Abstract
Genetic engineering of T lymphocytes is an attractive strategy to specifically redirect T-cell immunity toward viral infections and malignancies. We previously demonstrated redirected antileukemic reactivity of cytomegalovirus (CMV)–specific T cells by transfer of minor histocompatibility antigen HA-2–specific T-cell receptors (TCRs). HA-2–TCR-transferred CMV-specific T cells were potent effectors against HA-2–expressing leukemic cells, as well as CMV-expressing cells. Functional activity of these T cells correlated with TCR cell-surface expression. In the present study we analyzed which properties of transferred and endogenous TCRs are crucial for efficient cell-surface expression. We demonstrate that expression of the introduced TCR is not a random process but is determined by characteristics of both the introduced and the endogenously expressed TCR. The efficiency of TCR cell-surface expression is controlled by the intrinsic quality of the TCR complex. In addition, we demonstrate that chimeric TCRs can be formed and that efficiency of TCR expression is independent of whether TCRs are retrovirally introduced or naturally expressed. In conclusion, introduced, endogenous, and chimeric TCRs compete for cell-surface expression in favor of the TCR-CD3 complex with best-pairing properties.
Introduction
Adoptive immunotherapy with antigen-specific T lymphocytes can be effective in the treatment of chronic myeloid leukemia (CML)1 and melanoma2 or in the control of Epstein-Barr virus (EBV) and cytomegalovirus (CMV) diseases.3,4 For production of antigen-specific T cells for clinical purposes, T lymphocytes have been stimulated in vitro with malignant cells or with virus-infected or peptide-loaded stimulator cells in case of EBV or CMV infections. However, to obtain sufficient antigen-specific T cells for treatment, T cells have to be cultured for several weeks in vitro. Furthermore, antigenic specificity of in vitro–expanding T cells is difficult to control. Alternatively, transfer of T-cell receptors (TCRs) with defined antigen specificity into recipient T lymphocytes may simplify generation of antigen-specific T cells, because redirected T cells can be created within several days. Transfer of virus or tumor-specific TCRs resulted in the engineering of T cells with redirected antigen specificity.5-10 These TCR-transferred T cells have been demonstrated to be functionally active in mouse models.11,12
We recently demonstrated redirected antileukemic reactivity of unselected T cells as well as CMV-specific T cells using gene transfer of minor histocompatibility antigen (mHag) HA-2–specific TCRs.9,13 HA-2–TCR-modified T cells exerted specific cytolytic activity against HA-2–expressing target cells, including leukemic cells.9,13 Depending on the recipient T cells, between 20% and 40% of TCR-transduced T cells stained with HA-2 tetramer. On the basis of the correlation between HA-2 tetramer staining and specific cytolytic activity of HA-2–TCR-transduced T-cell clones,9 we concluded that the number of specific TCRs at the cell surface determines the functional avidity of TCR-transferred T cells.
Because the introduced TCR has to compete for cell-surface expression with the endogenous TCR as well as with potential chimeric TCRs formed, it is essential that transferred TCRs exhibit a high affinity for the specific peptide and HLA complex. Because of competition of these different TCR complexes for binding with CD3, the frequency of TCRs at the cell surface will be lower in TCR-transferred T cells than in the parental T-cell clone. However, by transferring high-affinity TCRs we speculate that the amount of TCRs is still sufficient to engineer high-avidity T cells. Recently, we and others demonstrated that TCRs with high affinity for their peptide and MHC complex produce high-avidity TCR-transferred T cells.13,14
Although selection of high-affinity TCRs for gene transfer appears to be critical, we also anticipate that the relative level of TCR membrane expression will be a crucial factor in influencing the avidity of engineered T cells and thus its function. If each individual TCR α- and β-chain pairs with similar affinity to each other TCR β- and α-chain, respectively, and no competition for cellular components between individual TCR α- and β-chains occurs, the introduced and endogenous TCR density on T cells would be approximately 25% of total TCR expression, and approximately 50% would consist of chimeric TCRs. Because part of the HA-2–TCR-transferred T cells preferentially expressed HA-2–TCRs, although others preferentially expressed endogenous TCRs, the TCR cell-surface composition might not be a random process but might be regulated by both the introduced TCR as well as characteristics of the recipient T cell. Previously, also others have demonstrated that, based on expression of the transferred TCR β-chain, only part of TCR-transferred T cells could bind the specific tetramer.10,14
In this study we investigated whether inherent properties of the endogenous and transferred TCR determined the efficiency of expression at the cell surface of TCR-transduced T cells. In addition, we investigated the biology of T cells expressing several TCR chains. We demonstrated that HA-2–TCR-transferred CMV-specific T cells preferentially expressing the HA-2–TCR expressed different endogenous TCR α- and β-chains than HA-2–TCR-transferred CMV T cells that predominantly expressed the endogenous TCR. By transferring high-affinity CMV-TCRs into αβ and γδ T cells, we demonstrated that the TCR makeup of TCR-transferred T cells is determined by intrinsic properties of the TCR-CD3 complexes.
Materials and methods
T-cell clones
The following T-cell clones were used in this study: HLA-A2–restricted HA-2–specific T-cell clones HA2.1, HA2.5, HA2.6, HA2.19, HA2.20, and HA2.27,15 HLA-A2–restricted alloreactive T-cell clones MBM15 and MBM13,7 HLA-B7–restricted mHag–specific T-cell clone 10G5,16 mHag-specific T-cell clones H and S,17 HLA-DQ5–restricted DBY-specific T-cell clone JBB4, and mHag-specific T-cell clone JBBun.18,19
Analysis of TCRAV and TCRBV transcripts and generation of retroviral vectors and virus supernatant
TCRAV and TCRBV gene usage of the different antigen-specific T-cell clones was determined as previously described.20 All TCR α- and β-chains derived from the different antigen-specific T-cell clones were cloned separately into retroviral vectors. The TCR α-chains were always combined with the marker eGFP, and the TCR β-chains were always combined with the truncated nerve growth factor receptor (ΔNGF-R).21 Retroviral vectors encoding eGFP or ΔNGF-R alone were used as control vectors. In addition, a retroviral vector encoding for pp65 of HCMV AD169 was used to transduce target cells. Using the Moloney murine leukemia virus–based retroviral vector LZRS and packaging cells φ-NX-A,22 viral supernatant was generated as previously described.23
Tetrameric HLA class I and peptide complexes, flow cytometric analyses, fluorescence-activated cell sorting (FACS), and reverse transcriptase–polymerase chain reaction (RT-PCR)
PE- or APC-conjugated tetrameric complexes were constructed as described24 with minor modifications. Tetrameric HLA-A2 molecules in complex with CMV pp65-derived peptide NLVPMVATV (CMVA2 tetramer) and HA-2–derived peptide YIGEVLVSV (HA-2A2 tetramer) were constructed. For flow cytometric analyses or cell sorting experiments, cells were labeled with tetramers for 2 hours at 4 °C. During the last 30 minutes mAbs directed against CD4 (FITC), CD8 (FITC or PECy5), CD40 (FITC), or NGF-R (PE or APC) (Becton Dickinson [BD], San Jose, CA) were added. Cell-surface staining with anti–TCR αβ (PECy5), anti-CD3 (PECy5), or mAbs specific for the BV2 and BV13 chains (PE) were performed for 30 minutes at 4°C. For cytoplasmatic staining the cells were incubated for 5 minutes at room temperature with 0.25% saponin, centrifuged, and labeled with anti–TCR αβ, anti-BV2, or isotype control mAbs for 30 minutes at 4°C.
Expression of TCR chains was quantified by real-time PCR (TaqMan) using primers specific for the C region (αTCR chain: forward primer, CCTGTGATGTCAAGCTGGTCG, reversed primer, AGCAGATTAAACCCGGCCA, probe TTGAAACAGATACGAACCTAAACTTTCAAAACCTGTCA; βTCR chain: forward primer, AGGATAGGGCCAAACCCGT, reversed primer, AGACAGGACCCCTTGCTGGT, probe, AGGCCTGGGGTAGAGCAGACTGTGGC).
Retroviral transduction of T lymphocytes and selection of transduced T cells
All studies were conducted with approval of the institutional review board at Leiden University Medical Center. Virus-specific T cells were isolated from peripheral-blood mononuclear cells (PBMCs) of CMV seropositive persons. After informed consent PBMCs were harvested and labeled with tetramers for 2 hours at 4°C in RPMI without phenol, supplemented with 2% FBS, washed 2 times, and sorted at 4°C using the FACS Vantage (BD). γδ T cells were isolated from healthy persons using FITC-labeled, γδ TCR–specific immunomagnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany), and the AutoMACS system (Miltenyi Biotec) according to the manufacturer's protocol. To obtain maximal purity, cells were subsequently sorted using the FACS Vantage (BD). Virus-specific T cells were stimulated with 1 × 106 cells/mL irradiated allogeneic PBMCs (50 Gy), 800 ng/mL PHA, and 100 IU/mL IL-2 (Chiron, Amsterdam, The Netherlands). γδ T cells were stimulated similarly with the addition of 10 ng/mL IL-15 (PeproTech, Rocky Hill, NJ). Total PBMCs were stimulated with 800 ng/mL PHA and 100 IU/mL IL-2. After 2 days of culture T cells were transduced with retroviral supernatant using recombinant human fibronectin fragments CH-296.23,25 Briefly, 0.5 × 106 T cells were cultured on CH-296–coated 24-well nonculture treated plates (Falcon, Franklin Lakes, NJ) together with 0.5 mL thawed retroviral supernatant for 6 hours or overnight at 37°C, washed, and transferred to 24-well tissue culture plates. HA-2–TCR and CMV-TCR–transduced T cells were sorted by FACS on bases of marker gene expression, cultured at 1 cell/well or 25 cells/well, and nonspecifically stimulated every 2 weeks.
Cytotoxicity assay
Target cells were labeled with 50 μCi (1.85 MBq) Na
Results
Cell-surface expression and function of the transferred TCR is strongly influenced by the endogenous TCR
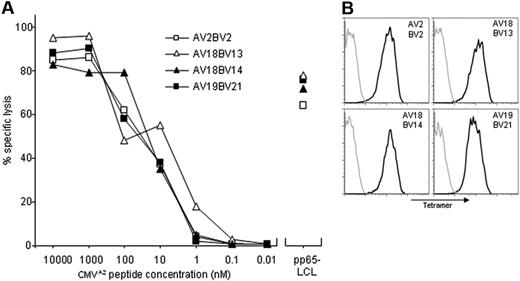
Previously, we demonstrated that after TCR transfer some TCR-modified T cells predominantly expressed the introduced TCR, whereas other TCR-modified T cells preferentially expressed the endogenous TCR. To investigate whether the endogenous TCR influences the cell-surface expression of the introduced TCR, we transferred the HA-2–TCR into well-characterized T-cell populations. We first characterized the TCR repertoire of a CMVA2-specific memory response of a CMV seropositive, HLA-A2–positive patient. CMV-specific T cells were isolated using CMVA2 tetramers and expanded, and 26 T-cell clones were analyzed for TCR usage. As shown in Table 1 the TCR repertoire of the CMV-specific memory response was restricted to 4 different TCRs. These 4 CMV-specific T-cell clones were of high avidity because these cells exerted half maximum lysis of CMV peptide-pulsed T2 targets at a concentration of approximately 20 nM and lysed pp65-positive targets efficiently (Figure 1A). In addition, all 4 clones stained similarly with CMVA2 tetramers (Figure 1B).
TCR α and β sequences of CMVA2-specific T-cell clones
| Clone (n)* . | AV† . | End AV . | CDR3 α . | Start AJ . | AJ . | BV . | End BV . | CDR3 β . | Start BJ . | BJ . |
|---|---|---|---|---|---|---|---|---|---|---|
| A (11) | 2S2 | CAM | NGARLM | FGD | 31 | 2S1 | CSA | RDVLAGGSDTQ | YFG | 2S3 |
| B (6) | 18S1 | CAR | NTGNQFY | FGT | 49 | 13S1 | CAS | SSVTGTGNYGY | TFG | 1S2 |
| C (6) | 18S1 | CAF | NFGNEKLT | FGT | 48 | 14S1 | CAS | SLFGGFTEA | FFG | 1S1 |
| D (3) | 19S1 | CAV | RSNFGNEKLT | FGT | 48 | 21S1 | CAS | SFWDRVSGANVL | TFG | 2S6 |
| Clone (n)* . | AV† . | End AV . | CDR3 α . | Start AJ . | AJ . | BV . | End BV . | CDR3 β . | Start BJ . | BJ . |
|---|---|---|---|---|---|---|---|---|---|---|
| A (11) | 2S2 | CAM | NGARLM | FGD | 31 | 2S1 | CSA | RDVLAGGSDTQ | YFG | 2S3 |
| B (6) | 18S1 | CAR | NTGNQFY | FGT | 49 | 13S1 | CAS | SSVTGTGNYGY | TFG | 1S2 |
| C (6) | 18S1 | CAF | NFGNEKLT | FGT | 48 | 14S1 | CAS | SLFGGFTEA | FFG | 1S1 |
| D (3) | 19S1 | CAV | RSNFGNEKLT | FGT | 48 | 21S1 | CAS | SFWDRVSGANVL | TFG | 2S6 |
Number of CMV-specific T-cell clones with identical TCR α- and β-chain.
Nomenclature according to Arden et al.26
High-affinity CMV-specific T cells isolated from memory response. (A) Cytolytic activity of CMV-specific T cells. T cells expressing different TCRs were isolated from the peripheral blood of CMV-seropositive persons and were tested at the effector-to-target (E/T) ratio of 10:1 against CMV-peptide–pulsed HLA-A2+ EBV-LCLs or pp65-transduced HLA-A2+ EBV-LCLs (pp65-LCL) in a 4-hour cytotoxicity assay. (B) Tetramer staining was performed with PE-conjugated CMVA2 tetramers (black line) and control PE-conjugated HA-2A2 tetramers (gray line) for 2 hours at 4°C on representative T-cell clones expressing 1 of the 4 different CMV-specific TCRs.
High-affinity CMV-specific T cells isolated from memory response. (A) Cytolytic activity of CMV-specific T cells. T cells expressing different TCRs were isolated from the peripheral blood of CMV-seropositive persons and were tested at the effector-to-target (E/T) ratio of 10:1 against CMV-peptide–pulsed HLA-A2+ EBV-LCLs or pp65-transduced HLA-A2+ EBV-LCLs (pp65-LCL) in a 4-hour cytotoxicity assay. (B) Tetramer staining was performed with PE-conjugated CMVA2 tetramers (black line) and control PE-conjugated HA-2A2 tetramers (gray line) for 2 hours at 4°C on representative T-cell clones expressing 1 of the 4 different CMV-specific TCRs.
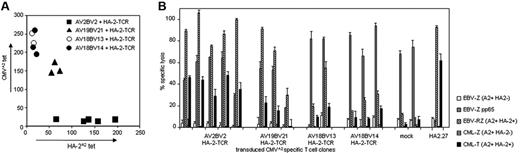
From the same original cell population, CMV-specific T cells were isolated and transduced with the HA-2–TCR.13 By double marker gene expression TCR-transferred T cells were sorted single cell and expanded by nonspecific stimulation. To exclude that variation in TCR cell-surface composition was due to differences in transgene expression, we selected clones that were highly positive for both marker genes. All T-cell clones showed similar overall TCR αβ expression (data not shown). TCR cell-surface makeup of the T-cell clones, measured by HA-2A2 and CMVA2 tetramers, did not change during months of culture. Interestingly, all CMV-specific T-cell clones transduced with the HA-2–TCR that predominantly expressed the HA-2–TCR at the cell surface expressed the same CMV-TCR composed of the AV2 and BV2 chain (Figure 2A) These T-cell clones most effectively lysed HA-2+ EBV-LCLs and leukemic cells, almost comparable to the cytolytic activity of the parental HA-2–specific T-cell clone (Figure 2B). Although CMVA2 tetramer staining was reduced to approximately 10% of mock-transduced cells (Figure 2A), and BV2 staining was reduced to approximately 25% of mock-transduced cells (data not shown), these HA-2–TCR CMV-specific T cells were able to exert pp65-specific cytotoxicity, although this reactivity was reduced compared with mock T cells. These data indicate that limited numbers of TCRs expressed at the cell surface are needed for the cytotoxic effector function of T cells. CMV-specific T cells with intermediate HA-2–TCR cell-surface expression all expressed the same endogenous CMV-TCR composed of the AV19 and BV21 chain (Figure 2A). Although these clones were capable of lysing HA-2+ EBV-LCLs, the T-cell clones exhibited only low recognition of HA-2+ leukemic cells. The pp65-specific cytotoxicity of these T cells was reduced compared with mock-transduced CMV-specific T cells. The HA-2–TCR-transferred CMV-specific T cells with low HA-2A2 tetramer and high CMVA2 tetramer staining expressed either the AV18 and BV13 chain or the AV18 and BV14 chain. These T-cell clones exerted only minor recognition of HA-2+ EBV-LCLs and leukemic cells, whereas these T-cell clones exhibited efficient recognition of pp65-positive EBV-LCLs. The sequences of the 4 different TCRs observed in the HA-2–TCR-transferred T cells were identical with the TCRs detected in the original non- (Table 1) and mock-transduced CMV-specific T-cell clones (data not shown). These data indicate that, although all HA-2–TCR-transferred T-cell clones exerted pp65-specific cytotoxicity, only those T-cell clones with high HA-2–TCR cell-surface expression efficiently eradicated leukemic cells expressing endogenously processed HA-2. Furthermore, our data demonstrated that TCR cell-surface composition in TCR-transferred T cells was not a random process but was strongly influenced by the endogenous TCR.
TCR repertoire analysis and functional analyses of the different types of HA-2–TCR-modified CMV-specific T clones. (A) The endogenous TCR repertoire was analyzed by RT-PCR and sequencing. To measure the expression level of HA-2 and CMV-TCR on the HA-2–TCR–transferred CMV-specific T-cell clones, the T cells were labeled with either PE-conjugated HA-2A2 tetramers or CMVA2 tetramers for 2 hours at 4°C. Four different types of HA-2–TCR–modified CMVA2-specific T clones were identified. (B) Representative T-cell clones of these 4 different types of HA-2–TCR–modified CMVA2-specific T cells, control-transduced CMVA2-specific T cells, and the original HA-2–specific T-cell clone HA2.27 were tested at an E/T ratio of 10:1 against HLA-A2+ HA-2− EBV-LCLs (EBV-Z), HLA-A2+ HA-2+ EBV-LCLs (EBV-RZ), EBV-Z transduced with the CMV pp65 gene (EBV-Z pp65), HLA-A2+ HA-2− CML cells (CML-Z), and HLA-A2+ HA-2+ CML cells (CML-T) in an 18-hour cytotoxicity assay. Error bars indicate SD of triplicate culture.
TCR repertoire analysis and functional analyses of the different types of HA-2–TCR-modified CMV-specific T clones. (A) The endogenous TCR repertoire was analyzed by RT-PCR and sequencing. To measure the expression level of HA-2 and CMV-TCR on the HA-2–TCR–transferred CMV-specific T-cell clones, the T cells were labeled with either PE-conjugated HA-2A2 tetramers or CMVA2 tetramers for 2 hours at 4°C. Four different types of HA-2–TCR–modified CMVA2-specific T clones were identified. (B) Representative T-cell clones of these 4 different types of HA-2–TCR–modified CMVA2-specific T cells, control-transduced CMVA2-specific T cells, and the original HA-2–specific T-cell clone HA2.27 were tested at an E/T ratio of 10:1 against HLA-A2+ HA-2− EBV-LCLs (EBV-Z), HLA-A2+ HA-2+ EBV-LCLs (EBV-RZ), EBV-Z transduced with the CMV pp65 gene (EBV-Z pp65), HLA-A2+ HA-2− CML cells (CML-Z), and HLA-A2+ HA-2+ CML cells (CML-T) in an 18-hour cytotoxicity assay. Error bars indicate SD of triplicate culture.
Formation of chimeric TCRs in TCR-transferred T cells
Because TCR transfer possibly leads to the formation of chimeric TCRs,27 we investigated whether introduction of single TCR α- or β-chains resulted in chimeric TCR formation, and whether the level of expression was also influenced by the endogenous TCR usage. Different CMV-specific T-cell subsets derived from the previously described cell population were selected by isolating CMVA2 tetramer–positive T cells and subsequent selection using mAbs directed against the relevant β-chains. MAb specific for the BV2 chain of the AV2BV2 T cells and mAb specific for the BV13 chain of AV18BV13 T cells were available. The 2 selected CMV-specific T-cell subsets were subsequently transduced with either the HA-2–AV15-eGFP or the HA-2–BV18-NGF-R or both HA-2–TCR encoding vectors (Figure 3A) On bases of high marker gene expression, the different transduced subsets were gated and analyzed. Introduction of either the HA-2–AV15 or the HA-2–BV18 into the CMV-AV2BV2 subset reduced the CMVA2 tetramer staining markedly to 42% and 32% of control cells, respectively. Introduction of both HA-2–TCR chains reduced the CMVA2 tetramer staining to 14% of control cells (Figure 3A). Introduction of either the HA-2–AV15 or HA-2–BV18 chain or both chains into the CMV-AV18BV13 T cells reduced the CMVA2 tetramer staining less pronounced to 85%, 75%, and 58% of control cells, respectively (Figure 3B). Analyses with the 2 mAbs directed against the endogenous CMV-TCR β-chains demonstrated that introduction of the HA-2–TCR BV18 chain reduced the endogenous TCR β-chain expression in both subsets (Figure 3C). Similar as observed with the CMVA2 tetramer staining the CMV-BV2 chain was more dramatically reduced than the CMV-BV13 chain. These results indicate that the decreased CMVA2 tetramer staining shown in Figure 3B was due to decreased cell-surface expression of the endogenous CMV-TCR chains (Figure 3C). Although the CMV-TCR expression decreased in both CMV-specific T-cell subsets by the introduction of a single TCR chain, no changes in total TCR αβ expression were observed (Figure 3D), illustrating that the introduced TCR α- or β-chains have the capability to form chimeric TCR complexes with the endogenous TCR chains. To exclude that differences in chimeric TCR formation were due to difference in RNA levels of endogenous TCRs, mRNA was isolated from AV2BV2 and AV18BV13 T cells, and TCR α- and β-chain message was quantified by RT-PCR and normalized to PBGD copies. No differences in endogenous TCR α and TCR β mRNA expression were observed between the AV2BV2 and AV18BV13 T-cell subsets (data not shown). Therefore, the more extended decrease in CMV-TCR expression in the CMV-AV2BV2 T cells compared with the CMV-AV18BV13 T cells after single TCR chain transfer indicated that chimeric TCRs are more prominently formed in CMV-AV2BV2 T cells compared with CMV-AV18BV13 T cells.
TCR transfer results in the formation of chimeric TCRs. Sorted AV2BV2 and AV18BV13 CMV-specific T cells (> 98% pure) were transduced with the HA-2–AV15-GFP and the HA-2–BV18-NGFR retroviral vectors. Seven days after transduction, the single- and double-transduced subsets were gated on bases of marker gene expression (A), the HA-2–AV15 single-positive cells were gated on bases of GFP+NGFR− (gate 1), the HA-2–BV18 single-positive cells were gated on bases of GFP−NGFR+ (gate 2) and HA-2–AV18BV13 double-positive cells were gated on bases of GFP+NGFR+ (gate 3). Cells were analyzed for CMV-TCR expression using CMVA2 tetramers (B) and mAbs directed against the CMV-TCR β-chains BV2 or BV13 (C); in addition, total TCRαβ cell-surface expression was measured (D). In the figures the MFI of the staining is indicated as the percentage of control-transduced cells (100%) of 2 independent experiments. Error bars indicate SD.
TCR transfer results in the formation of chimeric TCRs. Sorted AV2BV2 and AV18BV13 CMV-specific T cells (> 98% pure) were transduced with the HA-2–AV15-GFP and the HA-2–BV18-NGFR retroviral vectors. Seven days after transduction, the single- and double-transduced subsets were gated on bases of marker gene expression (A), the HA-2–AV15 single-positive cells were gated on bases of GFP+NGFR− (gate 1), the HA-2–BV18 single-positive cells were gated on bases of GFP−NGFR+ (gate 2) and HA-2–AV18BV13 double-positive cells were gated on bases of GFP+NGFR+ (gate 3). Cells were analyzed for CMV-TCR expression using CMVA2 tetramers (B) and mAbs directed against the CMV-TCR β-chains BV2 or BV13 (C); in addition, total TCRαβ cell-surface expression was measured (D). In the figures the MFI of the staining is indicated as the percentage of control-transduced cells (100%) of 2 independent experiments. Error bars indicate SD.
Competition of TCR cell-surface expression is independent of whether TCRs are retrovirally introduced or endogenously expressed
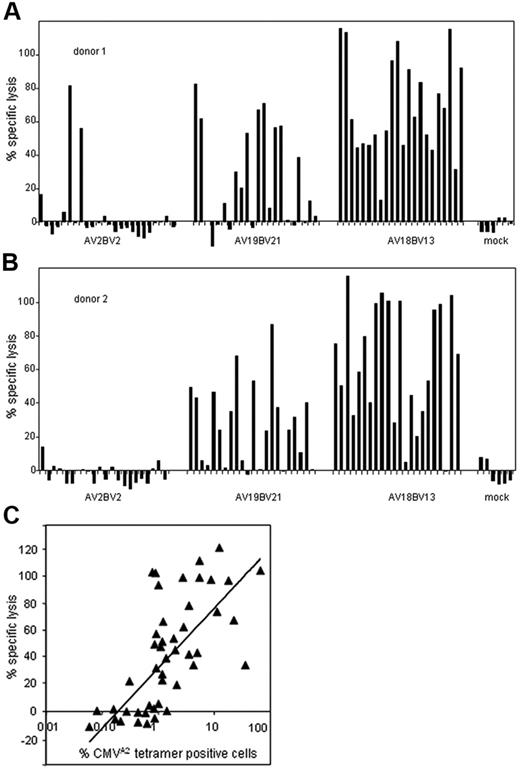
To show that competition for cell-surface expression is a general phenomenon, not restricted to HA-2–TCR versus CMV-TCR combinations and independent of differences in promotor activity of the introduced and endogenous TCRs, additional experiments were performed. PBMCs derived from 2 healthy persons were transferred with different CMV-TCRs identified from the earlier described memory response (Table 1). Retroviral vectors encoding 3 of the CMV-TCRs (AV2BV2, AV18BV13, and AV19BV21) were constructed. TCR α-chains were combined with eGFP, TCR β-chains were combined with NGF-R. One week after transduction the eGFP+NGF-R+ T cells were sorted 25 cell/well and expanded. From each CMV-TCR–transduced T-cell population 22 to 24 lines were established, which were analyzed for CMV-specific cytolytic activity and tetramer staining. T cells transferred with the CMV-AV2BV2-TCR showed low CMV-specific cytotoxicity. Only 2 of 46 transduced T-cell lines exerted CMV-specific cytotoxicity (Figure 4A-B), and no tetramer staining was shown (data not shown). T cells transduced with the CMV-AV19BV21-TCR showed intermediate CMV-specific cytotoxicity, because 50% of the cell lines lysed CMV-positive targets. Of the CMV-AV18BV13-TCR–transferred cell lines 95% exerted strong CMV-specific cytotoxicity. CMVA2 tetramer staining revealed that in TCR-transferred lines that exerted CMV-specific cytotoxicity the percentage of CMVA2 tetramer–positive T cells varied, ranging from 0.5% to 70%. By Pearson statistical analysis we determined that the cytolytic activity of the T-cell lines correlated with the percentage CMVA2 tetramer–positive T cells (P > .7) (Figure 4C). These data indicate that competition for cell-surface expression also occurred in the combination of CMV-TCRs and unknown TCRs expressed by unselected T cells. In addition, we demonstrate that introduced and endogenous TCRs shape the TCR cell-surface makeup independent of whether TCRs are retrovirally introduced or naturally expressed.
TCR cell-surface composition independent of whether the TCRs are retrovirally or endogenously expressed. Sorted CMV-TCR–transferred T-cell lines or mock-transduced T-cell lines (> 98% GFP+NGFR+) derived from 2 different healthy persons (A) and (B) were tested for cytolytic activity against HLA-A2+ EBV-LCLs loaded with 1 μM CMV peptide (black bars) or nonpeptide-loaded EBV-LCLs in a 4-hour cytotoxicity assay. The cytolytic activity against nonpeptide-loaded target cells was for all transduced T-cell lines less than 10%. All the transduced T-cell lines tested originated from wells that were seeded with 25 double marker gene–positive T cells. (C) The percentage of CMVA2 tetramer–positive CMV-TCR–transduced T cells is plotted against the cytolytic activity of the T cells toward HLA-A2+ EBV-LCLs loaded with 1 μM CMV peptide. Labeling with PE-conjugated CMVA2 tetramers was performed for 2 hours at 4°C. Our results show that the cytolytic activity of the T-cell lines is correlated with the percentage of tetramer-positive cells in these cultures (P > .7).
TCR cell-surface composition independent of whether the TCRs are retrovirally or endogenously expressed. Sorted CMV-TCR–transferred T-cell lines or mock-transduced T-cell lines (> 98% GFP+NGFR+) derived from 2 different healthy persons (A) and (B) were tested for cytolytic activity against HLA-A2+ EBV-LCLs loaded with 1 μM CMV peptide (black bars) or nonpeptide-loaded EBV-LCLs in a 4-hour cytotoxicity assay. The cytolytic activity against nonpeptide-loaded target cells was for all transduced T-cell lines less than 10%. All the transduced T-cell lines tested originated from wells that were seeded with 25 double marker gene–positive T cells. (C) The percentage of CMVA2 tetramer–positive CMV-TCR–transduced T cells is plotted against the cytolytic activity of the T cells toward HLA-A2+ EBV-LCLs loaded with 1 μM CMV peptide. Labeling with PE-conjugated CMVA2 tetramers was performed for 2 hours at 4°C. Our results show that the cytolytic activity of the T-cell lines is correlated with the percentage of tetramer-positive cells in these cultures (P > .7).
Intrinsic pairing properties of TCRs control the TCR makeup of TCR-transferred T cells
Although all 3 CMV-TCRs exhibited a similar high affinity for their cognate peptide-HLA ligand as shown in Figure 1, functional differences of the various CMV-TCR–transferred T cells were observed (Figure 4). To investigate which characteristics of TCRs were important for cell-surface expression, CMV-TCRs were transferred into γδ T cells. Because γδTCR complexes are unable to form heterodimers with αβTCRs,28,29 we were able to dissect whether low cell-surface expression of certain TCR combinations was due to favorable pairing with the endogenous TCR or due to reduced ability of the TCR chains to compete with endogenous TCR chains for binding to CD3. We isolated greater than 99% pure γδ T cells, all expressing the Vγ9Vδ1, and transduced the cells after 3 days of stimulation. Double-marker gene–expressing γδ T cells were sorted, expanded, and analyzed for TCR expression. As shown in Figure 5 the TCR αβ cell-surface expression of γδ T cells transduced with the 3 CMV-TCRs varied again considerably. The AV2BV2-TCR–transduced γδ T cells expressed significantly lower levels of TCR αβ compared with the other 2 CMV-TCR–transduced γδ T cells (Figure 5A; Table 2) In agreement with previous results (Figure 3), we observed in AV2BV2-TCR–transduced γδ T cells no or a marginal decrease of TCR γδ cell-surface expression, whereas the TCRγδ expression in the AV19BV21-TCR–transduced γδ T cells was reduced, and even a more dramatic reduction was observed for the AV18BV13-TCR–transduced cells. No change in CD3 cell-surface expression was observed between TCR- and control-transduced γδ T cells (data not shown). By quantitative RT-PCR no difference in TCR α and TCR β mRNA expression was observed between the different transduced γδ T cells (Figure 5C). Transfer of γδ T cells with all possible chimeric TCR combinations, consisting of the TCR α-chain of one antigen-specific TCR with the TCR β-chain of another TCR complex excluded that the low TCR αβ cell-surface expression of the CMV-AV2BV2-TCR was due to low protein expression of the separate TCR chains (Table 2), because chimeric TCRs containing either CMV-AV2 or CMV-BV2 could equally be expressed as other chimeric and wild-type TCRs.
Analyses of cell-surface TCRαβ and TCRγδ expression and intracellular TCRαβ expression of γδ T cells transduced with different CMV-TCRs. (A) Peripheral-blood–derived γδ T cells were transduced with 3 CMV-TCR complexes and with control retroviral vectors only expressing the marker genes. Double marker gene–positive cells were sorted and expanded, and the TCRαβ and TCRγδ cell-surface expression of the GFP+ NGF-R+ population was determined by FACS. The marker gene expression of the transduced γδ T cells is shown in the small dot-plots. (B left) Intracellular staining of TCRαβ complexes in γδ T cells transduced with control vector (filled histogram), original AV2BV2-TCR (black line), and original AV18BV13-TCR (gray line) with anti-TCRαβ mAb using saponin treatment. (B right) Intracellular staining of TCRαβ complexes in γδ T cells transduced with control vector (filled histogram), chimeric AV2BV13-TCR (black line), and chimeric AV18BV2-TCR (gray line) with anti-TCRαβ mAb using saponin treatment. (C) TCR β-chain mRNA from γδ T cells transduced with AV2BV2 (□), AV19BV21 (▪), or AV18BV13 (⊡) TCRs was quantified by RT-PCR and normalized to PBGD. Similar results were obtained for the TCR α-chain mRNA expression in these different TCR-transferred γδ T cells (data not shown). (D) Intracellular staining of TCR BV2 in γδ T cells transduced with control vector (filled histogram), original AV2BV2-TCR (black line), and the chimeric AV18BV2-TCR (gray line) with anti-BV2–specific mAb using saponin treatment.
Analyses of cell-surface TCRαβ and TCRγδ expression and intracellular TCRαβ expression of γδ T cells transduced with different CMV-TCRs. (A) Peripheral-blood–derived γδ T cells were transduced with 3 CMV-TCR complexes and with control retroviral vectors only expressing the marker genes. Double marker gene–positive cells were sorted and expanded, and the TCRαβ and TCRγδ cell-surface expression of the GFP+ NGF-R+ population was determined by FACS. The marker gene expression of the transduced γδ T cells is shown in the small dot-plots. (B left) Intracellular staining of TCRαβ complexes in γδ T cells transduced with control vector (filled histogram), original AV2BV2-TCR (black line), and original AV18BV13-TCR (gray line) with anti-TCRαβ mAb using saponin treatment. (B right) Intracellular staining of TCRαβ complexes in γδ T cells transduced with control vector (filled histogram), chimeric AV2BV13-TCR (black line), and chimeric AV18BV2-TCR (gray line) with anti-TCRαβ mAb using saponin treatment. (C) TCR β-chain mRNA from γδ T cells transduced with AV2BV2 (□), AV19BV21 (▪), or AV18BV13 (⊡) TCRs was quantified by RT-PCR and normalized to PBGD. Similar results were obtained for the TCR α-chain mRNA expression in these different TCR-transferred γδ T cells (data not shown). (D) Intracellular staining of TCR BV2 in γδ T cells transduced with control vector (filled histogram), original AV2BV2-TCR (black line), and the chimeric AV18BV2-TCR (gray line) with anti-BV2–specific mAb using saponin treatment.
TCR αβ cell-surface expression of γδ T cells transduced with original and chimeric TCRs
| TCR . | BV2S1 . | BV21S1 . | BV13S1 . | BV18S1 . |
|---|---|---|---|---|
| AV2S2 | 55 ± 4*† | 328 ± 36 | 328 ± 34 | 334 ± 16 |
| AV19S1 | 430 ± 23 | 383 ± 47† | 414 ± 40 | 361 ± 12 |
| AV18S1 | 361 ± 37 | 335 ± 120 | 411 ± 11† | 307 ± 34 |
| AV15S1 | 372 ± 62 | 343 ± 53 | 330 ± 31 | 374 ± 68† |
| TCR . | BV2S1 . | BV21S1 . | BV13S1 . | BV18S1 . |
|---|---|---|---|---|
| AV2S2 | 55 ± 4*† | 328 ± 36 | 328 ± 34 | 334 ± 16 |
| AV19S1 | 430 ± 23 | 383 ± 47† | 414 ± 40 | 361 ± 12 |
| AV18S1 | 361 ± 37 | 335 ± 120 | 411 ± 11† | 307 ± 34 |
| AV15S1 | 372 ± 62 | 343 ± 53 | 330 ± 31 | 374 ± 68† |
EGFP+ NGF-R+ γδ T cells were gated; TCRαβ expression has been indicated as mean fluorescence intensity (MFI). The MFI is shown as the mean of 3 independent experiments plus or minus SD.
MFI less than 100.
The TCR α and β combinations derived from the original T-cell clones.
Intracellular staining for TCR αβ demonstrated that CMV-AV2BV2-TCR–transferred γδ T cells expressed no or significantly lower levels of TCR αβ intracellularly compared with CMV-AV18BV13-TCR–transferred T cells or as compared with γδ T cells transferred with chimeric TCRs composed of AV2 with BV13 and BV2 with AV18 (Figure 5B). Intracellular staining using the specific TCR BV2 mAb demonstrated that all CMV-AV2BV2-TCR–transferred cells expressed the BV2 chain intracellularly (Figure 5D). BV2 expression, however, was lower compared with chimeric TCR-AV18BV2–transferred γδ T cells. This is probably because, if TCR α- and β-chains are not assembled with the CD3 subunits, these chains are highly susceptible to proteolysis in the ER and are lost from cells within hours of synthesis.30,31 We therefore assume that low TCR expression of CMV-AV2BV2-TCR–transduced γδ T cells is because AV2BV2-TCR loses the competition with the endogenous γδTCR for binding to the CD3 complex.
To investigate whether more combinations of TCR α- and β-chains were unable to compete with the γδTCR for cell-surface expression, we extended the panel of transferred TCR chains. γδ T cells were transduced with 16 different TCR α-chains in combination with 15 different TCR β-chains derived from mHag-, CMV-, or allo-reactive T-cell clones. The TCRαβ cell-surface expression of wild-type transferred TCRs varied between MFI of 124 to 687 (Table 3), with the exception of the AV2BV2 (MFI = 28). Most chimeric TCRs were expressed efficiently; however, 23 of the 224 (10%) chimeric TCRs tested were marginally expressed (MFI between 4 and 85). These data demonstrate that a significant proportion of TCRs vary in their ability to compete with other TCRs for efficient cell-surface expression. In addition, these data show that efficiency of expression is influenced by the V regions of both TCR α- and β-chains. Even subfamily differences influence expression levels, because, for example, the CMV-AV2S2 is expressed efficiently with the BV6S2 of the HA-2.1 and HA-2.20 T-cell clones, as well as with the BV6S7 of 10G5, whereas expression with the BV6S3 of JBBun is inefficient. In these combinations, however, we could not exclude that the difference in expression was due to differences in the CDR3 region, because the CDR3 regions of these β chains were different. That CDR3 regions have a significant role in the efficiency of expression is illustrated by transduction of 3 AV23S1 chains of MBM13, HA2.6, and HA2.20 with several different β-chains. Although only the N region in these 3 AV23S1 chains varied (Table 4), substantial differences in pairing efficiency could be observed. The AV23S1 chains were all 3 marginally expressed in combination with the BV15S1 of clone S and the BV6S3 of JBBun. In addition, the AV23S1 of MBM13 was marginally expressed with the CMV-BV2S1; however, the AV23S1 of HA2.6 and HA2.20 were efficiently expressed with this BV chain, indicating that 5 amino acids in the CDR3 region can influence the pairing properties dramatically, leading to reduced competition potential.
TCR αβ cell-surface expression of γδ T cells transferred with original and chimeric TCRs
| Clone . | ↓AV . | MBM15 . | 10G5 . | MBM13 . | Clone H . | CMV-A2 . | JBB4 . | Clone S . | JBBun . | CMV-A2 . | CMV-A2 . | HA2.1 . | HA2.5 . | HA2.6 . | HA-2.19 . | HA-2.20 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BV→ | — | 13S1 | 657 | 7S3 | 5S3 | 21S1 | 5S6 | 15S1 | 6S3 | 13S1 | 2S1 | 6S2 | 18S1 | 18S1 | 18S1 | 6S2 |
| MBM15 | 1S3 | 390† | 386 | 58* | 384 | 386 | 85* | 31* | 409 | 423 | 293 | 488 | 403 | 409 | 468 | 359 |
| 10G5 | 12S1 | 193 | 327† | 131 | 189 | 170 | 107 | 48* | 147 | 157 | 173 | 201 | 169 | 154 | 185 | 162 |
| MBM13 | 2S3 | 586 | 612 | 513† | 735 | 670 | 199 | 529 | 350 | 583 | 407 | 409 | 540 | 894 | 561 | 595 |
| MBM13 | 23S1 | 143 | 347 | 124† | 376 | 181 | 75* | 33* | 25* | 417 | 53* | 226 | 467 | 467 | 479 | 419 |
| Clone H | 20S1 | 589 | 272 | 256 | 616† | 394 | 266 | 296 | 247 | 613 | 520 | 973 | 540 | 706 | 690 | 610 |
| CMV-A2 | 19S1 | 174 | 189 | 83* | 30* | 368† | 55* | 4* | 65* | 241 | 153 | 159 | 281 | 275 | 473 | 201 |
| JBB4 | 1S4 | 311 | 454 | 366 | 665 | 574 | 204† | 76* | 296 | 434 | 181 | 515 | 553 | 512 | 585 | 422 |
| Clone S | 7S2 | 439 | 328 | 312 | 543 | 503 | 161 | 357† | 124 | 457 | 274 | 404 | 435 | 501 | 590 | 338 |
| JBBun | 14S1 | 535 | 647 | 445 | 776 | 783 | 484 | 367 | 583† | 497 | 622 | 503 | 548 | 565 | 632 | 572 |
| CMV-A2 | 18S1 | 394 | 426 | 278 | 208 | 324 | 196 | 214 | 213 | 309† | 220 | 232 | 411 | 362 | 400 | 256 |
| CMV-A2 | 2S2 | 180 | 396 | 71* | 233 | 203 | 59* | 40* | 4* | 324 | 28*† | 298 | 294 | 242 | 277 | 283 |
| HA2.1 | 4S1 | 581 | 611 | 340 | 422 | 606 | 230 | 232 | 236 | 560 | 311 | 483† | 671 | 567 | 605 | 720 |
| HA2.5 | 15S1 | 307 | 437 | 307 | 676 | 639 | 324 | 183 | 151 | 520 | 284 | 473 | 544† | 478 | 488 | 531 |
| HA2.6 | 23S1 | 442 | 692 | 308 | 447 | 401 | 411 | 14* | 71* | 554 | 233 | 469 | 698 | 687† | 707 | 572 |
| HA2.19 | 30S1 | 421 | 363 | 224 | 483 | 383 | 147 | 247 | 39* | 409 | 311 | 381 | 451 | 458 | 506† | 324 |
| HA2.20 | 23S1 | 437 | 564 | 221 | 401 | 321 | 195 | 54* | 69* | 446 | 201 | 368 | 595 | 528 | 450 | 490† |
| Clone . | ↓AV . | MBM15 . | 10G5 . | MBM13 . | Clone H . | CMV-A2 . | JBB4 . | Clone S . | JBBun . | CMV-A2 . | CMV-A2 . | HA2.1 . | HA2.5 . | HA2.6 . | HA-2.19 . | HA-2.20 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BV→ | — | 13S1 | 657 | 7S3 | 5S3 | 21S1 | 5S6 | 15S1 | 6S3 | 13S1 | 2S1 | 6S2 | 18S1 | 18S1 | 18S1 | 6S2 |
| MBM15 | 1S3 | 390† | 386 | 58* | 384 | 386 | 85* | 31* | 409 | 423 | 293 | 488 | 403 | 409 | 468 | 359 |
| 10G5 | 12S1 | 193 | 327† | 131 | 189 | 170 | 107 | 48* | 147 | 157 | 173 | 201 | 169 | 154 | 185 | 162 |
| MBM13 | 2S3 | 586 | 612 | 513† | 735 | 670 | 199 | 529 | 350 | 583 | 407 | 409 | 540 | 894 | 561 | 595 |
| MBM13 | 23S1 | 143 | 347 | 124† | 376 | 181 | 75* | 33* | 25* | 417 | 53* | 226 | 467 | 467 | 479 | 419 |
| Clone H | 20S1 | 589 | 272 | 256 | 616† | 394 | 266 | 296 | 247 | 613 | 520 | 973 | 540 | 706 | 690 | 610 |
| CMV-A2 | 19S1 | 174 | 189 | 83* | 30* | 368† | 55* | 4* | 65* | 241 | 153 | 159 | 281 | 275 | 473 | 201 |
| JBB4 | 1S4 | 311 | 454 | 366 | 665 | 574 | 204† | 76* | 296 | 434 | 181 | 515 | 553 | 512 | 585 | 422 |
| Clone S | 7S2 | 439 | 328 | 312 | 543 | 503 | 161 | 357† | 124 | 457 | 274 | 404 | 435 | 501 | 590 | 338 |
| JBBun | 14S1 | 535 | 647 | 445 | 776 | 783 | 484 | 367 | 583† | 497 | 622 | 503 | 548 | 565 | 632 | 572 |
| CMV-A2 | 18S1 | 394 | 426 | 278 | 208 | 324 | 196 | 214 | 213 | 309† | 220 | 232 | 411 | 362 | 400 | 256 |
| CMV-A2 | 2S2 | 180 | 396 | 71* | 233 | 203 | 59* | 40* | 4* | 324 | 28*† | 298 | 294 | 242 | 277 | 283 |
| HA2.1 | 4S1 | 581 | 611 | 340 | 422 | 606 | 230 | 232 | 236 | 560 | 311 | 483† | 671 | 567 | 605 | 720 |
| HA2.5 | 15S1 | 307 | 437 | 307 | 676 | 639 | 324 | 183 | 151 | 520 | 284 | 473 | 544† | 478 | 488 | 531 |
| HA2.6 | 23S1 | 442 | 692 | 308 | 447 | 401 | 411 | 14* | 71* | 554 | 233 | 469 | 698 | 687† | 707 | 572 |
| HA2.19 | 30S1 | 421 | 363 | 224 | 483 | 383 | 147 | 247 | 39* | 409 | 311 | 381 | 451 | 458 | 506† | 324 |
| HA2.20 | 23S1 | 437 | 564 | 221 | 401 | 321 | 195 | 54* | 69* | 446 | 201 | 368 | 595 | 528 | 450 | 490† |
Peripheral-blood–derived γδ T cells were transduced with 16 different TCR α-chains (AV) in combination with 15 different TCR β-chains (BV) derived from mHag-, CMV-, or allo-reactive T-cell clones. The transduction efficiency ranged between 28% and 64%, the percentage eGFP+ NGF-R+ γδ T cells ranged between 4% and 26%, these double positive γδ T cells were gated as indicated in Figure 3. TCRαβ expression is shown as MF1 of 3 independent FACS analyses, and results are representative of 2 other transduction experiments.
— indicates not applicable.
MFI less than 100.
The TCR α and β combinations derived from the original T-cell clones.
TCR α sequence of AV23S1-expressing T-cell clones
| Clone . | AV . | End AV . | CDR3 α . | Start AJ . | AJ . |
|---|---|---|---|---|---|
| HA2.6 | 23S1 | LCA | VRYKN | YGG | 42 |
| MBM13 | 23S1 | LCA | AP | YGG | 42 |
| HA2.20 | 23S1 | LCA | VRPGN | YGG | 42 |
| Clone . | AV . | End AV . | CDR3 α . | Start AJ . | AJ . |
|---|---|---|---|---|---|
| HA2.6 | 23S1 | LCA | VRYKN | YGG | 42 |
| MBM13 | 23S1 | LCA | AP | YGG | 42 |
| HA2.20 | 23S1 | LCA | VRPGN | YGG | 42 |
Discussion
In this study we demonstrated that efficiency of cell-surface expression of transferred TCRs is determined by both the introduced and the endogenous TCR. We demonstrated that the TCR cell-surface makeup of T cells expressing multiple TCR chains is controlled by the pairing properties of the different TCRs that can be formed. Because the TCR is expressed only at the cell surface when noncovalently bound to the CD3 complex composed of CD3γ, CDϵ, CD3δ, and CD3ζ, correct assembly of all these subunits with TCR α- and β-chains is required to assure optimal membrane expression of the TCR-CD3 complex in T cells.32-34 Single subunits and partial receptor complexes redundant for the assembly process retain in the ER where these products are highly susceptible to proteolysis.30,31
Our data demonstrated consistent differences in cell-surface expression between the various TCR complexes, although mRNA and protein expression were comparable. We speculate that the mechanism through which advantages may be conferred on some versus other chain pairs most likely is the interchain affinity of the TCR chains. Alternatively, folding efficiency and ability to capture CD3 may also explain these differences. On the basis of our results we hypothesize that TCR chains that exhibit high interchain affinity will preferentially assemble in the TCR-CD3 complexes and therefore will be expressed more favorably compared with TCRs that have a low or intermediate interchain affinity.
Transfer of TCR αβ complexes into both αβ as well as γδ T cells demonstrated that cell-surface expression of TCRs is independent of whether the TCR is retrovirally introduced or naturally expressed. TCRs that were favorably expressed on TCR transfer into peripheral-blood αβ and γδ T cells were hardly reduced after TCR transfer when naturally expressed. In contrast, CMV-AV2BV2-TCR loses the competition for cell-surface expression when this TCR was retrovirally introduced into peripheral blood–derived αβ and γδ T cells or when naturally expressed. Individual TCR chains of this complex however were able to pair with each other as shown by the prominent appearance of CMV-AV2BV2 T cells in the memory response of a CMV-exposed person. In addition, wild-type CMV-AV2BV2 T cells exhibited a similar high affinity for their cognate peptide-HLA ligand compared with other CMV-specific T cells present in the memory response (Figure 1). Therefore, in absence of other TCR chains most TCR complexes will be able to efficiently bind to CD3 and will be efficiently exposed on T cells. However, in the presence of more than one TCR the interactions between individual chains and the CD3 complex will eventually determine the TCR makeup. In TCR-transferred γδ T cells competition for association to CD3 will be restricted to the transferred TCR αβ complex and the endogenous TCRγδ complex. In TCR-transferred αβ T cells not only the introduced and endogenous TCR will shape TCR makeup, but also chimeric TCRs will play an important role (Figure 3).
Previously, it has been speculated that chimeric TCRs can be formed in TCR-transferred T cells. However, no data substantiating this assumption were provided thus far. Our data clearly demonstrate that chimeric TCRs can be formed, and that the level of chimeric TCRs is determined by the composition of the different TCR chains present in cells. Although no adverse allo- or self-reactive T-cell responses of TCR-transduced T cells have been observed thus far,35 we hypothesize that formation of unintended allo- or self-reactive TCR-transduced T cells cannot be excluded. Transfer of TCRs into oligoclonal T-cell populations (eg, virus-specific T cells) may minimize the risk.13
T cells naturally expressing 2 different TCRs have been discovered at a frequency of up to 30% of the normal repertoire in humans.36,37 These dual-receptor T cells are present because allelic exclusion of the TCR α-chain during thymic development is far from complete. Similar to TCR-transferred T cells, the TCR makeup of these T cells is also likely to be controlled by the intrinsic pairing properties of the different TCRs. Therefore, avidity of these dual-receptor T cells for their antigenic ligands will be determined not only by the TCR affinity but also by the number of specific TCRs at the cell surface. Thus, low-avidity T cells present in the T-cell repertoire of humans may be the result of either low-affinity TCRs or due to low cell-surface expression of that particular TCR because of competition with other TCRs expressed in the same cell.
The data presented here indicate that for purposes of TCR gene therapy it is not only important to select for high-affinity TCRs9,38 but also to search for TCRs that exhibit a high TCR-CD3 intrinsic affinity to generate T cells that preferentially express the transferred-TCR. Selection of TCRs for TCR transfer that fulfills these 2 criteria will most likely lead to the engineering of high-avidity T cells. In addition, we assume that for creation of high-avidity TCR-transferred T cells those host T cells are required that express an endogenous TCR exhibiting low competition potential compared with the introduced TCR. Transfer of HA-2–TCRs into CMV-AV2BV2 T cells resulted in optimal TCR makeup (Figure 2). Cells exhibited efficient antileukemic reactivity and preserved their endogenous TCR reactivity, important for persistence of these T cells in vivo. Selection of candidate TCRs for purposes of TCR gene transfer can be performed as has been described in this study. Several different TCRs specific for the antigen of interest have to be characterized from high-affinity T cells and transferred to different T-cell populations. Selection of useful TCRs for purposes of TCR transfer can be accomplished on bases of optimal TCR cell-surface makeup of the transferred T cells. Alternatively, selection of TCRs for purposes of gene transfer can be performed in a less time-consuming manner. T-cell lines with the interesting antigenic specificity could be transferred with strong competitor TCRs. T cells that do not dramatically lose their endogenous TCR cell-surface expression as well as their functionality on TCR transfer express TCRs with high competition potential and are therefore potentially useful for purposes of TCR transfer.
Our results demonstrated that the efficiency of cell-surface expression of the different transferred TCRs in the presence of an endogenous TCR varied dramatically. We have demonstrated that 10% of all original and chimeric TCR complexes tested were inefficiently expressed at the cell surface of TCR-transferred γδ T cells. Because the TCR α- and β-chains are extreme polymorphic proteins, with the exception of the C regions of both the α- and β-chain, it is difficult to reveal what determines the binding affinity between TCR α and β combinations. Our results suggest an important role for the V region of both the TCR α- as well as the β-chain. In addition, subfamily differences as well as the composition of the CDR3 region influence the binding affinity. We demonstrated striking differences in TCR cell-surface expression when 3 AV23S1 chains derived from different T-cell clones were transferred in combination with various β-chains, whereas the 3 AV23S1 chains were completely identical with the exception of 5 amino acids in the N region.
In conclusion, our results demonstrated that the TCR makeup of T cells expressing more than 1 TCR is determined by the intrinsic quality of the different TCR-CD3 complexes that can be formed. Those TCR-CD3 complexes with highest interchain-pairing properties will be preferentially expressed. Therefore, selection of high-affinity TCRs with superior intrinsic quality is crucial for successful clinical application of TCR transfer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: M.H.M.H. designed and performed the research, analyzed the data, and wrote the paper; R.S.H., M.A.W.G.v.d.H., and M.H. performed the research and analyzed the data; L.T.v.d.V. contributed vital analytic tools; M.G.D.K. contributed vital new reagents; R.W. wrote the paper; J.H.F.F. designed the research and wrote the paper.
Acknowledgments
We thank Reinier van der Linden for technical assistance and Frits Koning for critical reading of the manuscript.
This work was supported by the Dutch Cancer Society (grant 2001-2490).



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal