Abstract
Compared with fluorescence in situ hybridization (FISH), conventional metaphase cytogenetics play only a minor prognostic role in chronic lymphocytic leukemia (CLL) so far, due to technical problems resulting from limited proliferation of CLL cells in vitro. Here, we present a simple method for in vitro stimulation of CLL cells that overcomes this limitation. In our unselected patient population, 125 of 132 cases could be successfully stimulated for metaphase generation by culture with the immunostimulatory CpG-oligonucleotide DSP30 plus interleukin 2. Of 125 cases, 101 showed chromosomal aberrations. The aberration rate is comparable to the rate detected by parallel interphase FISH. In 47 patients, conventional cytogenetics detected additional aberrations not detected by FISH analysis. A complex aberrant karyotype, defined as one having at least 3 aberrations, was detected in 30 of 125 patients, compared with only one such case as defined by FISH. Conventional cytogenetics frequently detected balanced and unbalanced translocations. A significant correlation of the poor-prognosis unmutated IgVH status with unbalanced translocations and of the likewise poor-prognosis CD38 expression to balanced translocations and complex aberrant karyotype was found. We demonstrate that FISH analysis underestimates the complexity of chromosomal aberrations in CLL. Therefore, conventional cytogenetics may define subgroups of patients with high risk of progression.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world. The disease has a highly heterogeneous clinical course, with time to progression varying from months to many years. The classical clinical staging systems for CLL that were introduced 3 decades ago by Rai1 and Binet2 have been extremely useful in guiding disease management and treatment decisions. However, these staging systems have failed to predict the clinical course for individual patients at early-stage disease and to identify patients with poor prognosis.
More recently, the analysis of chromosomal aberrations has provided significant prognostic information.3,4 The use of metaphase cytogenetics, however, has turned out to be problematic, due to the low mitotic index of most CLL cells even in the presence of B-cell mitogens.5 But even when metaphases could be generated, the quality was often poor and aberrations escaped detection. Thus, clonal aberrations were detected in only 40% to 50% of cases.4 Regardless of these problems, metaphase cytogenetics and, later, comparative genomic hybridization6 could define the most common cytogenetic aberrations in CLL. Based on these results, the analysis of aberrant chromosomal regions with specific DNA probes by fluorescence in situ hybridization (FISH), which can be applied to interphase cells, resulted in the detection of clonal aberrations in more than 80% of CLL patients.7 Patients with 17p (7%) and 11q (17%) deletions had the shortest median survival, while deletions of 13q as the sole abnormality (36%) predicted a favorable outcome.
Recently, CD38 and the immunoglobulin variable heavy chain (IgVH) mutational status were identified as independent prognostic markers.8,9 Depending on different clinical studies, a cut-off for CD38 expression of 7%10 or 30%8 is made, with expression levels above this cut-off predicting poor survival. Somatic hypermutations in the IgVH region of more than 2% (mutated status), compared with the closest matching germ line variant, correlated with good prognosis in 2 pivotal studies.8,9 In multivariate analysis, a nonmutated IgVH status and deletions on chromosome 17p and 11q but not CD38 expression retained prognostic significance.10
It is presently not clear whether other aberrations that are not detected by the standard FISH panel have any impact on prognosis and disease progression. Therefore, an approach to generate high-quality metaphases in CLL still would be highly desirable.
The B-cell mitogen CD40-ligand (CD40L) was previously compared to conventional mitogens for metaphase induction in CLL, and these results were compared with those from standard FISH analysis.11,12 CD40L stimulation induced metaphases in 93% of cases, versus 78% with conventional methods.11 Even more important, CD40L stimulation resulted in the detection of aberrations in 89% of cases versus only 22% by conventional methods11 and confirmed all aberrations detected by FISH.11 In addition, so-called complex aberrations (that is, 3 or more aberrations), were detected in 41% of cases. Due to the need for a labor-intensive, cellular coculture system, CD40L-enhanced cytogenetics is hardly applicable for routine diagnostics.
We here suggest the use of the immunostimulatory CpG-oligonucleotide DSP30 in combination with IL-2 that has been reported to effectively induce cell cycle progression of CLL cells in vitro.13-15 Therefore, the objective of this study was to test the suitability of DSP30 as a B-cell mitogen for its use in metaphase cytogenetics by comparing these results with FISH analysis in a large set of CLL patients and to correlate both types of analysis with other prognostically important markers like the VH mutation status and CD38 expression.
Patients and methods
Patients
After informed written consent was obtained, consecutive peripheral blood or bone marrow samples, which were referred to our laboratory, were analyzed for cytogenetic aberrations. Our patient population consisted of 44 women (33%) and 88 men (67%), with a median age of 62 years (range, 34-79 years). At the time of analysis, CLL was diagnosed in 81 cases, and the disease had already been diagnosed in the residual 50 cases, when the samples were referred for follow-up investigations. No data about the time of diagnosis were available in one case. CLL was diagnosed based upon morphological criteria according to the World Health Organization classification and according to flow cytometric criteria where tumor cells expressed CD5, CD19, and CD23, but showed a weak Ig and CD22 expression.16 Approval for these studies was obtained from the Bavarian Medical Association.
Metaphase cytogenetics
For metaphase induction, 107 peripheral blood mononuclear cells were cultured in RPMI 1640 medium (Gibco, Gaithersburg, MD) with 20% fetal calf serum in the presence of the immunostimulatory CpG-oligonucleotide DSP30 (2 μM) (TIBMolBiol, Berlin, Germany) and interleukin 2 (IL-2) (200 U/mL) as described.15 After 48 hours, colcemid (Sigma, Munich, Germany), at a concentration of 0.15 μg/mL, was added for another 24 hours before chromosome preparation. In the initial phase of this study, parallel 12-O-tetradecanoyl-phorbol-13-acetate (TPA) cultures were set up in addition to the DSP30/IL-2 cultures (Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). In 3 cases, peripheral blood and bone marrow were available from the same patients, and culture with DSP30/IL-2 resulted in the detection of the same aberrations on metaphases from the different sources (data not shown). Chromosome preparation and staining was done according to standard protocols, as described elsewhere.11,17 The quality of the metaphases varied. In most cases, a resolution of 200 to 300 bands per haploid karyotype was reached. In case of translocations or complex aberrations, results of G-banding analysis were confirmed by 24-color FISH.18 In cases of unbalanced translocations resulting from gain as well as loss of chromosomal material, these unbalanced translocations were assessed as 2 aberrations. Chromosomes were classified according to the International System for Human Cytogenetic Nomenclature (ISCN).34
Fluorescence in situ hybridization
Interphase FISH was performed in all cases on peripheral blood or bone marrow smears, as described previously.11,17 The FISH panel included probes for the detection of trisomy 12, IGH rearrangements, and deletions of 6q21, 11q22.3 (ATM), 13q14 (D13S25 and D13S319), and 17p13 (TP53). Interphase FISH for the detection of t(11;14)(q13;q32) was routinely done on all samples. Samples with t(14;18)(q32;q21) in metaphase cytogenetics were confirmed by FISH analysis. All cases carrying these translocations could be confirmed with FISH probes on metaphases (n = 4). The hierarchical model to define genetic subgroups in FISH analysis is described elsewhere.7 The major categories are patients with del(17p), patients with del(11q) but not del(17p), patients with trisomy 12 but not del(11q) and not del(17p), del(13q) as sole abnormality, and normal karyotype.
Analysis of the IgVH status
We lysed 1 × 107 mononuclear cells from peripheral blood or bone marrow in 300 μL RLT buffer (Qiagen, Hilden, Germany) and stored them at –80°C until further processing. mRNA was extracted with MagnaPureLC mRNA Kit I (Roche Diagnostics, Mannheim, Germany). The cDNA synthesis from mRNA from an equivalent of 5 × 106 cells was performed using 300 U Superscript II (Gibco BRL/Invitrogen, Karlsruhe, Germany) and random hexamer primers (Pharmacia, Freiburg, Germany). VH genes were polymerase chain reaction (PCR)–amplified from cDNA using 6 different consensus framework region 1 (FR1) VH forward primers and 1 consensus joining region JH reverse primer as designed for the BIOMED-2 study.19 Clonally rearranged immunoglobulins were identified in 2 multiplex PCR reactions. Each reaction consisted of 3 differently fluorescence-labeled FR1-VH primers in combination with the unconjugated JH primer.20 PCR products were analyzed by GeneScan on a 3100-Avant Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany). The specific FR1-VH/JH primer combination was used to generate an independent PCR product in a second PCR reaction. This PCR product was agarose gel purified using the QIAquick Gel Extraction Kit (Qiagen). Sequencing reactions were carried out in both orientations using BigDye chemistry (Applied Biosystems) and analyzed on a 3100-Avant Genetic Analyzer. Sequences were aligned to immunoglobulin sequences in the V-BASE (http://vbase.mrc-cpe.cam.ac.uk/) and IMGT (http://imgt.cines.fr/) databases. The IgVH status was assigned as mutated when more than 2% somatic mutations, compared with the closest matching germ line sequence, could be identified.8,9
Immunophenotyping for CD38 expression
Samples were analyzed after Ficol gradient centrifugation. Cells were stained using a 4-color approach and analyzed on an FC500 cytometer (Beckman Coulter, Miami, FL). CD38 was analyzed using PC5-conjugated antibody clone A07780 (Immunotech, Marseille, France). The positivity for CD38 was determined in CD19-positive cells using isotype controls as cut-off. A sample was considered CD38 positive when a cut-off of 30% or more CD38-positive cells was reached.8
Statistics
The correlation of FISH or metaphase cytogenetic aberrations to the IgVH status or CD38 expression was assessed by chi-square test. For statistical analysis SPSS (version 13.0) software (SPSS, Chicago, IL) was used.
Results
Comparison of FISH and metaphase cytogenetics
Of the 132 samples that were included in this study, all cases could be successfully analyzed by interphase FISH, with a rate of 79% aberrations (Table 1).
Characterization of single FISH aberrations and their incidence in combination with other aberrations (n = 132 patient samples)
FISH . | All samples . | No additional abberations . | del (6q) . | del (11q) . | +12 . | del (13q) . | del(13q) biallelic . | del (17p) . | t(11;14) . | t(14;18) . | t(14q32)* . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| del(6q) | 9 | 1 | N/A | 0 | 0 | 7 | 1 | 2 | 0 | 0 | 0 |
| del(11q) | 21 | 7 | 0 | N/A | 1 | 13 | 1 | 0 | 0 | 0 | 0 |
| +12 | 17 | 10 | 0 | 1 | N/A | 2 | 0 | 0 | 1 | 2 | 1 |
| del(13q)† | 76 | 51 | 7 | 13 | 2 | N/A | 9 | 4 | 0 | 0 | 0 |
| del(13q) biallelic | 9 | 6 | 1 | 1 | 0 | 9 | N/A | 1 | 0 | 0 | 0 |
| del(17p) | 7 | 2 | 2 | 0 | 0 | 4 | 1 | N/A | 0 | 0 | 0 |
| t(11;14) | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | N/A | 0 | 0 |
| t(14;18) | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | N/A | 0 |
| t(14q32) | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
FISH . | All samples . | No additional abberations . | del (6q) . | del (11q) . | +12 . | del (13q) . | del(13q) biallelic . | del (17p) . | t(11;14) . | t(14;18) . | t(14q32)* . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| del(6q) | 9 | 1 | N/A | 0 | 0 | 7 | 1 | 2 | 0 | 0 | 0 |
| del(11q) | 21 | 7 | 0 | N/A | 1 | 13 | 1 | 0 | 0 | 0 | 0 |
| +12 | 17 | 10 | 0 | 1 | N/A | 2 | 0 | 0 | 1 | 2 | 1 |
| del(13q)† | 76 | 51 | 7 | 13 | 2 | N/A | 9 | 4 | 0 | 0 | 0 |
| del(13q) biallelic | 9 | 6 | 1 | 1 | 0 | 9 | N/A | 1 | 0 | 0 | 0 |
| del(17p) | 7 | 2 | 2 | 0 | 0 | 4 | 1 | N/A | 0 | 0 | 0 |
| t(11;14) | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | N/A | 0 | 0 |
| t(14;18) | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | N/A | 0 |
| t(14q32) | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
The number of samples per row can be greater than n, if several additional aberrations are present.
NA indicates not applicable.
Translocations involving 14q32 but not t(11;14) and not t(14;18).
Including del(13q) biallelic.
In comparison, 125 (95%) of the 132 cases could be successfully analyzed by cytogenetic banding techniques after culture with the immunostimulatory CpG-oligonucleotide DSP30 and IL-2. In these cases, at least 15 metaphases for each culture could be evaluated for chromosomal aberrations. According to international standards, structural aberrations and gains of chromosomes were included in the karyotype as clonal aberrations if present in at least 2 different metaphases; losses of chromosomes had to be observed in at least 3 different metaphases. Of the 125 samples, 101 (81%) showed chromosomal aberrations that involved all different chromosomes. Twenty-four samples displayed a normal karyotype, indicating that the culture conditions did not induce aberrations. In addition, no aberrant karyotype and no FISH aberration were induced in metaphases upon DSP30/IL-2 culture in healthy individuals (n = 5) (data not shown). For 6 patients, the DSP30/IL-2 culture was repeated at later time points when follow-up samples were available. The same aberrations were detected at both time points for each patient with clonal evolution in 2 cases (data not shown).
In detail, 10 single aberrations detected by FISH in 9 different samples could not be detected by cytogenetic banding techniques. The aberrations were del(6q) (n = 2), del(11q) (n = 1), and del(13q) (n = 7). However, in all but one of the samples the aberrant clone was detected by FISH in less than 30% of the interphase cell population.
On the other hand, a substantial number of aberrations, in addition to those detected by the FISH probes, were revealed by chromosome banding. Overall, 47 samples (38%) showed aberrations by chromosome banding analysis in addition to those covered by the FISH panel of probes (Table 2). Interestingly, 10 of 27 cases with no abnormalities detected by FISH analysis displayed aberrations in chromosome banding analysis. As these were found in areas not covered by the FISH panel, the true number of CLL patients with aberrations exceeds the 79% FISH-positive cases.
Characterization of samples with cytogenetic aberrations in addition to the FISH aberrations as detected on interphase cells
. | . | . | . | . | . | . | No. aberrations . | . | Translocation . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | VH status* . | Percent sequence homology . | VH-gene . | CD38, % positive† . | FISH‡ . | Karyotype§ . | At least 3 . | At least 3 unbalanced . | Unbalanced . | Balanced . | ||
| 1 | M | 91.8 | VH3-15 | 0 | del(13q), del(17p) | 44,XY,-9,der(9)t(9;9)(q34;?),del(13)(q12q21), der(15;17)(q10;q10), t(18;22)(q23;q11), [4], 43,idem,dic(1;3)(p36;q29) [2], 46,XY [15] | + | + | + | + | ||
| 2 | M | 92.6 | VH3-74 | 3 | +12, t(11;14) | 46,XY,t(11;14)(q13;q32) [1], 47,XY,t(11;14)(q13;q32), +12 [5], 46,XY,t(11;14)(q13;q32), +der(12)t(12;17)(p13;q21) [4] | + | + | + | + | ||
| 3 | M | 91.5 | VH4-59 | 64 | del(13q), +12 | 49,XY,+12,+18,+19 [8], 49,XY,+12,del(13)(q14q21),+18,+19 [2], 46,XY [3] | + | + | - | - | ||
| 4 | M | 87.6 | VH3-07 | 91 | +12 | 47,XY,+12,+18,+19 [8], 46,XY [6] | + | + | - | - | ||
| 5 | M | 97.3 | VH2-26 | 97 | del(6q), del(13q) | 46,XX,del(13)(q12q14) [7], 46,X,t(X;12)(p11.4;q24),del(13)(q12q14) [8]. 46,X,t(X;12)(p11.4;q24),del(6)(q21),del(13)(q12q14) [3], 46XX,del(11)(q11q22),del(13)(q12q14) [3] | + | + | - | + | ||
| 6 | M | 95.4 | VH3-15 | 97 | +12 | 48,XY,+12,+18 [9], 49,XY,+12,+18,+19 [8], 46,XY [3] | + | + | - | - | ||
| 7 | M | 91.0 | VH3-66 | ND | del(13q) | 46,XY,del(13)(q13q22) [3], 46,XY,del(7)(q11.2),der(8)t(8;10)(q24;q?)t(10;13) (q?;q?),der(10)t(10;13)(q11;q?)t(7;13)(q11.2;q?), der(13)t(13;16)(q11;p11), der(16)t(10;16)(?;p11)ins(10;13)(?;q?)t(10;13)(?;q?) [6], 46,XY [12] | + | + | + | + | ||
| 8 | M | 95.7 | VH3-23 | ND | del(17p) | 43,XX,t(1;7)(q21;q11.2),der(5)t(5;13)(p15;q?),-8, der(15;22)(q10;q10)t(8;22)(q11;q13),i(17)(q10),-21 [6], 46,XX [8] | + | + | + | + | ||
| 9 | ND | ND | ND | 0 | del(11q) | 46,XY,del(11)(q21q25) [8], 46,XY,del(11)(q21q25),der(18)t(2;18)(p11;p11) [12] | + | + | + | - | ||
| 10 | ND | ND | ND | 10 | del(11q), del(13q) | 46,XY,del(13)(q12q14) [4], 46,XY,del(11)(q21),del(13)(q12q14) [2], 46,XY,del(13)(q12),der(15)t(13;15)(q22;p11) [4], 46,XY [11] | + | + | + | - | ||
| 11 | ND | ND | ND | 87 | +12, del(13q) | 49,XY,t(1;10)(p22;q24),+12,del(13)(q12q14),+18,+19 [18], 46,XY [2] | + | + | - | + | ||
| 12 | ND | ND | ND | 91 | del(11q), del(13q) | 45,XY,inv(8)(p21q22),del(11)(q13q23),der(12)dup(12) (q24.3q11)t(12;22) (q11;q11),del(13)(q14q21),-22 [15], 46,XY [5] | + | + | + | + | ||
| 13 | UM | 100.0 | VH4-34 | 0 | Normal | 46,XY,inv(7)(p15q32),t(8;14)(p21;q32) [11], 46,XY,inv(7)(p15q32),t(8;14)(p21;q32),dup(9)(p13p24), der(19)t(11;19)(p11.2;p13) [14] | + | + | + | + | ||
| 14 | UM | 100.0 | VH1-69 | 0 | del(13q), del(17p) | 45,XY,der(9;17)(p10;q10),del(13)(q14q21) [8], 46,XY [6] | + | + | + | - | ||
| 15 | UM | 100.0 | VH3-11 | 0 | del(11q) | 46,XX,der(7)t(7;11)(p15;q23),der(11)t(7;11)(p?;q22) [20] | + | + | + | - | ||
| 16 | UM | 99.1 | VH3-21 | 2 | t(9;14) | 47,XY,+1,+3,dic(1;4)(p11;p16),t(9;14)(p13;q32) [7], 46,idem,ins(2;19)(q13;p11p13.1) [3], 46,XY [6] 46,idem,ins(2;19)(q13;p11p13.1) [3], 46,XY [6] | + | + | + | + | ||
| 17 | UM | 100.0 | VH3-13 | 14 | del(11q) | 46,XY,del(11)(q21) [8], 46,XY,der(6;18)dic(6;18)(p21;p11.3)del(6)(q15q21), der(14)t(7;14)(p11;p11), [4], 46,XY [10] | + | + | + | - | ||
| 18 | UM | 100.0 | VH1-69 | 26 | del(13q), del(17p) | 45,XY,del(13)(q14q21),der(17;18)(q10;q10) [4], 44,XY,-5,del(13)(q14q21),der(17;22)(q10;q10), der(18)t(5;18) (q11;p11.3) [14] | + | + | + | - | ||
| 19 | UM | 99.6 | DP-58 | 28 | del(13q) | 46,XX,der(1)t(1;13)(q44;q22),del(11)(q23q25), del(13) (q14) [7], 46,XX [16] | + | + | + | - | ||
| 20 | UM | 98.7 | VH3-21 | 34 | del(11q), del(13q) | 46,XX,del(11)(q13),del(13)(q12q14) [6], 46,XX,del(13)(q12q14),der(18)t(2;18)(p11;p11) [3], 46,XX [11] | + | + | + | - | ||
| 21 | UM | 100.0 | VH1-18 | 61 | del(6q), del(17p) | 46,XX,del(6)(q21),der(17)t(2;17)(p13;p11) [17] 46,XX [1] | + | + | + | - | ||
| 22 | UM | 100.0 | VH1-69 | 68 | del(11q), del(13q) | 46,XX,der(1)t(1;11)(q32;q13)del(1)(p34),der(6)t(6;13) (p22;q32),t(7;12)(p13;p13), del(11)(q22),der(11)t(7;11)(p15;q13)t(7;13)(p22;q?), der(13)t(6;13)(p22;q14) [cp5], 46,XX [8] | + | + | + | + | ||
| 23 | UM | 99.1 | VH3-49 | ND | Normal | 46-51,XY,+Y,der(2)t(2;21)(q21;q11), +3,+del(5)(q?q?),der(8)t(8;12)(q24;?),+11, -13, der(14)t(13;14)(q?;q?)inv(14)(p?q?),+del(14)(q22q32), +15, der(18)t(2;18)(?;q21),+19,-20 [cp14], 46,XY [1] | + | + | + | + | ||
| 24 | UM | 100.0 | VH1-e | ND | Normal | 46,XY,der(3)ins(3;11)(q21;q13q25),der(6)t(6;11)(q13;p11), der(11)del(11)(p11)del(11)(q13) [15], 46,XY [2] | + | + | + | - | ||
| 25 | M | 61.0 | VH1-3 | 0 | Normal | 46,XY,t(18;22)(q21;q11) [10], 46,XY [14] | - | - | - | + | ||
| 26 | M | 89.8 | VH3-23 | 0 | del(13q) | 46,XY,ins(13;2)(q14;q?q?) [8], 46,XY [12] | - | - | - | - | ||
| 27 | M | 93.8 | VH5-51 | 0 | del(13q) | 45,XX,-13 [4], 46,XX [16] | - | - | - | - | ||
| 28 | M | 91.6 | VH1-02 | 1 | del(13q) | 46,XX,del(13)(q12q14) [8]46,XX,inv(1)(p36q25),del(13)(q12q14) [3] 46,XX [4] | - | - | - | + | ||
| 29 | M | 96.0 | VH3-21 | 19 | Normal | 46,XX,t(4;11)(q23;q13) [2], 46,XX [3] | - | - | - | + | ||
| 30 | ND | ND | ND | 26 | +12, t(14;19) | 47,XX,+12,t(14;19)(q32;q13) [9] 47,XX,t(1;6)(p35;p23),+12,t(14;19)(q32;q13) [12] | + | - | - | + | ||
| 31 | ND | ND | ND | ND | del(13q) | 46,XY,del(13)(q12q14),der(13)t(13;17)(q14;q21), der(17)t(13;17)(q31;q21) [7], 46,XY [11] | - | - | + | - | ||
| 32 | UM | 100.0 | VH1-02 | 0 | Normal | 46,XY,del(11)(q23) [8], 46,XY [12] | - | - | - | - | ||
| 33 | UM | 100.0 | VH5-a | 2 | del(11q), del(13q) | 46,XY,del(11)(q21),del(13)(q12q14) [3] 46,XY,der(8)t(2;8)(p13;p23),del(11)(q21) [19] | + | + | + | - | ||
| 34 | UM | 100.0 | VH1-69 | 2 | del(13q) | 46,XX,t(10;13)(q24;q14) [4], 46,XX [9] | - | - | - | + | ||
| 35 | UM | 99.6 | VH3-33 | 7 | +12, t(14;18) | 47,XX,-4,der(7)t(4;7)(q21;q22),+der(7)t(4;7)(q21;q22), +12,t(14;18)(q32;q21) [2], 46,XX [11] | - | - | - | - | ||
| 36 | UM | 100.0 | VH-1-e | 16 | del(17p) | 45,XY,der(2)t(2;17)(q37;q21),-17 [6], 46,XY [18] | - | - | + | - | ||
| 37 | UM | 100.0 | VH1-69 | 19 | del(13q) | 46,XY,del(13)(q12q14) [8], 46,XY,del(11)(q22),del(13)(q12q14) [2], 46,XY [10] | - | - | - | - | ||
| 38 | UM | 100.0 | VH3-09 | 41 | del(13q) | 46,XX,t(10;13)(q24;q14) [14], 46,XY [6] | - | - | - | + | ||
| 39 | UM | 100.0 | VH3-23 | 44 | del(13q) | 46,XX,t(2;13)(q37;q14) [10], 46,XX [13] | - | - | - | + | ||
| 40 | UM | 99.6 | VH1-69 | 53 | Normal | 46,XY,der(20)t(2;20)(p11;p11) [13], 46,XY [7] | - | - | + | - | ||
| 41 | UM | 99.5 | VH1-69 | 68 | del(11q), del(13q) | 46,XY,del(11)(q21q24),t(13;15)(q14;q25) [11], 46,XY [4] | - | - | - | + | ||
| 42 | UM | 99.1 | VH4-39 | 73 | +12 | 47,XX,+12 [4], 47,XX,del(9)(p21),+12,t(16;17)(q24;q23) [4], 46,XX [13] | + | - | - | + | ||
| 43 | UM | 100.0 | VH3-33 | 78 | del(11q), del(13q) | 46,XY,t(1;15;13)(q21;q15;q14),t(9;20)(q34;q11) [8], 46,XY,t(1;11)(q23;q22.3),t(4;20)(q31;p11) [12], 46,XY [5] | + | - | - | + | ||
| 44 | UM | 99.6 | VH1-58 | 83 | Normal | 46,XY,del(14)(q24q32) [14], 46,XY [6] | - | - | - | - | ||
| 45 | UM | 100.0 | VH4-61 | 93 | del(11q), +12 | 46,XY,t(7;16)(p11;q24) [2], 47,XY,t(7;16)(p11;q24), +12 [12], 47,XY,t(7;16)(p11;q24),del(11)(q13q24),+12 [3] | + | - | - | + | ||
| 46 | UM | 99.0 | VH4-34 | ND | Normal | 46,XY,der(18)t(2;18)(p11;p11) [5], 46,XY [74] | - | - | + | - | ||
| 47 | UM | 100.0 | VH1-69 | ND | Normal | 46,XY,del(14)(q32) [13], 46,XY [7] | - | - | - | - | ||
. | . | . | . | . | . | . | No. aberrations . | . | Translocation . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. . | VH status* . | Percent sequence homology . | VH-gene . | CD38, % positive† . | FISH‡ . | Karyotype§ . | At least 3 . | At least 3 unbalanced . | Unbalanced . | Balanced . | ||
| 1 | M | 91.8 | VH3-15 | 0 | del(13q), del(17p) | 44,XY,-9,der(9)t(9;9)(q34;?),del(13)(q12q21), der(15;17)(q10;q10), t(18;22)(q23;q11), [4], 43,idem,dic(1;3)(p36;q29) [2], 46,XY [15] | + | + | + | + | ||
| 2 | M | 92.6 | VH3-74 | 3 | +12, t(11;14) | 46,XY,t(11;14)(q13;q32) [1], 47,XY,t(11;14)(q13;q32), +12 [5], 46,XY,t(11;14)(q13;q32), +der(12)t(12;17)(p13;q21) [4] | + | + | + | + | ||
| 3 | M | 91.5 | VH4-59 | 64 | del(13q), +12 | 49,XY,+12,+18,+19 [8], 49,XY,+12,del(13)(q14q21),+18,+19 [2], 46,XY [3] | + | + | - | - | ||
| 4 | M | 87.6 | VH3-07 | 91 | +12 | 47,XY,+12,+18,+19 [8], 46,XY [6] | + | + | - | - | ||
| 5 | M | 97.3 | VH2-26 | 97 | del(6q), del(13q) | 46,XX,del(13)(q12q14) [7], 46,X,t(X;12)(p11.4;q24),del(13)(q12q14) [8]. 46,X,t(X;12)(p11.4;q24),del(6)(q21),del(13)(q12q14) [3], 46XX,del(11)(q11q22),del(13)(q12q14) [3] | + | + | - | + | ||
| 6 | M | 95.4 | VH3-15 | 97 | +12 | 48,XY,+12,+18 [9], 49,XY,+12,+18,+19 [8], 46,XY [3] | + | + | - | - | ||
| 7 | M | 91.0 | VH3-66 | ND | del(13q) | 46,XY,del(13)(q13q22) [3], 46,XY,del(7)(q11.2),der(8)t(8;10)(q24;q?)t(10;13) (q?;q?),der(10)t(10;13)(q11;q?)t(7;13)(q11.2;q?), der(13)t(13;16)(q11;p11), der(16)t(10;16)(?;p11)ins(10;13)(?;q?)t(10;13)(?;q?) [6], 46,XY [12] | + | + | + | + | ||
| 8 | M | 95.7 | VH3-23 | ND | del(17p) | 43,XX,t(1;7)(q21;q11.2),der(5)t(5;13)(p15;q?),-8, der(15;22)(q10;q10)t(8;22)(q11;q13),i(17)(q10),-21 [6], 46,XX [8] | + | + | + | + | ||
| 9 | ND | ND | ND | 0 | del(11q) | 46,XY,del(11)(q21q25) [8], 46,XY,del(11)(q21q25),der(18)t(2;18)(p11;p11) [12] | + | + | + | - | ||
| 10 | ND | ND | ND | 10 | del(11q), del(13q) | 46,XY,del(13)(q12q14) [4], 46,XY,del(11)(q21),del(13)(q12q14) [2], 46,XY,del(13)(q12),der(15)t(13;15)(q22;p11) [4], 46,XY [11] | + | + | + | - | ||
| 11 | ND | ND | ND | 87 | +12, del(13q) | 49,XY,t(1;10)(p22;q24),+12,del(13)(q12q14),+18,+19 [18], 46,XY [2] | + | + | - | + | ||
| 12 | ND | ND | ND | 91 | del(11q), del(13q) | 45,XY,inv(8)(p21q22),del(11)(q13q23),der(12)dup(12) (q24.3q11)t(12;22) (q11;q11),del(13)(q14q21),-22 [15], 46,XY [5] | + | + | + | + | ||
| 13 | UM | 100.0 | VH4-34 | 0 | Normal | 46,XY,inv(7)(p15q32),t(8;14)(p21;q32) [11], 46,XY,inv(7)(p15q32),t(8;14)(p21;q32),dup(9)(p13p24), der(19)t(11;19)(p11.2;p13) [14] | + | + | + | + | ||
| 14 | UM | 100.0 | VH1-69 | 0 | del(13q), del(17p) | 45,XY,der(9;17)(p10;q10),del(13)(q14q21) [8], 46,XY [6] | + | + | + | - | ||
| 15 | UM | 100.0 | VH3-11 | 0 | del(11q) | 46,XX,der(7)t(7;11)(p15;q23),der(11)t(7;11)(p?;q22) [20] | + | + | + | - | ||
| 16 | UM | 99.1 | VH3-21 | 2 | t(9;14) | 47,XY,+1,+3,dic(1;4)(p11;p16),t(9;14)(p13;q32) [7], 46,idem,ins(2;19)(q13;p11p13.1) [3], 46,XY [6] 46,idem,ins(2;19)(q13;p11p13.1) [3], 46,XY [6] | + | + | + | + | ||
| 17 | UM | 100.0 | VH3-13 | 14 | del(11q) | 46,XY,del(11)(q21) [8], 46,XY,der(6;18)dic(6;18)(p21;p11.3)del(6)(q15q21), der(14)t(7;14)(p11;p11), [4], 46,XY [10] | + | + | + | - | ||
| 18 | UM | 100.0 | VH1-69 | 26 | del(13q), del(17p) | 45,XY,del(13)(q14q21),der(17;18)(q10;q10) [4], 44,XY,-5,del(13)(q14q21),der(17;22)(q10;q10), der(18)t(5;18) (q11;p11.3) [14] | + | + | + | - | ||
| 19 | UM | 99.6 | DP-58 | 28 | del(13q) | 46,XX,der(1)t(1;13)(q44;q22),del(11)(q23q25), del(13) (q14) [7], 46,XX [16] | + | + | + | - | ||
| 20 | UM | 98.7 | VH3-21 | 34 | del(11q), del(13q) | 46,XX,del(11)(q13),del(13)(q12q14) [6], 46,XX,del(13)(q12q14),der(18)t(2;18)(p11;p11) [3], 46,XX [11] | + | + | + | - | ||
| 21 | UM | 100.0 | VH1-18 | 61 | del(6q), del(17p) | 46,XX,del(6)(q21),der(17)t(2;17)(p13;p11) [17] 46,XX [1] | + | + | + | - | ||
| 22 | UM | 100.0 | VH1-69 | 68 | del(11q), del(13q) | 46,XX,der(1)t(1;11)(q32;q13)del(1)(p34),der(6)t(6;13) (p22;q32),t(7;12)(p13;p13), del(11)(q22),der(11)t(7;11)(p15;q13)t(7;13)(p22;q?), der(13)t(6;13)(p22;q14) [cp5], 46,XX [8] | + | + | + | + | ||
| 23 | UM | 99.1 | VH3-49 | ND | Normal | 46-51,XY,+Y,der(2)t(2;21)(q21;q11), +3,+del(5)(q?q?),der(8)t(8;12)(q24;?),+11, -13, der(14)t(13;14)(q?;q?)inv(14)(p?q?),+del(14)(q22q32), +15, der(18)t(2;18)(?;q21),+19,-20 [cp14], 46,XY [1] | + | + | + | + | ||
| 24 | UM | 100.0 | VH1-e | ND | Normal | 46,XY,der(3)ins(3;11)(q21;q13q25),der(6)t(6;11)(q13;p11), der(11)del(11)(p11)del(11)(q13) [15], 46,XY [2] | + | + | + | - | ||
| 25 | M | 61.0 | VH1-3 | 0 | Normal | 46,XY,t(18;22)(q21;q11) [10], 46,XY [14] | - | - | - | + | ||
| 26 | M | 89.8 | VH3-23 | 0 | del(13q) | 46,XY,ins(13;2)(q14;q?q?) [8], 46,XY [12] | - | - | - | - | ||
| 27 | M | 93.8 | VH5-51 | 0 | del(13q) | 45,XX,-13 [4], 46,XX [16] | - | - | - | - | ||
| 28 | M | 91.6 | VH1-02 | 1 | del(13q) | 46,XX,del(13)(q12q14) [8]46,XX,inv(1)(p36q25),del(13)(q12q14) [3] 46,XX [4] | - | - | - | + | ||
| 29 | M | 96.0 | VH3-21 | 19 | Normal | 46,XX,t(4;11)(q23;q13) [2], 46,XX [3] | - | - | - | + | ||
| 30 | ND | ND | ND | 26 | +12, t(14;19) | 47,XX,+12,t(14;19)(q32;q13) [9] 47,XX,t(1;6)(p35;p23),+12,t(14;19)(q32;q13) [12] | + | - | - | + | ||
| 31 | ND | ND | ND | ND | del(13q) | 46,XY,del(13)(q12q14),der(13)t(13;17)(q14;q21), der(17)t(13;17)(q31;q21) [7], 46,XY [11] | - | - | + | - | ||
| 32 | UM | 100.0 | VH1-02 | 0 | Normal | 46,XY,del(11)(q23) [8], 46,XY [12] | - | - | - | - | ||
| 33 | UM | 100.0 | VH5-a | 2 | del(11q), del(13q) | 46,XY,del(11)(q21),del(13)(q12q14) [3] 46,XY,der(8)t(2;8)(p13;p23),del(11)(q21) [19] | + | + | + | - | ||
| 34 | UM | 100.0 | VH1-69 | 2 | del(13q) | 46,XX,t(10;13)(q24;q14) [4], 46,XX [9] | - | - | - | + | ||
| 35 | UM | 99.6 | VH3-33 | 7 | +12, t(14;18) | 47,XX,-4,der(7)t(4;7)(q21;q22),+der(7)t(4;7)(q21;q22), +12,t(14;18)(q32;q21) [2], 46,XX [11] | - | - | - | - | ||
| 36 | UM | 100.0 | VH-1-e | 16 | del(17p) | 45,XY,der(2)t(2;17)(q37;q21),-17 [6], 46,XY [18] | - | - | + | - | ||
| 37 | UM | 100.0 | VH1-69 | 19 | del(13q) | 46,XY,del(13)(q12q14) [8], 46,XY,del(11)(q22),del(13)(q12q14) [2], 46,XY [10] | - | - | - | - | ||
| 38 | UM | 100.0 | VH3-09 | 41 | del(13q) | 46,XX,t(10;13)(q24;q14) [14], 46,XY [6] | - | - | - | + | ||
| 39 | UM | 100.0 | VH3-23 | 44 | del(13q) | 46,XX,t(2;13)(q37;q14) [10], 46,XX [13] | - | - | - | + | ||
| 40 | UM | 99.6 | VH1-69 | 53 | Normal | 46,XY,der(20)t(2;20)(p11;p11) [13], 46,XY [7] | - | - | + | - | ||
| 41 | UM | 99.5 | VH1-69 | 68 | del(11q), del(13q) | 46,XY,del(11)(q21q24),t(13;15)(q14;q25) [11], 46,XY [4] | - | - | - | + | ||
| 42 | UM | 99.1 | VH4-39 | 73 | +12 | 47,XX,+12 [4], 47,XX,del(9)(p21),+12,t(16;17)(q24;q23) [4], 46,XX [13] | + | - | - | + | ||
| 43 | UM | 100.0 | VH3-33 | 78 | del(11q), del(13q) | 46,XY,t(1;15;13)(q21;q15;q14),t(9;20)(q34;q11) [8], 46,XY,t(1;11)(q23;q22.3),t(4;20)(q31;p11) [12], 46,XY [5] | + | - | - | + | ||
| 44 | UM | 99.6 | VH1-58 | 83 | Normal | 46,XY,del(14)(q24q32) [14], 46,XY [6] | - | - | - | - | ||
| 45 | UM | 100.0 | VH4-61 | 93 | del(11q), +12 | 46,XY,t(7;16)(p11;q24) [2], 47,XY,t(7;16)(p11;q24), +12 [12], 47,XY,t(7;16)(p11;q24),del(11)(q13q24),+12 [3] | + | - | - | + | ||
| 46 | UM | 99.0 | VH4-34 | ND | Normal | 46,XY,der(18)t(2;18)(p11;p11) [5], 46,XY [74] | - | - | + | - | ||
| 47 | UM | 100.0 | VH1-69 | ND | Normal | 46,XY,del(14)(q32) [13], 46,XY [7] | - | - | - | - | ||
The VH status is designated as mutated (M) or unmutated UM); percent sequence homology of ≥ 98% characterized an unmutated VH status; nd = not determined.
Percent CD38-positive cells compared to the appropriate isotype control.
FISH was performed on interphase cells.
In case of translocations or complex aberrations, results of G-banding analysis were confirmed by 24-color FISH.
It was remarkable that 30 (64%) of the 47 cases showing aberrations in addition to those detected by FISH had at least 3 aberrations, including 26 cases (55%) that had 3 or more unbalanced aberrations (Table 2), while FISH analysis showed only one case with 3 or more aberrations. The same group of 47 samples also was characterized by a high frequency of samples with balanced (45%) and unbalanced (53%) translocations (Table 2).
Cytogenetic characterization of FISH del(13q)
Detection of chromosomal deletions of 13q14 as the sole abnormality by FISH with the probes D13S25 and D13S319 has been reported as a favorable prognostic marker in CLL.7 Recent studies have indicated that this deletion is often cryptic and escapes detection by conventional cytogenetic studies.21,22 In our cohort of 132 samples, 76 were characterized as del(13q) by FISH, including 9 samples with a homozygous del(13q). No metaphases could be generated in 5 of the 76 cases. The remaining 71 cases consisted of 52 interstitial deletions, 3 unbalanced translocations, 5 reciprocal translocations, and 1 case with loss of chromosome 13. In 7 cases, del(13q) could not be detected by G-banding, and in another 3 cases with a homozygous del(13q) the deletions on the second allele were cryptic. In 2 of these cryptic cases, the deletion was confirmed by FISH on metaphases, as detailed under “Submicroscopic loss of genetic material.”
Characterization of aberrations by chromosome banding
In this study, we could confirm trisomy 12 by FISH and by chromosome banding as the most frequent gain of genetic material (n = 17).23 With the latter technique, however, 4 cases with trisomy 12 displayed an additional trisomy of the chromosomes 18 and 19, which emerged exclusively in this combination. Another recurrent chromosomal gain was trisomy 3 in 2 patients, again occurring in combination with the gain of additional albeit nonrecurrent other chromosomes. A different mechanism of gain of chromosomal material was by means of unbalanced translocations. This mechanism was seen in 18 of 25 patients with unbalanced translocations (Table 2). Of note are gains of the short arm of chromosome 2(2)(p11pter) (n = 4) and (2)(p13pter) (n = 2) by this mechanism. The gain of genetic material by mechanisms such as duplications or insertions was rare and nonrecurrent.
Chromosome banding confirmed the results of FISH analysis in detecting del(6)(q21), del(11)(q22.3), del(13)(q14), and del(17)(p13) as the most frequent aberrations.23 However, these deletions, characterized by FISH probes—which only bind in a specific area of the genome—were characterized by a very variable loss of genetic material as detected by chromosome banding. The spectrum ranged from deletions of just the specific chromosomal band mentioned above to loss of the whole chromosome (Figure 1).
A very common mechanism for deletions was the occurrence of unbalanced translocations (Table 2). Of the 135 single deletions that were detected by chromosomal banding, 42 (31%) were affected by unbalanced translocations. When characterizing the mechanism of deletion of the different FISH groups—del(6)(q21), del(11)(q22.3), del(13)(q14), and del(17)(p13)—by chromosome banding, the following results were obtained: no unbalanced translocation was detected for the 6q21 deletions (n = 5), 2 unbalanced translocations were detected for the 11q22.3 deletions (n = 19), 3 unbalanced translocations were detected for the 13q14 deletions (n = 64), and 4 unbalanced translocations were detected for the 17p13 deletions (n = 6). The remaining 31 of a total of 40 single unbalanced translocations (in 25 patients) were involved in deletions not described by the standard FISH panel of probes; they were nonrecurrent and located in a variety of different chromosomes (Table 2).
There were 23 samples with balanced translocations, with a total number of 30 single events. Similar to the unbalanced translocations, balanced translocations were in most cases nonrecurrent except for t(10;13)(q24;q14), t(11;14)(q13;q32), and t(14; 18)(q32;q21), with 2 cases each.
The analysis of breakpoints, which were generated by balanced and unbalanced translocations, showed some more recurrent features. Translocations involving the band 14q32 are among the best characterized in leukemia. This breakpoint emerged from 7 samples with 5 different partner chromosomes in our cohort of patients. Another very common breakpoint was 13q14 (n = 7). Regardless of the type of translocation, balanced or unbalanced, this breakpoint always was associated with a del(13q) in the respective FISH analysis. Additional recurrent break-points are listed in Table 3. Many of these are known in leukemia with the respective candidate genes.
Breakpoints . | No. . | Genes . |
|---|---|---|
| 1p36 | 2 | MDS2 |
| 1q21 | 2 | AF1q, BCL9, IRTA1, MUC1 |
| 2p13 | 2 | BCL11A |
| 2q37 | 2 | Unknown |
| 7p11 | 2 | Unknown |
| 7q11.2 | 2 | HIP1 |
| 8p21 | 2 | Unknown |
| 8q24 | 2 | C-MYC |
| 9q34 | 2 | ABL, AF9q34, CAN, FBP17, NOTCH1, PRRX2 |
| 12p13 | 2 | ETV6 |
| 13q22 | 2 | Unknown |
| 15q10 | 2 | Unknown |
| 17q23 | 2 | CLTC, MSI2 |
| 18p11.3 | 2 | Unknown |
| 19q13 | 2 | BCL3 |
| 20p11 | 2 | Unknown |
| 22q10 | 2 | Unknown |
| 7p15 | 3 | HOXA9, HOXA10 |
| 10q24 | 3 | HOX 11 |
| 16q24 | 3 | CBFA2T3 |
| 17q10 | 3 | Unknown |
| 17q21 | 3 | RARA |
| 18p11 | 3 | Unknown |
| 18q21 | 3 | BCL2, MALT1 |
| 20q11 | 3 | TOP1 |
| 2p11 | 4 | Unknown |
| 11q13 | 4 | CCND1 |
| 13q14 | 7 | MIRN15A, MIRN16-1 |
| 14q32 | 7 | IGH |
Breakpoints . | No. . | Genes . |
|---|---|---|
| 1p36 | 2 | MDS2 |
| 1q21 | 2 | AF1q, BCL9, IRTA1, MUC1 |
| 2p13 | 2 | BCL11A |
| 2q37 | 2 | Unknown |
| 7p11 | 2 | Unknown |
| 7q11.2 | 2 | HIP1 |
| 8p21 | 2 | Unknown |
| 8q24 | 2 | C-MYC |
| 9q34 | 2 | ABL, AF9q34, CAN, FBP17, NOTCH1, PRRX2 |
| 12p13 | 2 | ETV6 |
| 13q22 | 2 | Unknown |
| 15q10 | 2 | Unknown |
| 17q23 | 2 | CLTC, MSI2 |
| 18p11.3 | 2 | Unknown |
| 19q13 | 2 | BCL3 |
| 20p11 | 2 | Unknown |
| 22q10 | 2 | Unknown |
| 7p15 | 3 | HOXA9, HOXA10 |
| 10q24 | 3 | HOX 11 |
| 16q24 | 3 | CBFA2T3 |
| 17q10 | 3 | Unknown |
| 17q21 | 3 | RARA |
| 18p11 | 3 | Unknown |
| 18q21 | 3 | BCL2, MALT1 |
| 20q11 | 3 | TOP1 |
| 2p11 | 4 | Unknown |
| 11q13 | 4 | CCND1 |
| 13q14 | 7 | MIRN15A, MIRN16-1 |
| 14q32 | 7 | IGH |
Submicroscopic loss of genetic material
The comparison of FISH and metaphase cytogenetics suggested discordant results in some cases where deletions could be detected by FISH but not by chromosomal banding. Six of these cases with del(13)(q14) in FISH analysis displayed no such visible deletion in chromosomal banding but rather looked like a balanced translocation involving 13q14 (n = 5) or an insertion into 13q14. While there was no microscopic loss of genetic material in these cases, a submicroscopic deletion was confirmed by FISH with the 13q14 FISH probes D13S25 and D13S319 on metaphases after G banding in all cases tested (n = 4). The presence of submicroscopic deletions was further confirmed in 2 cases with a homozygous 13q14 deletion in FISH but with only a visible deletion in one chromosome 13 in chromosomal banding. FISH analysis on metaphases revealed the deletion also on the second chromosome, which looked cytogenetically normal. Another locus that was affected by deletion, which appeared cytogenetically as a balanced translocation, was the ATM locus. In this case, a sample with a FISH del(11q) was characterized by a t(1;11)(q23; q22.3) in chromosomal banding. These cases suggest a loss of submicroscopic amounts of genetic material that escapes detection by chromosomal banding.
Correlation of the IgVH status with cytogenetic aberrations
Of the 125 samples that were successfully analyzed by metaphase cytogenetics, the status of somatic mutations in the variable region of the immunoglobulin heavy chain (IgVH) gene was available in 110 cases. Fifty-two cases (47%) displayed a mutated IgVH status (M) (37 men, 15 women), while the residual 58 cases (53%) had an unmutated IgVH (UM) (38 men, 20 women). Sixty-six cases were analyzed at the time of diagnosis for CLL. The median age at diagnosis of these patients with a mutated IgVH (n = 30) was 61 years (range, 34-78 years) and was not significantly different from the median age of 60 years (range, 39-79 years) of the patients with an unmutated IgVH (n = 36).
The presence of samples with mutated or unmutated IgVH status was correlated with the different categories of FISH anomalies (see “Fluorescence in situ hybridization” under “Patients and methods”). This analysis showed a significantly different distribution, as already reported,10 of the IgVH status in various groups (Table 4). While samples with del(13q) as sole abnormality were overrepresented in the group of mutated IgVH (P = .008), samples with del(11q) had a higher abundance in the group of unmutated IgVH (P = .003) (Table 4).10,24 All other FISH groups showed no significant different distribution. In a second step, the number of FISH groups was reduced according to the correlation to the IgVH status. The FISH groups with no significant correlation to the IgVH status (normal, del(6q), del(17p) and +12) were compared against the single FISH groups del(11q) and del(13q) by chi-square test. This analysis resulted in a P value of .002.
Aberrations as detected by FISH are correlated to the IgVH status and CD38 expression
. | IgVH . | . | . | CD38* . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | No. Patients . | . | . | No.Patients . | . | . | ||||
| FISH . | M . | UM . | P . | Negative . | Positive . | P . | ||||
| Normal | 10 | 15 | .409 | 17 | 5 | .414 | ||||
| del(6q) | 4 | 2 | .326 | 3 | 1 | .830 | ||||
| del(11q) | 2 | 14 | .003 | 8 | 11 | .003 | ||||
| del(13q) sole | 26 | 15 | .008 | 34 | 6 | .009 | ||||
| del(17p) | 2 | 4 | .483 | 4 | 1 | .623 | ||||
| +12 | 7 | 7 | .823 | 7 | 7 | .076 | ||||
. | IgVH . | . | . | CD38* . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | No. Patients . | . | . | No.Patients . | . | . | ||||
| FISH . | M . | UM . | P . | Negative . | Positive . | P . | ||||
| Normal | 10 | 15 | .409 | 17 | 5 | .414 | ||||
| del(6q) | 4 | 2 | .326 | 3 | 1 | .830 | ||||
| del(11q) | 2 | 14 | .003 | 8 | 11 | .003 | ||||
| del(13q) sole | 26 | 15 | .008 | 34 | 6 | .009 | ||||
| del(17p) | 2 | 4 | .483 | 4 | 1 | .623 | ||||
| +12 | 7 | 7 | .823 | 7 | 7 | .076 | ||||
For IgVH M, n = 51; for UM, n = 58. For CD38-negative, n = 73; for CD38-positive, n = 31.
The cut-off for the separation of CD38-positive and -negative samples was 30% by flow cytometry.
The set of chromosomal aberrations detected by metaphase cytogenetics is very heterogeneous in CLL. To correlate the IgVH status to specific cytogenetic features, we employed 5 different cytogenetic subgroups. The results of this analysis are described in Table 5. More complex aberrant karyotypes, characterized by the presence of at least 3 unbalanced aberrations or at least 3 aberrations did not correlate significantly with the IgVH status (P = .349 and P = .122, respectively). However, the presence of translocations (irrespective of unbalanced or balanced) resulted in a significant correlation to an unmutated IgVH status (P = .006). When this group was further subdivided into samples with balanced or unbalanced translocations, only samples with unbalanced translocations retained their highly significant correlation with an unmutated IgVH status (P = .004), indicating that unbalanced translocation might predict per se for unfavorable prognosis.
Correlation of metaphase cytogenetics to the IgVH status and CD38 expression
. | IgVH . | . | . | CD38 . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | No. Patients . | . | . | No. Patients . | . | . | ||||
| Metaphase cytogenetics . | M . | UM . | P . | Negative . | Positive . | P . | ||||
| At least 3 aberrations | 8 | 16 | .122 | 13 | 12 | .018 | ||||
| At least 3 unbalanced aberrations | 8 | 13 | .35 | 12 | 9 | .126 | ||||
| Translocations* | 9 | 24 | .006 | 19 | 14 | .045 | ||||
| Unbalanced translocations† | 4 | 17 | .004 | 14 | 5 | .757 | ||||
| Balanced translocations† | 9 | 11 | .82 | 10 | 11 | .009 | ||||
. | IgVH . | . | . | CD38 . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | No. Patients . | . | . | No. Patients . | . | . | ||||
| Metaphase cytogenetics . | M . | UM . | P . | Negative . | Positive . | P . | ||||
| At least 3 aberrations | 8 | 16 | .122 | 13 | 12 | .018 | ||||
| At least 3 unbalanced aberrations | 8 | 13 | .35 | 12 | 9 | .126 | ||||
| Translocations* | 9 | 24 | .006 | 19 | 14 | .045 | ||||
| Unbalanced translocations† | 4 | 17 | .004 | 14 | 5 | .757 | ||||
| Balanced translocations† | 9 | 11 | .82 | 10 | 11 | .009 | ||||
For IgVHM, n = 52; for UM, n = 58. For CD38-negative, n = 75; for CD38-positive, n = 31.
Nine samples had balanced as well as unbalanced translocations.
Including samples with balanced or unbalanced translocations in parallel.
Correlation of the CD38 expression with cytogenetic aberrations
In addition to the IgVH status, CD38 expression is an independent prognostic marker in CLL. In most studies, a cut-off of at least 30% CD38-positive cells in flow cytometry describes a CD38-positive status, which is associated with unfavorable clinical course.8 In this study, CD38 expression was available for 104 patients. About two thirds of the samples were characterized by having less than 30% CD38 expression (n = 73), while 31 samples were CD38 positive. CD38 expression significantly correlated with IgVH status (P = .048), which was mainly affected by a prevalence of unmutated IgVH (n = 20) in the CD38-positive group, compared with only 9 mutated IgVH. There was an almost equal distribution of mutated (n = 34) and unmutated (n = 30) IgVH in the CD38-negative group.
Similar to the IgVH status, CD38 expression significantly correlated to the FISH aberrations del(11q) and del(13q) as sole abnormality (P = .003 and P = .009, respectively; Table 4). Similar to the IgVH analysis, the FISH groups with no significant correlation to CD38 expression (normal, del(6q), del(17p), and +12) were compared against the single FISH groups del(11q) and del(13q) by chi-square test. This analysis resulted in a P value of .003.
However, compared with the IgVH status, CD38 expression showed a completely different correlation to metaphase cytogenetics (Table 5). While the IgVH status significantly correlated to unbalanced translocations, CD38 expression significantly correlated to balanced translocations (P = .009) and to the cytogenetic subgroup with 3 or more aberrations (P = .018) (Table 5). In both cases, CD38-positive patients were more likely to carry the aberration.
Discussion
Cytogenetic aberrations play an important role as prognostic factors in CLL.7 Due to the low mitotic index of CLL B cells in vitro,5 analysis of a set of the most commonly known aberrations is usually done by FISH on interphase cells, which detects aberrations in about 80% of CLL samples.7,10
The use of metaphase cytogenetics, which provides an unsupervised insight into the chromosomal aberrations of a specific sample, has been limited in CLL because of technical issues. Even though 90% of patient samples could be evaluated cytogenetically, aberrations could be detected in only 40% to 50% of cases,4 in contrast to FISH analysis.
In this study, we first present a method for the efficient induction of metaphases in CLL B cells in a cohort of 132 patient samples. This method relies on the addition of the immunostimulatory oligonucleotide DSP30 plus IL-2 as a CLL B-cell–specific mitogen to the cultures, which was originally developed for immunotherapeutic applications.13-15 In contrast to earlier reports on metaphase cytogenetics in CLL that employed different B-cell mitogens such as TPA, lipopolysaccharide, and others,5 here we could generate metaphases for a successful analysis in 95% of all CLL samples that were subjected to the DSP30/IL-2 culture. More importantly, aberrations could be detected by chromosomal banding in more than 80% of these samples, a number that is similar to FISH analysis in our patient population and that shows an almost 2-fold increase relative to earlier studies. Only the use of CD40 ligand as a B-cell stimulus in 2 recent studies produced comparable results with respect to metaphase generation and chromosomal aberrations.11,12 However, while CD40 ligand stimulation is very elaborate, the DSP30/IL-2 stimulation is suitable for large numbers of samples in routine diagnostics.
In our study, the comparison of FISH and metaphase cytogenetics revealed some important differences and limitations between the 2 techniques. The total numbers of percent aberrant samples are similar between FISH (79%) and metaphase cytogenetics (81%). However, in 10 samples with no apparent abnormalities in FISH analysis, aberrations could be detected only by chromosomal banding. In these cases, aberrations were located in chromosomal regions different from the FISH probes. Adding these 10 samples to the 104 samples with FISH aberrations results in a total number of 114 samples (86%) with cytogenetic aberrations.
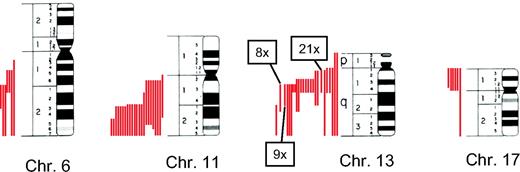
Characterization of FISH deletions by metaphase cytogenetics. Samples that had been characterized by FISH as del6q21, del11q22.3, del13q14, or del17p13 were analyzed by conventional cytogenetics for the extent of deletion. Metaphase chromosomes 6, 11, 13, and 17 are shown schematically with the respective deletions indicated as a red line on the left of each chromosome. Each red line represents a single deletion. Due to space limitations, the numbers of some frequently occurring deletions on chromosome 13 are combined.
Characterization of FISH deletions by metaphase cytogenetics. Samples that had been characterized by FISH as del6q21, del11q22.3, del13q14, or del17p13 were analyzed by conventional cytogenetics for the extent of deletion. Metaphase chromosomes 6, 11, 13, and 17 are shown schematically with the respective deletions indicated as a red line on the left of each chromosome. Each red line represents a single deletion. Due to space limitations, the numbers of some frequently occurring deletions on chromosome 13 are combined.
Overall, chromosomal banding detected aberrations in 47 samples in addition to those found by parallel FISH analysis, indicating that FISH analysis underestimates the complexity and heterogeneity of chromosomal aberrations in CLL. Only one FISH case in our analysis had 3 aberrations; however, in 30 cases from the same patient population, chromosomal banding found 3 or more aberrations, including 26 cases with at least 3 unbalanced aberrations. Since many of these aberrations are heterogeneous and nonrecurrent, the prognostic impact for the individual patients still has to be determined. Samples with 3 or more aberrations have been described in acute myeloid leukemia as so-called complex aberrant karyotypes and are associated in acute myeloid leukemia with an unfavorable prognosis.25 We hypothesize that such a complex aberrant karyotype might also predict an unfavorable prognosis in CLL, analogous to the situation in acute leukemia. However, prospective clinical studies are needed to test this hypothesis.
Even though the aberrations in metaphase cytogenetics that are detected beyond the FISH aberrations are in most cases nonrecurrent, we could define a panel of recurrent breakpoints. Examples of such breakpoints that are known from other leukemias and where candidate genes are known include 2p11 (IGK), 7p15 (HOXA9, HOXA10), 10q24 (HOX11), 11q13 (CCND1), 13q14 (MIRN15A, MIRN16-1), 14q32 (IGH), and 17q21 (RARA). Since little is known about genes involved in the pathogenesis of CLL, information regarding such breakpoints might be helpful in defining new candidate genes.
The characterization of an aberration as del(13q) in FISH can have very different characteristics when the same sample is analyzed by cytogenetic banding. In our cohort of samples, del(13q) included a range of deletions, from submicroscopic to loss of the whole chromosome 13 and also translocations with the breakpoint in 13q14. Almost the same is true for del(6q), del(11q), and del(17p). This raises the question of whether the extent of a deletion in the respective chromosomal regions has an impact on the prognosis of the individual patient.
Especially interesting are the cases with del(17p). This deletion, when detected by interphase FISH, is associated with poor prognosis and resistance to chemotherapy.7,26 Analysis of the del(17p) samples by chromosomal banding showed a recurrent loss of at least the short arm of chromosome 17 in all samples (n = 6). The mechanism of deletion was an unbalanced translocation (n = 4), an isochromosome i(17q) (n = 1), or loss of the whole chromosome 17 (n = 1), indicating that genes in addition to TP53 might be factors for the poor prognosis of this aberration.
The loss of submicroscopic amounts of genetic material at the breakpoints of reciprocal translocations is known from different acute and chronic myeloid leukemias (CML). In CML, the incidence of these deletions is 9% to 16%, and they are significantly associated with inferior survival27 and resistance to interferon therapy.28 A similar loss of genetic material was observed in all cytogenetically balanced reciprocal translocations involving 13q14 (n = 5) and in a cytogenetically balanced reciprocal translocation involving 11q22.3 (n = 1). In all cases, the respective FISH analysis indicated a del(13q) or a del(11q). Unlike the situation in CML, where the reciprocal translocation t(9;22) leads to the BCR-ABL oncogene, it is not clear whether translocations involving 13q14 or 11q22.3 are relevant for the pathogenesis of CLL. Therefore, further clinical studies should address this type of aberration to get some clues about the prognostic information of these translocations.
A second type of submicroscopic deletions was those in which metaphases appeared karyotypically normal. Two such FISH del(13q) cases were confirmed by FISH on metaphases after G banding, again indicating that both techniques complement each other. In contrast to cases with large deletions surrounding this locus, these specific deletions might point to a pathogenetically important gene in this chromosomal area. This notion is further supported by recent studies indicating a loss of the micro-RNA genes miR-15 and miR-16 by del(13q).29 These micro-RNAs negatively regulate expression of BCL2,30 an antiapoptotic gene that is highly expressed in CLL.31
Cytogenetic aberrations as detected by interphase FISH have been shown as independent prognostic factors in CLL.7 However, the correlation to other relevant prognostic factors like the IgVH status and CD38 expression is necessary to evaluate whether they provide redundant or independent prognostic information. It has been shown previously10,32 and also in this study that while the overall incidence of FISH aberrations is equally distributed in the different IgVH subsets, various FISH aberrations are significantly differently distributed in the IgVH mutated and unmutated groups (Table 4). In this study, the poor prognosis FISH marker del(11q) is clearly overrepresented in patients with unmutated IgVH (P = .003) compared with the good prognosis del(13q) as sole abnormality with an overrepresentation in the mutated IgVH group (P = .008).
Cytogenetic aberrations detected by chromosomal banding in addition to FISH aberrations were heterogeneous and in many cases nonrecurrent. To summarize these data and correlate them to the other prognostic markers, we employed 5 different cytogenetic subgroups, which were characterized by the number of aberrations in a specific sample or by the type of translocation (Table 5). The cutoff point for group separations was 3 or more aberrations/unbalanced aberrations. This number of aberrations defines the so-called “complex aberrant” karyotype in acute myeloid leukemia, which is associated with poor prognosis in this disease entity.25 On the other hand, translocations often lead to the formation of fusion genes that might be involved in the pathogenesis of the disease. When these 5 different subgroups were correlated to the IgVH status of the samples, the presence of translocations significantly correlated with an unmutated IgVH status (P = .006). However, a further subdivision of the group with translocations into the subgroups with balanced or unbalanced translocations revealed only a significant correlation of unbalanced translocations to an unmutated IgVH (P = .004). This significant correlation reflects the fact that 80% of samples with unbalanced translocations were associated with an unmutated IgVH status. In contrast, the poor-prognosis marker, CD38 expression, was not significantly correlated to unbalanced but to balanced translocations (P = .009) and to a complex aberrant karyotype with 3 or more aberrations (P = .018). On the basis of these correlations, the subgroups of patients with unbalanced translocations plus unmutated IgVH status and with CD38 expression plus balanced translocations or at least 3 aberrations might represent patient populations with poor prognosis. From a recent study it is also known that CLL patients with balanced or unbalanced translocations or complex aberrant karyo-type have a significantly decreased treatment-free survival.12
In conclusion, the data of the present study in CLL indicate that the immunostimulatory oligonucleotide DSP30 in combination with IL-2 is an easy and efficient stimulus in metaphase generation for chromosomal banding. This technique allows for a more comprehensive chromosome analysis compared to FISH and resulted in the detection of additional aberrations in 38% of patient samples in this study.
Furthermore, the presence of unbalanced translocations significantly correlated to an unmutated IgVH status of CLL samples as well as at least 3 aberrations and balanced translocations to a positive CD38 expression status. We therefore suggest that chromosomal banding analysis can define further subsets of CLL with complex aberrant karyotype and translocations with poor prognosis, which only in part overlap with other prognostic markers such as CD38 expression or IgVH status.
Prepublished online as Blood First Edition Paper, July 13, 2006; DOI 10.1182/blood-2006-02-005322.
F.D. performed IgVH analysis, data analysis, and writing; S.S. performed research; T.H. evaluated morphology and contributed to writing; W.K. evaluated flow cytometry and statistical analysis; C.S. designed research, evaluated FISH and cytogenetics, and contributed to writing.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This study was conducted in part in the Laboratory for Leukemia Diagnostics in the Medical Department III of the Ludwig-Maximilians-University Munich. The authors would like to thank all coworkers at the Munich Leukemia Laboratory for approaching together many other aspects in the field of leukemia diagnostics and research. Our special thanks are directed to all referring centers for sending patient samples to our laboratory.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal