority of IMF patients and a minority of PV patients. The sequence traces of granulocytes and T, NK, and B cells from 2 IMF (patients IMF9 and IMF7) and 2 PV patients (patients PV2 and PV8) are shown; – indicates that the mutation was not detected. Red arrows indicate the presence of a mutant peak. Semiquantitative estimate of the percentage of mutated allele is presented on each sequence trace.
JAK2 V617F genotyping of blood and bone marrow fractionated cell populations
| Patient . | Prognostic Lille score . | Peripheral myeloid cells . | Peripheral lymphoid cells . | Bone marrow mononuclear cells . | CD34+* . | CD34+ CD38+* . | CD34+ CD38−* . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gr . | Pl . | Mo . | B . | NK . | T . | ||||||
| IMF1 | Low | ++ | ++ | ++ | ++ | − | − | ND | ND | ND | ND |
| IMF2 | High | ++ | +++ | ++ | ++ | + | − | ND | ++ | ++ | ++ |
| IMF3 | High | ++ | ++ | ++ | ++ | ++ | +low | ND | ND | ND | ND |
| IMF4 | Int | ++ | ++ | ++ | + | − | − | ND | + | + | + |
| IMF5 | Low | + | + | + | − | ND | ND | ND | ND | ND | ND |
| IMF6 | Int | ++ | ++ | ++ | − | + | ND | ND | ++ | ++ | ++ |
| IMF7 | High | ++ | ++ | ++ | + | ++ | +low | ND | + | + | + |
| IMF8 | Int | + | + | + | − | ND | − | ND | ND | ND | ND |
| IMF9 | Int | + | + | + | − | − | − | ND | + | + | + |
| IMF10 | Int | ++ | ++ | ++ | ++ | ++ | − | ND | ++ | ++ | ++ |
| PV1 | NA | ++ | ++ | + | − | − | − | ++ | ++ | ND | ND |
| PV2 | NA | ++ | + | + | − | − | − | ND | ND | ND | ND |
| PV3 | NA | ++ | ++ | ++ | − | − | − | ++ | ++ | ++ | − |
| PV4 | NA | ++ | ++ | ++ | − | − | − | +++ | ++ | ++ | − |
| PV5 | NA | ++ | ++ | ++ | − | − | − | +++ | ND | ND | ND |
| PV6 | NA | ++ | ++ | + | − | − | − | ++ | + | ++ | − |
| PV7 | NA | +++ | ++ | ++ | − | − | − | +++ | ND | ND | ND |
| PV8 | NA | +++ | ++ | ++ | − | + | − | ND | ND | ND | ND |
| PV9 | NA | +++ | ++ | ++ | + | − | − | ++ | ND | ND | ND |
| PV10 | NA | +++ | ++ | ++ | − | − | − | +++ | ++ | ++ | ++ |
| PV11 | NA | ++ | ND | ND | ND | ND | ND | ++ | ++ | ++ | + |
| PV12 | NA | +++ | ND | ND | ND | ND | ND | +++ | ND | ND | ND |
| Patient . | Prognostic Lille score . | Peripheral myeloid cells . | Peripheral lymphoid cells . | Bone marrow mononuclear cells . | CD34+* . | CD34+ CD38+* . | CD34+ CD38−* . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gr . | Pl . | Mo . | B . | NK . | T . | ||||||
| IMF1 | Low | ++ | ++ | ++ | ++ | − | − | ND | ND | ND | ND |
| IMF2 | High | ++ | +++ | ++ | ++ | + | − | ND | ++ | ++ | ++ |
| IMF3 | High | ++ | ++ | ++ | ++ | ++ | +low | ND | ND | ND | ND |
| IMF4 | Int | ++ | ++ | ++ | + | − | − | ND | + | + | + |
| IMF5 | Low | + | + | + | − | ND | ND | ND | ND | ND | ND |
| IMF6 | Int | ++ | ++ | ++ | − | + | ND | ND | ++ | ++ | ++ |
| IMF7 | High | ++ | ++ | ++ | + | ++ | +low | ND | + | + | + |
| IMF8 | Int | + | + | + | − | ND | − | ND | ND | ND | ND |
| IMF9 | Int | + | + | + | − | − | − | ND | + | + | + |
| IMF10 | Int | ++ | ++ | ++ | ++ | ++ | − | ND | ++ | ++ | ++ |
| PV1 | NA | ++ | ++ | + | − | − | − | ++ | ++ | ND | ND |
| PV2 | NA | ++ | + | + | − | − | − | ND | ND | ND | ND |
| PV3 | NA | ++ | ++ | ++ | − | − | − | ++ | ++ | ++ | − |
| PV4 | NA | ++ | ++ | ++ | − | − | − | +++ | ++ | ++ | − |
| PV5 | NA | ++ | ++ | ++ | − | − | − | +++ | ND | ND | ND |
| PV6 | NA | ++ | ++ | + | − | − | − | ++ | + | ++ | − |
| PV7 | NA | +++ | ++ | ++ | − | − | − | +++ | ND | ND | ND |
| PV8 | NA | +++ | ++ | ++ | − | + | − | ND | ND | ND | ND |
| PV9 | NA | +++ | ++ | ++ | + | − | − | ++ | ND | ND | ND |
| PV10 | NA | +++ | ++ | ++ | − | − | − | +++ | ++ | ++ | ++ |
| PV11 | NA | ++ | ND | ND | ND | ND | ND | ++ | ++ | ++ | + |
| PV12 | NA | +++ | ND | ND | ND | ND | ND | +++ | ND | ND | ND |
Prognostic score of IMF patients (Lille score)24 is indicated.
Gr indicates granulocytes; Pl, platelets; Mo, monocytes; +++, samples with > 50% of mutated allele; ++, samples with mutated allele ranging from 25% to 50%; +, samples with 5% to 25% of mutated allele; +low, positive samples with mutated allele at the threshold of detection; −, absence of the JAK2 V617F mutation; ND, no data; NA, not applicable.
CD34+, CD34+CD38+, and CD34+CD38– cells were separated from whole blood in IMF patients, and from bone marrow samples in PV patients.
Genotyping of CD34+ and CD34+ CD38− cells
To study the presence of the JAK2 V617F mutation in more immature cells, CD34+CD38+ and CD34+CD38− cells were purified by a combined immunomagnetic (CD34) and flow cytometric method on the basis of CD34 and CD38 expression. These cells were isolated either from blood circulating cells (IMF) or bone marrow aspirate samples (PV) with a 95% purity as assessed by cytometric analysis. Sequence analysis revealed the presence of the mutation in CD34+ cells from all IMF (n = 6) and PV (n = 6) patients. JAK2 V617F was detected in CD34+CD38+ and CD34+CD38− purified fractions from all IMF samples. The level of mutated allele was in the same order of magnitude as that observed in mature granulocytes. In 5 PV patients, similar levels of mutated JAK2 were found in granulocytes and bone marrow CD34+CD38+ cells, but in contrast, the mutation was detected in CD34+CD38− cells for only 2 of these 5 PV patients (Table 1). In this assay we used the real-time quantitative PCR technique, which has a sensitivity of about 2% and thus immature progenitors harboring the mutation were either absent or present but at a very low frequency.
In vitro B/NK/myeloid differentiation from PV and IMF CD34+CD38− cells
To more precisely ascertain that JAK2 mutation may be present in lymphomyeloid progenitors, CD34+CD38− cells from 3 healthy bone marrows, 3 IMF, and 5 PV patients including 3 PV patients in which the mutation was not detected in this cell population were purified and grown at one cell per well on the murine stromal MS-5 cell line in the presence of a combination of 7 cytokines (see “Patients, materials, and methods”). The mean frequency ± SD of proliferating clones (> 200 cells) at 4 to 6 weeks in normal, IMF, and PV samples was 35.8% ± 5.7%, 12.6% ± 4.1%, and 26.6% ± 3.2%, respectively. These results are in the same order of magnitude as previously reported with the same technique.20 Myeloid, B, and NK potentialities were then analyzed by flow cytometry in each proliferating clone. Although patient-dependent heterogeneous results were observed, similar percentages of myeloid, NK/myeloid, and B/NK/myeloid clones were present overall in the patient and control samples (Figure 2) All types of potentialities except pure B cells were found in the patient samples (Figure 2). Clones with B/myeloid and B/NK potentialities were infrequent, and low proportions of B cells (< 10%) were present in B/NK/myeloid clones from both patient and control samples. When comparing the size, the variety, and the composition of the clones differentiated in vitro, no significant differences between controls and patients were noticed (Student t test, P > .05; Figures 2 and S2).
Assessment of B/NK/myeloid potentialities in IMF and PV cell fractions. Analysis of the progeny of single CD34+CD38− cells from normal bone marrow (N), PV bone marrow, and IMF peripheral blood. Histograms represent 76 positive clones in 576 plated wells from 3 IMF patients, 386 positive clones in 1482 plated wells from 5 PV patients, and 372 positive clones in 1056 plated wells from 3 normal bone marrows. The experiment was performed once for each patient or control sample. The mean percentage of clones containing one (B, NK, myeloid [M]), 2 (B/M, NK/M, B/NK), or 3 (B/NK/M) lineages per total number of clones analyzed after triple-staining by flow cytometry are shown. Myeloid cells correspond to CD15+ cells, B cells to CD19+ cells, and NK cells to CD56+ cells. Error bars indicate SEM.
Assessment of B/NK/myeloid potentialities in IMF and PV cell fractions. Analysis of the progeny of single CD34+CD38− cells from normal bone marrow (N), PV bone marrow, and IMF peripheral blood. Histograms represent 76 positive clones in 576 plated wells from 3 IMF patients, 386 positive clones in 1482 plated wells from 5 PV patients, and 372 positive clones in 1056 plated wells from 3 normal bone marrows. The experiment was performed once for each patient or control sample. The mean percentage of clones containing one (B, NK, myeloid [M]), 2 (B/M, NK/M, B/NK), or 3 (B/NK/M) lineages per total number of clones analyzed after triple-staining by flow cytometry are shown. Myeloid cells correspond to CD15+ cells, B cells to CD19+ cells, and NK cells to CD56+ cells. Error bars indicate SEM.
Genotyping characterization of B/NK/myeloid monopotent and multipotent IMF and PV clones
Proliferating clones were submitted to immunophenotypic and genotypic analysis by sequencing or by TaqMan SNP detection real-time PCR (Figures 3 and S2). Lack of PCR product occurred in 5 of 76 samples for IMF clones but in none of the 386 PV clones. The 3 IMF patients analyzed harbored different patterns of heterozygosity/homozygosity in their different clones. One patient presented a heterozygous mutation in all analyzed clones (Figure 3Bi), one patient had a mixed pattern of homozygous and heterozygous clones without detectable normal clones (Figure 3Bii), and the third patient exhibited a more heterogeneous pattern with the presence of nonmutated, heterozygous, and homozygous clones (Figure 3Biii). In contrast, a high proportion of nonmutated clones ranging from 52% to 97% was observed in all 5 PV patients. Among them, 2 had a heterozygous mutation that was detected in 9 of 84 and 3 of 89 clones (Figure 3Biv-v). Among the 3 PV patients with a homozygous mutation, different results were observed. In the first patient, only 4 of 72 clones had the mutation and with only a homozygous pattern (Figure 3Bvi). In the 2 remaining patients, we observed the coexistence of, respectively, 4 and one homozygous, 29 and 4 heterozygous, 66 and 36 normal clones (Figure 3Bvii-viii). When analyzing the mutational status with respect to the immunophenotype of the clones, we observed that all patients had at least one clone with lymphomyeloid potentialities harboring the mutation. Indeed, the JAK2 V617F mutation was detected in B/NK/myeloid clones from 2 of 3 patients with IMF (Figure 3Bii-iii) and 2 of 5 patients with PV (Figure 3Bv,vii). The 4 remaining patients had mutated NK/myeloid clones (Figure 3Bi,iv, vi,viii). In addition, 2 patients with IMF and 3 patients with PV had NK/myeloid or B/NK/myeloid clones with a homozygous mutation (Figure 3Bii-iii,vi-viii), indicating that the subsequent mitotic recombination also occurred in a lymphomyeloid progenitor cell. Altogether, these results demonstrate that the JAK2 mutation is present in lymphomyeloid progenitors in patients with IMF or PV, with a heterogeneous patient-dependent pattern.
Genotyping analysis of B/NK/myeloid individual clones derived from PV and IMF purified CD34+CD38minus]. (A) Immunophenotypic analysis of B/NK/myeloid clones from a healthy bone marrow, an IMF patient, and a PV patient. The isotype control scattergrams are shown in the top panel. The B, NK, and myeloid potentials were assessed by the presence of CD19+, CD56+, and CD15+ cells, respectively, after 5 weeks of B, NK, and myeloid differentiation condition culture. The JAK2 G1849T homozygous sequence traces of the IMF and the PV clones are shown. Red arrows indicate the mutant T peak. (B) Genotype analysis of single CD34+CD38− cell culture-derived clones from 3 IMF (i-iii) and 5 PV patients (iv-viii) with respect to their monopotent B, NK, myeloid (M), bipotent B/M, B/NK, M/NK, or tripotent B/NK/M immunophenotypic characterization (clone type).
Genotyping analysis of B/NK/myeloid individual clones derived from PV and IMF purified CD34+CD38minus]. (A) Immunophenotypic analysis of B/NK/myeloid clones from a healthy bone marrow, an IMF patient, and a PV patient. The isotype control scattergrams are shown in the top panel. The B, NK, and myeloid potentials were assessed by the presence of CD19+, CD56+, and CD15+ cells, respectively, after 5 weeks of B, NK, and myeloid differentiation condition culture. The JAK2 G1849T homozygous sequence traces of the IMF and the PV clones are shown. Red arrows indicate the mutant T peak. (B) Genotype analysis of single CD34+CD38− cell culture-derived clones from 3 IMF (i-iii) and 5 PV patients (iv-viii) with respect to their monopotent B, NK, myeloid (M), bipotent B/M, B/NK, M/NK, or tripotent B/NK/M immunophenotypic characterization (clone type).
T-lymphoid potential and genotyping of PV and IMF CD34+ cells
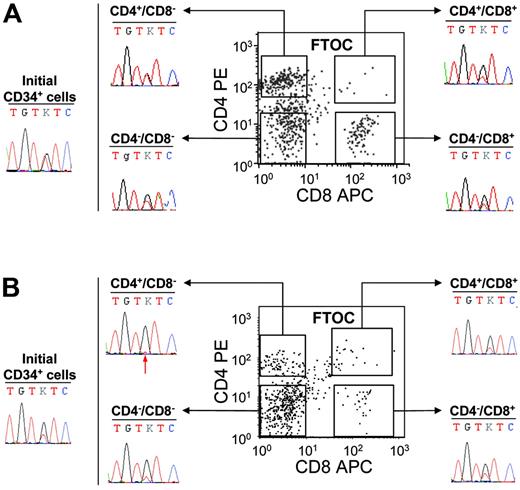
These experiments demonstrated that the JAK2 mutation is present in IMF and PV cells with B or NK lymphoid potential. However, it remained to be determined if the CD34+ cells harboring the JAK2 mutation also had a T-cell potential or lost their T-cell potential during T-cell differentiation. C57/Bl6 embryonic thymus can be used to reveal the T-cell potential of human CD34+ cells. Using this strategy, we investigated the T-cell potential of the CD34+ cells from 5 IMF and 2 PV patients. First, 50 000 cells were cultured in the hanging-drop procedure in individual thymic lobes. After 3 weeks of culture, cells were recovered and human T-cell differentiation was assessed by flow cytometry using human anti-CD45, anti-CD4, and anti-CD8 antibodies. In these conditions, CD34+ cells generated human (CD45+) T cells (CD4+CD8−, CD4+CD8+, CD4−CD8+, CD4−CD8−), which were sorted for further sequencing (Figure 4) The mutation was detected in CD4+CD8−, CD4−CD8+, and CD4−CD8− human CD45+ fractions of all patients analyzed (Figure 4). The double-positive (CD4+CD8+) cell population was rare and was sorted in 5 IMF patients and one PV patient. Very low numbers of cells (< 3000 cells) were recovered and could be studied in 3 of these 6 samples owing to sufficient material. In all 3 analyzed cases (3 IMF), the presence of the G1849T mutation was detected (Figure 4). Interestingly, the mutant allele was present at a quite high level in some FTOC-recovered T-cell populations from IMF patients (Figure 4). However, the level of JAK2 G1849T was similar to that found in initial CD34+ cells from the same patients (Figure 4). Altogether these results show that some progenitor cells with a T-cell potentiality also harbor the JAK2 mutation.
Detection of the JAK2 V617F mutation in FTOC-derived T-cell subpopulations from PV and IMF CD34+ cells. CD34+ cells (50 000) were cultured in fetal thymic lobes for 3 weeks as indicated in “Patients, materials, and methods.” At the end of the culture, cells collected from each thymic lobe were labeled by FITC-conjugated anti–human CD45, PE anti–human CD4, and APC anti–human CD8 monoclonal antibodies and analyzed by flow cytometry. Human CD45+ CD4+CD8−, CD4−CD8+, CD4−CD8−, and CD4+CD8+ fractions were subsequently sorted and sequenced. The immunophenotypic scattergrams and sequence traces of initial CD34+ cells and FTOC T-cell fractions from patients IMF6 (A) and IMF7 (B) are shown. The red arrow indicates the presence of a small mutant T peak at the threshold of detection.
Detection of the JAK2 V617F mutation in FTOC-derived T-cell subpopulations from PV and IMF CD34+ cells. CD34+ cells (50 000) were cultured in fetal thymic lobes for 3 weeks as indicated in “Patients, materials, and methods.” At the end of the culture, cells collected from each thymic lobe were labeled by FITC-conjugated anti–human CD45, PE anti–human CD4, and APC anti–human CD8 monoclonal antibodies and analyzed by flow cytometry. Human CD45+ CD4+CD8−, CD4−CD8+, CD4−CD8−, and CD4+CD8+ fractions were subsequently sorted and sequenced. The immunophenotypic scattergrams and sequence traces of initial CD34+ cells and FTOC T-cell fractions from patients IMF6 (A) and IMF7 (B) are shown. The red arrow indicates the presence of a small mutant T peak at the threshold of detection.
Discussion
It has been well established that CML arises in a multipotent HSC that contains the Philadelphia chromosome. In most MPDs, indirect evidence indicates that the oncogenic process occurs at the same cellular level although it is not presently clear if lymphoid lineages, especially the T-cell lineage, are involved. The evidence is based on the presence of nonrecurrent cytogenetic abnormalities or on genetic studies in women heterozygous for a X-linked gene in PV, ET, or IMF in different cell lineages.3,5,12,13,14 Nevertheless, these approaches have 2 limitations, namely, cytogenetic abnormalities are common in these MPDs but there may be late secondary events during clonal development, and most of these diseases occur in patients older than 50 years in whom the X-linked approach is not easy to interpret.
In the majority of PV, ET, and IMF cases, a JAK2 mutation (JAK2 V617F) leading to a gain of function has been found. Recent works with mouse models using transduction of JAK2 V617F into HSCs via a retrovirus showed us that this overexpression induced a disease that recapitulates PV with its evolution toward a spent phase.15 Therefore, this approach evidently demonstrates that this single mutation is sufficient to induce a PV disease although we cannot exclude that additional genetic or epigenetic events may occur during disease progression. However, it remains possible that a pre–JAK2 V617F stage may exist even in PV because in familial forms of MPDs no germline transmission of the JAK2 mutation has been observed.25 The role of JAK2 V617F in the pathogenesis of ET and IMF is still a matter of debate and it has been suggested that this mutation may be only a secondary event occurring during the progression of the disease.
In the murine model the JAK2 V617F cDNA is transduced in a multipotent HSC capable of long-term and secondary reconstitution.15 Therefore, it could be hypothesized that phenotype heterogeneity induced by the mutated JAK2 could be related to the precise stage at which the clonal process occurred: multipotent stem/progenitor cell versus common myeloid progenitor. We tested this hypothesis in both PV and IMF by first analyzing the JAK2 status in the different circulating mature cell populations. The JAK2 V617F mutation, as expected, was found in all myeloid peripheral blood cells including monocytes, platelets, and neutrophils, further demonstrating that this mutation occurs at least in a common myeloid progenitor. However, this mutation could be detected in the majority of IMF cases in peripheral blood B and NK cell compartments although at a lower frequency than in myeloid cells. In 2 cases the mutation was also detected in T cells, but at the threshold of detection. Although we cannot completely exclude that this was related to a contamination of T cells by myeloid cells, this seems unlikely for 2 reasons, specifically, the purity of the T-cell fraction after a double purification is over 98% and the sensitivity of the sequencing technique is estimated at about 5% to 10%. In contrast, the mutation was detected in peripheral blood B or NK cells from 2 of 10 patients with PV. Altogether these data strongly suggest that a lymphomyeloid stem/progenitor cell could be JAK2 mutated at least in some IMF cases but results were much less striking in PV.
However, the presence of a clonal mutation in peripheral blood leukocytes depends not only on the cellular level at which the clonal event has occurred but also on the proliferative advantage given by the oncogene to the stem cell progeny. Therefore, there was a need to directly analyze the stem/progenitor compartment. A first approach was to study immature stem cells based on differentiation membrane antigens. Analysis of the JAK2 status in CD34+ cells and the CD34+CD38− cell population, which is enriched in lymphomyeloid progenitors, was then performed and the JAK2 mutation was detected in CD34+CD38+ cells from all IMF and PV samples when the mutation was present in granulocytes in agreement with a previous study.17 Interestingly, we were able to find the mutation in CD34+CD38− cells from all IMF patients and from only half the patients with PV. Therefore, to analyze more precisely the lymphomyeloid potential of these CD34+CD38− cells, a clonal analysis was performed using culture conditions that support myeloid, B, and NK cell differentiation.
In all cases of PV and IMF we could find at least one mutated clone that was able to raise myeloid, B, and NK or myeloid and NK differentiation. This clearly demonstrates that in both cases the disease is driven by a lymphomyeloid progenitor. However, this culture system does not support T-cell differentiation. Thus, in parallel we performed a FTOC assay using CD34+ cells from PV and IMF patients and in all cases we could demonstrate that the mutation was also present in a T-cell progenitor able to give rise to double-positive (CD4+CD8+ cell) or single-positive CD4+ or CD8+ cells. Interestingly, some patients with IMF had similar levels of mutated JAK2 in the FTOC-derived T-cell population and in their circulating CD34+ cells. These levels of mutated JAK2 were obviously higher than those found in mature T cells (Figures 1 and 4B). We hypothesize that these T cells generated ex vivo arose from a compartment of mutated CD34+ cells, which was not as important or even present at the time of the generation of the long-lived mature circulating T cells. Altogether these results afford strong evidence that as in CML, PV and IMF occur at the level of a lymphomyeloid stem/progenitor cell. Because ET positive for JAK2 V617F is characterized by a low JAK2 V617F/JAK2 wild-type ratio, we expected very low frequencies of mutated lymphoid cells in ET samples. Indeed, we were unable to detect the mutation in peripheral lymphoid cells from 6 patients with ET (data not shown). However, we identified one patient with ET as having 40% of mutated JAK2 in the granulocytes, as determined by the real-time quantitative assay. We detected the mutation in both bone marrow CD34+CD38+ and CD 34+CD38− cells from this patient and in his B/NK/myeloid CD34+CD38−-derived clones (data not shown). These observations suggest that the JAK2 V617F mutation also occurs, at least in some patients with ET, at the level of a lymphomyeloid stem/progenitor cell. This hypothesis is strengthened by the fact that a fraction of ET cases evolve to PV and that the acquired JAK2 V617F mutation could be detected in B, T, and NK cells of familial forms of IMF, PV, and ET.25
In this study we have observed 2 new findings concerning both the homozygosity of the mutation and differences between PV and IMF. It has been previously shown that about one third of the PV and IMF patients have a homozygous JAK2 V617F mutation associated with a loss of heterozygosity of the short arm of chromosome 9 and due to a mitotic recombination. By clonal analysis we could demonstrate that this secondary event also occurs at the level of a lymphomyeloid stem/progenitor cell because in this compartment coexist nonmutated and mutated clones either heterozygous or homozygous for JAK2 V617F. Due to the low number of cases studied it is yet impossible to determine if the homozygous mutation gives a proliferative advantage to the lymphomyeloid progenitor or later during differentiation. Only patients with more than 50% of mutated JAK2 allele in granulocytes can be considered as presenting a homozygous clone. The presence of nonmutated, heterozygous, and homozygous clones in the same patient makes it difficult to determine the heterozygous versus homozygous status of the mutation in patients. The use of a ratio greater than 50% in granulocytes to assess homozygosity will certainly underestimate the frequency of patients with some homozygous clones. Therefore, defining a disease prognosis relying on the homozygous versus heterozygous status of JAK2 by DNA or RNA-quantitative-PCR on peripheral blood leukocytes becomes questionable.
Surprisingly, we found marked differences in the proportion of lymphomyeloid mutated progenitors between PV and IMF samples. Indeed, the frequency of mutated lymphomyeloid progenitors was much higher in IMF than in PV samples. Presently we do not know if the JAK2 V617F is capable of really expanding the HSC compartment. In CML it has been shown that in chronic phase the BCR-ABL fusion protein only gives a proliferative advantage to progenitors. Therefore, based on this model, we can suggest that IMF and PV are 2 stages of the same disease but in IMF other genetic events have occurred that have favored the proliferative advantage of the lymphomyeloid stem progenitor cell compartment. Whether this event only modifies the constitutive activity of the JAK2 kinase or is independent leading to the acquisition of new stem cell functions remains to be determined. Studies on NOD/SCID repopulating cells are warranted to clearly identify that both diseases have occurred in a true HSC capable of hematopoietic reconstitution in vivo and to precisely quantify the exact proportion of HSC belonging to the clonal process in these 2 disorders.
In conclusion, this study has provided the first evidence that the JAK2 mutation in PV and IMF drives a lymphomyeloid stem/progenitor cell and that the phenotype of the disease is probably related to the proliferative advantage given essentially to the myeloid series leading thus to a pure myeloproliferative and not to a lymphomyeloid proliferative disorder. Defining the precise signaling defect induced by the JAK2 mutation may allow a better determination of the pathogenesis of these MPDs and help to define new prognostic markers.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: F.D. and S.G. designed the study, conducted cellular and molecular experiments, analyzed the data, and wrote the paper; S.D. and A.M. performed cellular experiments and genotyping analysis; I.G. performed FTOC assays and reviewed the manuscript; C.T., J.P.L.C., and P.S. performed sequence and genotyping analyses; N.D. contributed to cellular experiments; N.C. contributed to the design of the study and the recruitment of the patients; and W.V. designed the study, contributed to the recruitment of patients, and wrote the paper.
S.D., C.T., and A.M. contributed equally to this work.
We thank Yann Lécluse and Frédéric Larbret for their contribution to cell sorting experiments. The authors are grateful to Caroline Lefebvre for the helpful discussions and for improving the English manuscript.
This work was supported by grants from the INSERM, la Ligue Nationale contre le Cancer “équipe labellisée 2004,” and from the European Hematology Association (fellowship no. 2005/27; F.D.).

![Figure 2. Assessment of B/NK/myeloid potentialities in IMF and PV cell fractions. Analysis of the progeny of single CD34+CD38− cells from normal bone marrow (N), PV bone marrow, and IMF peripheral blood. Histograms represent 76 positive clones in 576 plated wells from 3 IMF patients, 386 positive clones in 1482 plated wells from 5 PV patients, and 372 positive clones in 1056 plated wells from 3 normal bone marrows. The experiment was performed once for each patient or control sample. The mean percentage of clones containing one (B, NK, myeloid [M]), 2 (B/M, NK/M, B/NK), or 3 (B/NK/M) lineages per total number of clones analyzed after triple-staining by flow cytometry are shown. Myeloid cells correspond to CD15+ cells, B cells to CD19+ cells, and NK cells to CD56+ cells. Error bars indicate SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/1/10.1182_blood-2006-03-007146/4/m_zh80010705760002.jpeg?Expires=1767712085&Signature=JXx8hLHMcRNXSJ2zmnUL0kYFI1uc8BPZEA2R6AYbJOAPbdnKThtje8rO3RAIS9uHtlBEtHgGCsMH3edtsi4yKjza2iT45JKPAQ2FEvgD6E5DtqyY6frsvzHMlt4CgaA3ypPqBhunq8b21lW2U57QhjrTnhigPA2v5WefzpDmKyRmpvzOJa0rzaqSLc4AXzgkvRRUciZ-4MttB8uEJ6JW0rgbKhf4BLxYHzbOXdggnD2Jbt8gB4EpT7BHkn0a1xgAESMqGo7~P2lo6VuObXQfXhfiWrkOt~8zq~H~ZxIhPc-0xwXa-9Yn1ztjvxHM4l~16J~R9Ir3a9pf~zrfpo3~Pw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Genotyping analysis of B/NK/myeloid individual clones derived from PV and IMF purified CD34+CD38minus]. (A) Immunophenotypic analysis of B/NK/myeloid clones from a healthy bone marrow, an IMF patient, and a PV patient. The isotype control scattergrams are shown in the top panel. The B, NK, and myeloid potentials were assessed by the presence of CD19+, CD56+, and CD15+ cells, respectively, after 5 weeks of B, NK, and myeloid differentiation condition culture. The JAK2 G1849T homozygous sequence traces of the IMF and the PV clones are shown. Red arrows indicate the mutant T peak. (B) Genotype analysis of single CD34+CD38− cell culture-derived clones from 3 IMF (i-iii) and 5 PV patients (iv-viii) with respect to their monopotent B, NK, myeloid (M), bipotent B/M, B/NK, M/NK, or tripotent B/NK/M immunophenotypic characterization (clone type).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/1/10.1182_blood-2006-03-007146/4/m_zh80010705760003.jpeg?Expires=1767712085&Signature=AYw7YEXHz1y~Junn6NL4XG-ZufDXwqXoZ7kRbDP443zYe8AiaVHsnW7WPd2kovnLDvq~UjTjVkzfaeyvfqCQxKF~-qvq~Se72QDp6TmIVwCTLThGJ~jpth1jeykULzMdMCG7rJa~ZYz-XVrBQmFrxVUDAbylfqORLeLOAbolXgb8XYuyvR0S1MM1Ukb-U~9XuxeK0NkZfiTEZy7DAuA2LxvhRvByX-P7y9dS24VWdaGv~cDLCwkwMb2~E001rG4KxbCfngw4asVczWIiGFR9YWWoErx89xeGSf5LNk4LpW93Eo7704fgO2C6Mxofd59q1U2SLM1W24pFFEVN4Ze71g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal