Abstract
STAT5 regulates definitive (adult stage) erythropoiesis through its ability to transduce signals from the erythropoietin receptor. A function for STAT-dependent signaling during primitive (embryonic) erythropoiesis has not been analyzed. We tested this in the Xenopus system, because STAT5 is expressed at the right time and place to regulate development of the embryonic primitive ventral blood island. Depletion of STAT5 activity results in delayed accumulation of the first globinexpressing cells, indicating that the gene does regulate primitive erythropoiesis. Our results suggest that in this context STAT5 functions as a repressor, since forced expression of an activator isoform blocks erythropoiesis, while embryos expressing a repressor isoform develop normally. The erythroid phenotype caused by the activator isoform of STAT5 resembles that caused by overexpression of fibroblast growth factor (FGF). We show that STAT5 isoforms can function epistatic to FGF and can be phosphorylated in response to hyperactivated FGF signaling in Xenopus embryos. Therefore, our data indicate that STAT5 functions in both primitive and definitive erythropoiesis, but by different mechanisms.
Introduction
The first blood cells differentiate in Xenopus embryos at approximately 1 day after fertilization (dpf). These embryonic or “primitive” erythrocytes develop within the ventral blood island (VBI), which is an early embryonic structure analogous to the mammalian extra-embryonic yolk sac.1 Over the next day, primitive erythrocytes differentiate in a rostral to caudal wave, so that by 2 dpf the VBI extends along the full ventral aspect of the tailbud embryo comprised of differentiated primitive erythroid cells that express larval globin genes.2 These erythrocytes move into circulation as a transient cell lineage that is replaced eventually by “definitive” erythrocytes derived from lateral plate mesoderm,3 a process that is analogous to the aorta-gonad-mesonephric origin of mammalian adult hematopoietic stem cells.
VBI development is regulated by several known signaling pathways. There is a gradient of bone morphogenetic protein (BMP) signaling that is important for the generation of ventral mesoderm,4-6 and the VBI is a ventral mesoderm derivative. In addition, recent fate mapping studies7-10 showed that VBI progenitors are derived more generally from the leading edge mesoderm that is located circumferentially and most vegetal along the marginal zone. Therefore, the animal-vegetal axis plays a major role in specifying those cells that contribute to the VBI anlage.11 This axis is regulated by a gradient of FGF activity.12 The field of VBI progenitors is marked at the vegetal side of this axis by the expression of the nodal-related gene Xnr-2, and the exclusion of Xbra transcripts, which mark instead the progenitors of somites and more dorsal derivatives. Overexpression of a dominant-negative FGF receptor expands the Xnr-2 expression domain with concomitant expansion of the VBI,12 while forced expression of eFGF inhibits blood-island development. Although establishment of the Xnr-2 expression domain is BMP independent, BMP signaling is still required for the development of the VBI,13-15 perhaps acting on mesoderm made competent to respond by an earlier endoderm-derived activin-like signal.16 There is evidence that the function of BMPs involves, either directly or indirectly, a signal coming from ectoderm that eventually overlies the VBI mesoderm.17-19 Using an inducible inhibitor of Smad signaling, we showed that BMP signaling continues to be required for differentiation of the primitive erythroid program after the establishment of the VBI anlage.20 This is consistent with a requirement for BMPs to activate lineage-specific regulatory genes including Scl, Lmo2, and Gata2.21,22 Therefore, the combination of early FGF, nodal and/or activin signaling with subsequent BMP signals ensures the proper temporal and spatial organization of the VBI.
These early developmental signaling pathways appear to also regulate progenitor-cell biology for fetal and adult stages of hematopoiesis (Larsson and Karlsson23 ). Less is known about whether the well-characterized adult-stage hematopoietic cytokine signaling pathways are also involved in early embryonic hematopoiesis. Xenopus provides an experimental advantage for such analysis, since the embryos can survive defects in early hematopoiesis that would be early embryonic lethal in mice. Here we investigate whether STAT5 regulates VBI development. STAT5 is a known regulator of erythropoiesis, since it is one of the downstream mediators of erythropoietin receptor (EpoR) signaling.24 Furthermore, STAT5 is expressed at the right time and place to regulate VBI development.25 Our data suggest that STAT5 functions during embryonic blood-island development as a repressor, perhaps to attenuate signaling pathways that normally inhibit the initiation of erythropoiesis.
Materials and methods
Xenopus embryo culture
Freshly laid Xenopus eggs were obtained by standard gonadotropin induction. Eggs were collected in 1 × high salt modified Barth solution (MBS), fertilized in vitro using macerated testes, and dejellied in 2% cysteine (pH 7.9). All injections were done with embryos cultured in 0.1 × MBS + 5% ficoll (Sigma, St Louis, MO). Embryos were staged according to Nieuwkoop and Faber.26
Morpholinos
Morpholinos were purchased from Gene Tools, LLC (Philomath, OR). The sequences are MO1: 5′-TCCAAACAGCCATGGTATCACTACC (TE1306) or MO2: 5′-CATGGTATCACTACCTCAGGG (TE1307). A control nonspecific morpholino is complementary to sequences in the pBluescript polylinker. The in vitro transcription/translation experiments were carried out using TnT coupled reticulocyte lysate kit from Promega (Madison, WI) and 1 μg of template DNA. For experiments testing activity in vivo, the embryos were co-injected with 1 ng of xSTAT5 RNA. These embryos were harvested at stage 12 to 13 and lysates analyzed for levels of STAT5 protein by Western blotting using an antibody to STAT5, as described in “Immunoprecipitation and Western blot analysis.” For the phenotype analysis, embryos were injected into both blastomeres at the 2-cell stage with 20 ng of each morpholino and fixed at stage 24 for whole mount in situ hybridization, or at various stages for isolation of RNA used in qRT-PCR assays.
Injection of mRNA
The injected mRNAs were transcribed in vitro from linear template using SP6 polymerase. LacZ mRNA was transcribed using T7 polymerase (Table 1).
Information on expression constructs
mRNA . | Plasmid template . | Enzyme . | ng RNA/injection . |
|---|---|---|---|
| lacZ | pGEM-HE-lacZ | HindIII | Varied |
| EF-1α | pTRI-Xef | N/A | Varied |
| xSTAT5 | pSP64T-xSTAT5 | Xbal | 1.0 |
| xSTAT5YF695 | pSP64T-xSTAT5YF695 | Xbal | 1.0 |
| xSTAT5-VP16 | pSP64T-xSTAT-VP16-myc | Xbal | 0.5-0.7 |
| xSTAT5-EnR | pCS2MT-xSTAT5-EnR | Kpnl | 0.8-1.0 |
| myc-xSTAT5 | pCS2MT-xSTAT5 | Kpnl | 1.0 |
| torso-FGFR1 | pSP64T-myc-torso-FGFR1 | Sacl | 1.0 |
| eFGF | pSP64T-eFGF | EcoRl | 0.0005 |
| XFD | pSP64T-XFD | EcoRl | 1.0 |
mRNA . | Plasmid template . | Enzyme . | ng RNA/injection . |
|---|---|---|---|
| lacZ | pGEM-HE-lacZ | HindIII | Varied |
| EF-1α | pTRI-Xef | N/A | Varied |
| xSTAT5 | pSP64T-xSTAT5 | Xbal | 1.0 |
| xSTAT5YF695 | pSP64T-xSTAT5YF695 | Xbal | 1.0 |
| xSTAT5-VP16 | pSP64T-xSTAT-VP16-myc | Xbal | 0.5-0.7 |
| xSTAT5-EnR | pCS2MT-xSTAT5-EnR | Kpnl | 0.8-1.0 |
| myc-xSTAT5 | pCS2MT-xSTAT5 | Kpnl | 1.0 |
| torso-FGFR1 | pSP64T-myc-torso-FGFR1 | Sacl | 1.0 |
| eFGF | pSP64T-eFGF | EcoRl | 0.0005 |
| XFD | pSP64T-XFD | EcoRl | 1.0 |
N/A indicates not applicable.
Purified RNAs were injected individually or in combination into both blastomeres of 2-cell–stage embryos or into the 2 ventral blastomeres of 4-cell–stage embryos. Variable amounts of lacZ or xEF-1α RNA were used as filler RNA to ensure that equal amounts of total RNA were injected.
Expression constructs
The Xenopus STAT5 (xSTAT5) coding sequence was inserted into the EcoRV site of pSP64TBX or cloned immediately after the 3′ end of the myc tag sequence in pCS2MT using an EcoR1 site. The substitution of a tyrosine by a phenylalanine at position 695 was generated by polymerase chain reaction (PCR)–based mutagenesis using the following primer: 5′-AAAACAGACGGCTTTGTAAAACCAC. The chimeric constructs xSTAT5-EnR and xSTAT5-VP16 were obtained by fusing xSTAT5 truncated at amino acid 720 to the EnR repressor domain or the VP16 activator domain, respectively. A ClaI restriction site was inserted in the xSTAT5 sequence in order to subclone PCR products corresponding to EnR or VP16 followed by a myc epitope. Torso-R1 is as described.27 The pKS-xSCL construct was generated by ligating a 1.9 kb EcoRI fragment from pSCL22 into pBluescript KS+.
Hemoglobin and transcript analysis
Immunoprecipitation and Western blot analysis
Protein extracts were prepared from embryos injected with mRNA for lacZ, myc-xSTAT5, myc-xSTAT5 + myc-torso-FGFR1, myc-xSTAT5 + eFGF or myc-xSTAT5 + XFD and cultured in 0.1 × MBS until stage 12. Embryos were homogenized in lysis buffer (20 embryos for each sample in 1 mL) adapted from Faure et al33 (20 mM Tris [tris(hydroxymethyl)aminomethane]-HCl pH 8.0, 50 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 1% NP-40, 1 mM Na3Vo4 and protease inhibitor cocktail [Roche Diagnostics, Indianapolis, IN]) using Kontes pestles in 1.5 mL microfuge tubes. Following homogenization, extracts were centrifuged at 15 000 g at 4°C for 30 minutes. After centrifugation, cleared extracts were divided in half and incubated with either 5 μg of 9E10 anti–c-myc monoclonal antibody (Sigma) or 5 μg of control IgG at 4°C. After an hour, a 50% slurry of 3 times washed (wash buffer is the same as lysis buffer without protease inhibitor cocktail). Protein G sepharose beads were added to each sample and incubated overnight at 4°C. The beads were then washed 5 times in wash buffer and 1 time in distilled water to dilute out NP-40. Bound protein was eluted off the beads in 4 × running buffer (Invitrogen, Carlsbad, CA) by incubating at 65°C for 15 minutes followed by centrifugation at 1000g for 5 minutes. The eluate was spun through a slit in the bottom of the microfuge tube and collected in another microfuge tube to remove the beads. After heating, 15 μL protein per sample were size separated on a 10% NuPage Novex Bis-Tris gel (Invitrogen).
Electroblotting and Western blot analysis were as described.20 The mammalian polyclonal anti–phospho-STAT5A/B antibody from Sigma (S-5058) cross-reacts with overexpressed (but not endogenous) Xenopus STAT5 and was used at a 1:500 dilution for Western blot analysis. An anti–rabbit-horseradish peroxidase secondary antibody from Jackson Labs (Bar Harbor, ME) was used at a dilution of 1:25 000. For detection, enhanced chemiluminescence (ECL) Western blotting detection reagents from Amersham Biosciences (Buckinghamshire, England) were used according to manufacturer's instructions. Blots were subsequently eluted of antibodies and reprobed similarly using antibodies to recognize total STAT5 protein (Sigma S-6183, 1:500) or total myc-tagged protein (using the 9E10 antibody).
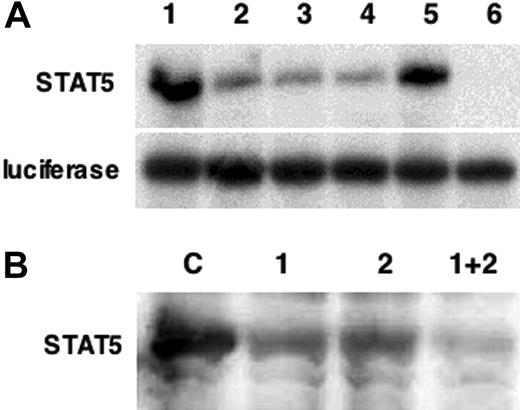
Characterization of STAT5 morpholinos. (A) Two distinct morpholinos (MO1 and MO2) were designed to target sequences around the ATG initiation site of xSTAT5. The ability of each to inhibit STAT5 translation was tested first in vitro. STAT5 protein (top panel) or as a control luciferase protein (bottom panel) was transcribed and translated in vitro using rabbit reticulocyte lysates either alone or in the presence of MO1, MO2, or both. Lanes are 1) DNA template alone, 2) 100 ng MO1, 3) 100 ng MO2, 4) 100 ng MO1 + 100 ng MO2, 5) 200 ng nonspecific control morpholino, and 6) 250 ng MO1 + 250 ng MO2. Translation lysates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by fluorography. (B) The activity of the morpholinos was next tested in vivo. Shown is a representative Western blot for xSTAT5, following injection of RNA encoding xSTAT5 alone (control, C) or co-injected with either MO1 or MO2 alone (1 or 2, 10 ng) or both MO1 and MO2 together (1 + 2, 10 ng each). Under these conditions the morpholinos reproducibly result in a substantial inhibition of STAT5 expression.
Characterization of STAT5 morpholinos. (A) Two distinct morpholinos (MO1 and MO2) were designed to target sequences around the ATG initiation site of xSTAT5. The ability of each to inhibit STAT5 translation was tested first in vitro. STAT5 protein (top panel) or as a control luciferase protein (bottom panel) was transcribed and translated in vitro using rabbit reticulocyte lysates either alone or in the presence of MO1, MO2, or both. Lanes are 1) DNA template alone, 2) 100 ng MO1, 3) 100 ng MO2, 4) 100 ng MO1 + 100 ng MO2, 5) 200 ng nonspecific control morpholino, and 6) 250 ng MO1 + 250 ng MO2. Translation lysates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by fluorography. (B) The activity of the morpholinos was next tested in vivo. Shown is a representative Western blot for xSTAT5, following injection of RNA encoding xSTAT5 alone (control, C) or co-injected with either MO1 or MO2 alone (1 or 2, 10 ng) or both MO1 and MO2 together (1 + 2, 10 ng each). Under these conditions the morpholinos reproducibly result in a substantial inhibition of STAT5 expression.
Semiquantitative and quantitative RT-PCR
The semiquantitative reverse transcriptase–polymerase chain reaction reverse transcriptase–polymerase chain reaction (RT-PCR) assay used to measure endogenous levels of STAT5 RNA were carried out as described15 using 0.25 μg (1 ×) or 0.5 μg (2 ×) total RNA and PCR for 28 cycles. Under these conditions the accumulation of PCR product was approximately linear. Real-time quantitative RT-PCR was used to measure accurately endogenous α-globin transcript levels. For each sample, total RNA was isolated (TRI REAGENT; Molecular Research Center, Cincinnati, OH) from 50 stage-matched embryos. First-strand cDNA synthesis was performed (Superscript III First-Strand Synthesis System for RT-PCR, Invitrogen) using 10 μg RNA and for each reaction 0.5% of the RT reaction was subjected to quantitative PCR analysis (Opticon DNA Engine 2; MJ Research, Watertown, MA). For each independent experiment (each time done in triplicate), the data were processed using the 2-ΔΔCT method.34 The median of each sample was normalized first to any minor changes in the levels of the housekeeping control gene ornithine decarboxylase (ODC),35 and then normalized to its respective stage-matched control. The median average from 3 independent experiments was graphed as fold change in RNA expression. The optimized primers were tested using primer design from frodo.wi.mit.edu. Primers used for PCR were xSTAT5 forward: 5′ GCCTGGAAATTTGAATTGCC(TE1443);xSTAT5reverse:5′CTGTGCGTGAGGGATCCATTG (TE1444); α-globin forward: 5′ GACCTGCATGCCTACAACCT (TE1427); α-globin reverse: 5′ AAGTGGATGGCCAGAGTCAC (TE1428); ODC forward: 5′ CAACGTGTGATGGGCTGGAT (TE1431); ODC reverse: 5′ CATAATAAAGGGTTGGTCTCTGA(TE1432).
Results
Reduction of xSTAT5 activity delays differentiation of the ventral blood island
Previous studies25 had shown that STAT5, the putative down-stream mediator of erythropoietin signaling, is expressed ubiquitously up through the end of gastrulation, and then also expressed like EpoR36 on the ventral side of the embryo during VBI development, beginning around stage 28. However, the function of STAT5 in Xenopus has not been analyzed. To test if Xenopus STAT5 (xSTAT5) regulates primitive erythropoiesis, we used sequence-specific morpholinos to reduce the endogenous xSTAT5 levels during embryogenesis. Two independent morpholinos were designed that target distinct sequences of xSTAT5 mRNA around the initiation ATG and should deplete xSTAT5 protein levels by inhibiting translation of the mRNA. Currently available antibodies are insufficient to measure directly the endogenous xSTAT5 protein. However, we showed that each morpholino is capable of inhibiting translation of xSTAT5 RNA in vitro (Figure 1A). When co-injected with RNA encoding xSTAT5 into developing embryos, each morpholino was able to reduce the levels of expressed protein as determined in Western blotting experiments, and injection of both morpholinos had an additive inhibitory effect (Figure 1B).
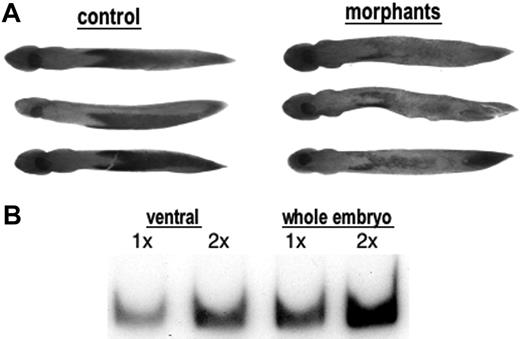
STAT5 activity is required for normal blood island development. (A) Shown are representative embryos that were either uninjected (control) or injected with both MO1 and MO2 (20 ng each) into both blastomeres at the 2-cell stage. Embryos were harvested at stage 25 and processed by whole mount in situ hybridization to detect transcript levels for embryonic α-globin, which are substantially reduced in the morphant embryos. Embryos are positioned ventral side up and anterior to the left. This experiment was repeated 3 times, and although the number of affected embryos varies, the statistics provided under “Results” are representative. (B) RNA was isolated at stage 25 from presumptive VBI regions (ventral) or whole embryos and tested for the presence of xSTAT5 RNA using semiquantitative RT-PCR. Samples for the RT reaction contained either 0.25 μg(1×) or 0.5 μg(2×) of total RNA. The xSTAT5 RNA was detected in this example using 28 cycles of PCR and still showing dependence on the amount of RNA in the original RT reaction. This experiment was repeated 3 times with similar results.
STAT5 activity is required for normal blood island development. (A) Shown are representative embryos that were either uninjected (control) or injected with both MO1 and MO2 (20 ng each) into both blastomeres at the 2-cell stage. Embryos were harvested at stage 25 and processed by whole mount in situ hybridization to detect transcript levels for embryonic α-globin, which are substantially reduced in the morphant embryos. Embryos are positioned ventral side up and anterior to the left. This experiment was repeated 3 times, and although the number of affected embryos varies, the statistics provided under “Results” are representative. (B) RNA was isolated at stage 25 from presumptive VBI regions (ventral) or whole embryos and tested for the presence of xSTAT5 RNA using semiquantitative RT-PCR. Samples for the RT reaction contained either 0.25 μg(1×) or 0.5 μg(2×) of total RNA. The xSTAT5 RNA was detected in this example using 28 cycles of PCR and still showing dependence on the amount of RNA in the original RT reaction. This experiment was repeated 3 times with similar results.
Injection of STAT5-specific morpholinos did not cause any obvious gross temporal or structural developmental abnormalities. The embryos appeared normal and survived embryogenesis. However, many of the embryos injected with the combination of xSTAT5-specific morpholinos showed abnormal development of the VBI, assessed at stage 25 by whole mount in situ hybridization for expression of larval α-globin transcripts (Figure 2A). In a representative experiment we found that 78% of morpholino-injected embryos (n = 27) demonstrated an observable decrease in globin transcript levels compared to controls. While most embryos eventually recover by stage 36, the levels of globin RNA are still reduced at stage 32 in 64% of morpholino-injected embryos (n = 28). Measurement of globin transcripts by quantitative RT-PCR indicates that morphant embryos at stage 25 are depleted for α-globin transcripts to approximately 40% normal levels (not shown here, but discussed under “The morphant phenotype is rescued by STAT5-EnR expression”).
Although the morpholino approach is by nature a transient knockdown, globin levels begin to normalize in the morphants at a time (within the first 48 hpf) that morpholinos are expected to remain effective. This suggests that the requirement for STAT5 in the VBI might be limited to early stages of hematopoiesis. The restoration of normal globin transcript levels is interesting since the STAT5 mRNA is only clearly detected in the VBI pattern by in situ hybridization at around stage 28, and this is the time when embryos are recovering from the effect of the morpholinos. To determine if STAT5 is expressed at earlier tailbud stages, we isolated RNA from the VBI region of stage 25 embryos and used semiquantitative RT-PCR to measure transcript levels. Although the levels are lower than samples prepared from whole embryos at that stage (consistent with higher expression levels of xSTAT5 in dorsal anterior regions25 ), we detected easily transcripts for STAT5 in the VBI samples (Figure 2B). At this time we cannot determine if the embryos recover because the morpholinos become ineffective as STAT5 transcript levels rise in the VBI, or because STAT5 is just not essential at later stages. Regardless, depletion of STAT5 does cause a significant delay in embryonic erythropoiesis, indicating that the protein does have a regulatory function.
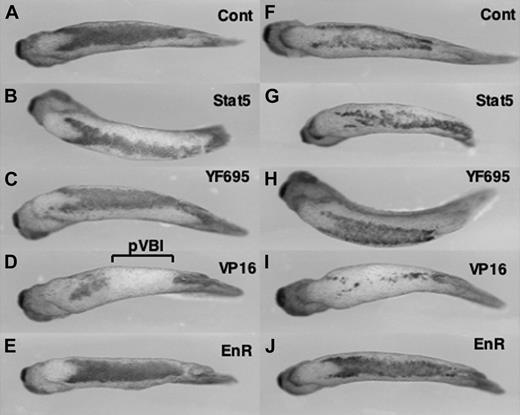
Expression of an activator isoform of STAT5 inhibits primitive erythropoiesis in the VBI. Embryos were injected at the 4-cell stage into the 2 presumptive “ventral” blastomeres, to target RNA to the presumptive posterior VBI. Shown are representative embryos processed by in situ hybridization at stage 35 with probes for embryonic α-globin (left panels, A-E) or SCL (right panels, F-J). Embryos were injected with RNA encoding lacZ as a control (A, F), STAT5 (B, G), a mutated form of STAT5 that should be inactive (C, H), a constitutively activated isoform STAT5-VP16 (D, I), or a repressor isoform STAT5-EnR (E, J). The region of the pVBI that is inhibited for erythropoiesis by STAT5-VP16 is indicated in panel D. Views are ventral, with anterior to the left. The data are consistent with a normal function for STAT5 as a repressor of genes that inhibit globin expression. While the transcript patterns can vary somewhat from embryo to embryo, the phenotypes seen were reproducible in at least 3 independent experiments. The embryos shown here are derived from one experiment, where the expression pattern shown is most representative from a batch of (n) embryos: A, 12 of 12; B, 11 of 11; C, 10 of 15; D, 7 of 9; E, 13 of 13; F, 17 of 20; G, 10 of 13; H, 8 of 13; I, 9 of 12; J, 10 of 14.
Expression of an activator isoform of STAT5 inhibits primitive erythropoiesis in the VBI. Embryos were injected at the 4-cell stage into the 2 presumptive “ventral” blastomeres, to target RNA to the presumptive posterior VBI. Shown are representative embryos processed by in situ hybridization at stage 35 with probes for embryonic α-globin (left panels, A-E) or SCL (right panels, F-J). Embryos were injected with RNA encoding lacZ as a control (A, F), STAT5 (B, G), a mutated form of STAT5 that should be inactive (C, H), a constitutively activated isoform STAT5-VP16 (D, I), or a repressor isoform STAT5-EnR (E, J). The region of the pVBI that is inhibited for erythropoiesis by STAT5-VP16 is indicated in panel D. Views are ventral, with anterior to the left. The data are consistent with a normal function for STAT5 as a repressor of genes that inhibit globin expression. While the transcript patterns can vary somewhat from embryo to embryo, the phenotypes seen were reproducible in at least 3 independent experiments. The embryos shown here are derived from one experiment, where the expression pattern shown is most representative from a batch of (n) embryos: A, 12 of 12; B, 11 of 11; C, 10 of 15; D, 7 of 9; E, 13 of 13; F, 17 of 20; G, 10 of 13; H, 8 of 13; I, 9 of 12; J, 10 of 14.
Xenopus STAT5 functions as a repressor
Based on the finding that loss of STAT5 delays primitive erythropoiesis, we tested whether overexpression of xSTAT5 during embryogenesis might promote or expand erythropoiesis. Although STAT proteins are regulated by phosphorylation, overexpression assays in Xenopus can often obviate this requirement and lead to a gain-of-function phenotype (as seen, for example, with forced expression of Smad537 ). For this purpose, a set of STAT5 isoforms was generated and expressed ectopically in developing embryos. In addition to wild-type STAT5, isoforms were created containing fusions at the C-terminus with either the VP16 acidic activation domain (xSTAT5-VP16, which should activate target genes) or the strong repressor domain derived from the Drosophila engrailed gene (xSTAT5-EnR, which should function to repress STAT5 target genes). Another isoform was generated with a mutation at the key JAK-dependent phosphorylation site (Y695F) and is therefore expected to be inactive in terms of Epo-dependent signaling. RNA encoding wild-type xSTAT5 or variant isoforms were injected into the ventral blastomeres of 4-cell embryos. This targets expression of the protein to the presumptive posterior ventral blood island (pVBI), which is normally delayed in differentiation compared to the anterior VBI. Thus, the experiment tests whether forced expression of STAT5 enhances erythropoiesis in the pVBI.
To evaluate VBI development, we again used in situ hybridization to analyze the expression pattern of embryonic α-globin. Compared to control embryos (Figure 3A, Cont), forced expression of xSTAT5 in the pVBI does not obviously enhance or expand α-globin levels (Figure 3B, Stat5). Likewise, forced expression of the mutant isoform of xSTAT5 that cannot be phosphorylated fails to affect globin expression significantly (Figure 3C, YF695). In contrast, forced expression of the isoform of STAT5 that is fused to a strong transcriptional activator completely ablates globin expression (VP16, Figure 3D). These results indicate that xSTAT5 functions normally as a repressor, since the “activator” VP16-fused isoform generates a phenotype that resembles STAT5 depletion rather than STAT5 overexpression. To test this we expressed the “repressor” form of STAT5 fused to the EnR domain. Consistent with a normal function of STAT5 as a repressor, xSTAT5-EnR had no marked effect on globin transcript levels (Figure 3E). Stem-cell leukemia (SCL) is another marker for embryonic erythroid cells, and correspondingly similar changes in SCL expression are seen when analyzed in the injected embryos (Figure 3F-J).
Expression of STAT5 isoforms does not correlate with effects modulating BMP signaling
Transcripts for the erythropoietin receptor36 do not accumulate until early tadpole stages (eg, stage 32). This fact, and the observation that STAT5 functions as a repressor for VBI development, make it unlikely that STAT5 regulates embryonic globin expression by the same mechanism described for definitive erythropoiesis. One possibility is that alteration of STAT5 signaling disrupts globin expression indirectly caused by defects in normal axis formation or generation of ventral mesoderm. For example, the phenotypes caused by STAT5 depletion (or STAT5-VP16 expression) could be caused by inhibition of the bone morphogenetic protein (BMP) signaling pathway or by increased levels of FGF signaling. Although this is not strongly supported by the morphologic analysis (the embryos are essentially normal), the embryos expressing xSTAT-VP16 (and in a minority of cases, xSTAT5-EnR) were mildly smaller in size, and the effects might be subtle. Therefore, we first considered if STAT5 could be modifying the generation of ventral mesoderm, which would implicate it as functioning downstream of the BMP signaling pathway. For this purpose we analyzed embryonic patterning at the molecular level by examining expression patterns for key markers of the dorsalventral axis in embryos manipulated for STAT5 activity.
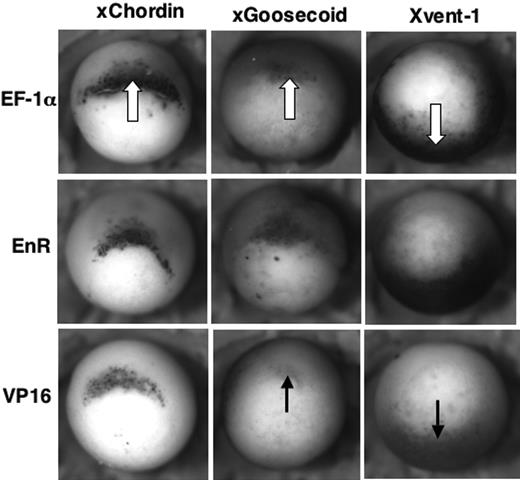
Modulation of STAT5 activity does not correlate with altered BMP signaling. Shown are representative embryos processed by whole mount in situ hybridization at stage 10.5 for the dorsal-anterior mesoderm marker xChordin (left column panels), the Spemann Organizer marker xGoosecoid (middle column panels), or the ventral-posterior marker Xvent-1 (right column panels). All embryos are shown with a dorsal lip view, anterior at the top. Embryos were injected with EF-1α as a control (top row) or had been injected with RNA encoding STAT5-EnR (middle row) or STAT5-VP16 (bottom row), as indicated. The white arrows point out the normal expression domains for these early markers (seen in this experiment for the majority of at least 12 embryos). As indicated by the black arrows, STAT5-VP16 reduces modestly the expression levels for both xGoosecoid (seen in this experiment for 7 of 12 embryos) and Xvent-1 (seen in this experiment for 10 of 16 embryos) but not xChordin (13 embryos).
Modulation of STAT5 activity does not correlate with altered BMP signaling. Shown are representative embryos processed by whole mount in situ hybridization at stage 10.5 for the dorsal-anterior mesoderm marker xChordin (left column panels), the Spemann Organizer marker xGoosecoid (middle column panels), or the ventral-posterior marker Xvent-1 (right column panels). All embryos are shown with a dorsal lip view, anterior at the top. Embryos were injected with EF-1α as a control (top row) or had been injected with RNA encoding STAT5-EnR (middle row) or STAT5-VP16 (bottom row), as indicated. The white arrows point out the normal expression domains for these early markers (seen in this experiment for the majority of at least 12 embryos). As indicated by the black arrows, STAT5-VP16 reduces modestly the expression levels for both xGoosecoid (seen in this experiment for 7 of 12 embryos) and Xvent-1 (seen in this experiment for 10 of 16 embryos) but not xChordin (13 embryos).
The xChordin (xChd30 ) and xGoosecoid (xGsc32 ) genes are expressed at stage 10.5 (late gastrulation) in the cells of the dorsal lip. At this same stage, Xvent-1 transcripts are expressed in a pattern complementary to xChd, in cells of the ventral marginal zone.31 Embryos were injected into both cells at the 2-cell stage with RNA encoding EF-1α (as a control), xSTAT5-EnR, or xSTAT5-VP16, harvested at stage 10.5, and processed for in situ hybridization using probes for xChd, xGsc, or Xvent-1 (Figure 4). If alterations in STAT5 signaling interfered with BMP signaling, it is expected that this would be reflected in a corresponding expansion of the xChd and xGsc patterns. In contrast, the xChd pattern during gastrulation is normal in most embryos expressing xSTAT5-EnR or xSTAT5-VP16. In embryos expressing xSTAT5-EnR, the xGsc and Xvent-1 transcript patterns also appear normal. In many of the embryos expressing xSTAT5-VP16, the early expression levels of both xGsc and xVent-1 are reduced. The delay in globin expression cannot be explained by these changes, since by stage 12.5 the Xvent-1 pattern is essentially normal (data not shown). These gene-expression patterns are inconsistent with modulation of the BMP signaling pathway being the cause of the globin phenotype.
The morphant phenotype is rescued by STAT5-EnR expression
Because of the strong phenotype of the STAT5-VP16 isoform compared to the morphant embryos, we were concerned if the morphant phenotype could be explained by a nonspecific delay in embryonic development. Although the embryos were staged carefully and similar phenotypes on globin expression were not seen using control morpholinos, we tested directly the ability to rescue globin transcript levels by specifically restoring levels of STAT5 repressor activity. For this purpose embryos were again injected with the STAT5-specific morpholinos, either alone or with RNA encoding the STAT5-EnR isoform. The in situ hybridization analysis described previously shows the spatial pattern of gene expression but is not very quantitative. Therefore, we developed a quantitative RT-PCR assay, to measure the effect on globin expression. Embryos were either control injected or were injected with STAT5-specific morpholinos, with or without inclusion of mRNA encoding STAT5-EnR. Embryos were then cultured to defined stages of development, and RNA was purified from batches of 50 embryos for analysis by quantitative RT-PCR using primers specific for larval alpha globin, or as a control the housekeeping gene ornithine decarboxylase (ODC). Consistent with the in situ hybridization data shown previously, samples generated from embryos injected with the STAT5-specific morpholinos showed a reproducible and significant reduction in globin transcript levels when analyzed at stage 25 (Figure 5). The globin transcript levels were reduced to around 40% of the normal level. In contrast, globin levels were restored to normal when the morpholinos were co-injected with RNA encoding STAT5-EnR. This experiment shows that the morphant embryos are not inherently deficient in the ability to express normal globin levels, but that normal globin transcript levels are dependent on STAT5 repressor activity.
xSTAT5 can act downstream of FGF
FGF is also important for mesoderm development and normally restricts the primitive blood-island domain. The loss of globin transcripts in embryos expressing STAT5-VP16 is similar to the phenotype of embryos forced to express eFGF, although eFGF also causes morphologic disturbances.38 Therefore, we next tested if xSTAT5 can modulate the effect of FGF signaling with respect to VBI development. For this purpose an epistasis study was designed, to test whether expression of xSTAT5-EnR can rescue the block to globin expression caused by eFGF, and if xSTAT5-VP16 expression inhibits erythropoiesis in embryos expressing a dominant-negative FGF receptor (XFD, which when expressed alone expands the blood island). The RNAs encoding LacZ (as a control), xSTAT5-VP16, xSTAT5-EnR, eFGF, or XFD were injected individually or in combination into the ventral blastomeres (from which the pVBI progenitors are derived) of 4-cell embryos, followed by benzidine staining to detect differentiated blood cells at stage 35.
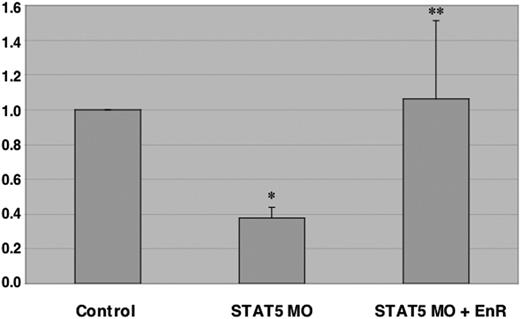
Globin transcript levels are dependent on STAT5 repressor activity. Embryos were injected with a control morpholino or the STAT5 morpholino (STAT5 MO) and, in addition, 1 ng RNA encoding STAT5-EnR, or RNA encoding lacZ as a control. Embryos were harvested at stage 24-25 and RNA processed for quantitative RT-PCR assays. Each independent sample consisted of a pool of 50 embryos. The median of each sample was normalized to its respective ODC control, and the median average from 4 independent experiments was graphed as fold change in RNA expression. Error bars indicate standard error of the mean. The asterisk indicates that the change in α-globin transcript levels comparing control injected and STAT5 MO injected (first and second samples) is statistically significant (P < .001). The double asterisk indicates that the rescue with STAT5-EnR RNA compared to MO co-injected with lacZ RNA (second and third samples) is statistically significant (P < .03).
Globin transcript levels are dependent on STAT5 repressor activity. Embryos were injected with a control morpholino or the STAT5 morpholino (STAT5 MO) and, in addition, 1 ng RNA encoding STAT5-EnR, or RNA encoding lacZ as a control. Embryos were harvested at stage 24-25 and RNA processed for quantitative RT-PCR assays. Each independent sample consisted of a pool of 50 embryos. The median of each sample was normalized to its respective ODC control, and the median average from 4 independent experiments was graphed as fold change in RNA expression. Error bars indicate standard error of the mean. The asterisk indicates that the change in α-globin transcript levels comparing control injected and STAT5 MO injected (first and second samples) is statistically significant (P < .001). The double asterisk indicates that the rescue with STAT5-EnR RNA compared to MO co-injected with lacZ RNA (second and third samples) is statistically significant (P < .03).
Control embryos injected with RNA encoding LacZ develop with normal morphology and normal blood islands (Figure 6A). As shown above, embryos expressing xSTAT5-EnR have an essentially normal pattern of primitive blood development (Figure 6B), whereas in embryos expressing xSTAT5-VP16 the benzidine staining pattern of the pVBI is completely ablated (Figure 6C). In agreement with published results,38 embryos overexpressing eFGF have shortened tails and lack benzidine-staining blood cells (Figure 6D), whereas XFD-expressing embryos lack axial structures, but have an expanded VBI (Figure 6E). Co-expression of xSTAT5-EnR with eFGF rescues the body axis and provides at least a partial rescue of VBI development (Figure 6F). Therefore, the repressor isoform of STAT5 abrogates the pathway activated by eFGF. This indicates that xSTAT5 can act downstream of eFGF signaling and is consistent with the notion that STAT5 is a repressor. Also consistent with this interpretation, embryos co-expressing xSTAT5-VP16 and XFD still lack axial structures, but now also display a loss in benzidine staining (Figure 6G). Thus, the xSTAT5 isoforms can act epistatic to FGF signaling.
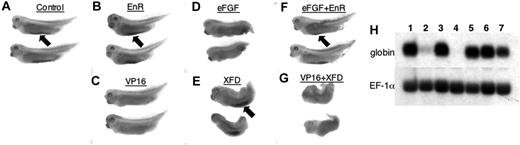
STAT5 activity can interact with FGF signaling in Xenopus embryos. Shown are representative embryos fixed and stained with benzidine to detect differentiated globin-expressing erythroid cells. Embryos had been injected into both blastomeres at the 2-cell stage with RNA encoding (A) lacZ as a control, (B) STAT5-EnR, (C) STAT5-VP16, (D) eFGF, (E) the dominant-negative FGFR isoform XFD, (F) eFGF + STAT5-EnR, (G) STAT5-VP16 + XFD. Benzidine-positive blood islands are indicated by the black arrows. Note that STAT5-VP16 and eFGF each inhibit VBI development (C-D). Restoring STAT5 repressor activity by STAT5-EnR is sufficient to rescue at least partially the repressive effect of eFGF (F), while XFD is unable to rescue VBI development that is repressed by STAT5-VP16 (G). Views are lateral with anterior to the left. The patterns shown were reproducible in at least 3 independent experiments. The embryos shown here are most representative of the staining patterns from one experiment representing the phenotype of (n) embryos: A, 12 of 19; B, 14 of 27; C, 12 of 12; D, 8 of 19; E, 23 of 26; F, 18 of 32; G, 21 of 29. (H) Shown is a representative Northern blot probed for RNA encoding embryonic α-globin (top panel) or as a loading control, the same blot reprobed for the housekeeping gene EF-1α (bottom panel). Samples are derived from pools of at least 25 embryos that had been injected with RNA encoding (1) lacZ as a control, (2) STAT5-VP16, (3) STAT5-EnR, (4) eFGF, (5) XFD, (6) eFGF + STAT5-EnR, (7) XFD + STAT5-VP16. Note that STAT5 repressor activity rescues much of the inhibition of globin transcription by eFGF (lane 6), while STAT5-VP16 is still partially repressive even in the presence of XFD (lane 7).
STAT5 activity can interact with FGF signaling in Xenopus embryos. Shown are representative embryos fixed and stained with benzidine to detect differentiated globin-expressing erythroid cells. Embryos had been injected into both blastomeres at the 2-cell stage with RNA encoding (A) lacZ as a control, (B) STAT5-EnR, (C) STAT5-VP16, (D) eFGF, (E) the dominant-negative FGFR isoform XFD, (F) eFGF + STAT5-EnR, (G) STAT5-VP16 + XFD. Benzidine-positive blood islands are indicated by the black arrows. Note that STAT5-VP16 and eFGF each inhibit VBI development (C-D). Restoring STAT5 repressor activity by STAT5-EnR is sufficient to rescue at least partially the repressive effect of eFGF (F), while XFD is unable to rescue VBI development that is repressed by STAT5-VP16 (G). Views are lateral with anterior to the left. The patterns shown were reproducible in at least 3 independent experiments. The embryos shown here are most representative of the staining patterns from one experiment representing the phenotype of (n) embryos: A, 12 of 19; B, 14 of 27; C, 12 of 12; D, 8 of 19; E, 23 of 26; F, 18 of 32; G, 21 of 29. (H) Shown is a representative Northern blot probed for RNA encoding embryonic α-globin (top panel) or as a loading control, the same blot reprobed for the housekeeping gene EF-1α (bottom panel). Samples are derived from pools of at least 25 embryos that had been injected with RNA encoding (1) lacZ as a control, (2) STAT5-VP16, (3) STAT5-EnR, (4) eFGF, (5) XFD, (6) eFGF + STAT5-EnR, (7) XFD + STAT5-VP16. Note that STAT5 repressor activity rescues much of the inhibition of globin transcription by eFGF (lane 6), while STAT5-VP16 is still partially repressive even in the presence of XFD (lane 7).
The interpretation of these benzidine staining patterns was confirmed by Northern blotting analysis of embryonic globin transcripts (Figure 6H). Compared to controls, embryos expressing either eFGF or xSTAT5-VP16 show a striking loss of globin transcript levels (compare lane 1 with lanes 2 and 4, respectively). Embryos coexpressing xSTAT5-EnR are rescued for globin transcript levels compared to those expressing eFGF alone (compare lane 6 with lane 4). On the other hand, coexpression of xSTAT5-VP16 caused a significant decrease in globin expression compared with expression of XFD alone (compare lane 7 with lane 5). Together, these data suggest that xSTAT5 is a repressor that can modulate the inhibitory activity of eFGF with respect to transcriptional programs during embryogenesis.
Activated FGF signaling results in xSTAT5 phosphorylation
Since STAT5 isoforms can act downstream of FGF signaling, we tested whether a constitutively active FGF receptor (torso-FGFR1) or eFGF could induce tyrosine phosphorylation of xSTAT5. Because the available antisera are ineffective at immunoprecipitation of endogenous xSTAT5 protein, embryos were injected with RNA encoding myc-tagged xSTAT5, either alone or in combination with RNA encoding torso-FGFR1 or eFGF. Embryos were harvested at stage 12, and total STAT5 protein was immunoprecipitated from lysates using antibodies that recognize the myc-tag (or as a control, isotype-matched antibodies), followed by Western blotting using an anti–phospho-STAT5 antibody that recognizes specifically phosphorylated tyrosine 694 (which, in the Xenopus protein, is actually at position 695).
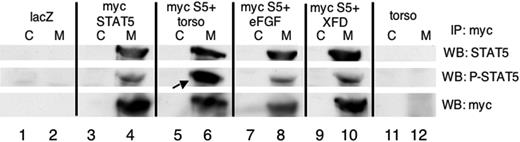
A representative Western blot analyzing levels of phospho-STAT5 is shown in Figure 7. The sample derived from embryos expressing myc-xSTAT5 contains a reproducibly detectable signal for phospho-STAT5 (lane 4) compared to the lacZ-injected control embryos (lane 2). When the activated torso-FGFR1 is co-injected, more myc-xSTAT5 becomes tyrosine phosphorylated (lane 6). Control Western blotting experiments using antibodies that recognize total STAT5 protein (upper panel) and total myc-tagged protein (lower panel) show that this change is not due to increased levels of STAT5 protein, but represents increased phosphorylation. Although coexpression of eFGF did in some cases increase phospho-STAT5 levels (not shown), this was always less than with torso, as shown here in lane 8 (normalized to total myc-tagged protein). Therefore, low levels of increased FGF signaling that are sufficient to affect morphology are less effective at changing phosphorylation of STAT5. Furthermore, forced expression of the dominant-negative FGF receptor isoform (XFD) fails to reduce the basal level of phospho-STAT5 (lane 10), indicating that phosphorylation of STAT5, although increased by hyperactivated FGF signaling, is not dependent on FGF signaling. These results are consistent with a function for STAT5 that is not directly controlled by FGF, but that STAT5 function could be indirectly stimulated by FGFR signaling.
Expression of an activated FGF receptor results in increased phosphorylation of STAT5 in Xenopus embryos. Shown are representative Western blotting experiments on samples that had been immunoprecipitated with antibodies specific for the myc epitope (M) or equivalent samples that were processed similarly using an isotype-matched control antibody (C). Lysates were derived from pools of 20 stage 12 embryos that had been injected with RNA encoding LacZ as a control (lanes 1, 2), myc-tagged xSTAT5 (myc STAT5, lanes 3, 4), the constitutively activate isoform of the FGFR1 (torso) + myc-STAT5 (myc S5, lanes 5, 6), eFGF + myc STAT5 (lanes 7, 8), myc-STAT5 + the dominant-negative FGFR1 (XFD, lanes 9, 10), or the constitutively active isoform of the FGFR1 (torso) alone (lanes 11, 12). Blotted samples were probed sequentially using antibodies for total STAT5 protein (top panel), STAT5 phosphorylated specifically at tyrosine 695 (P-STAT5, middle panel), or total myc-tagged proteins (myc, bottom panel). The phospho-STAT5 levels are increased over baseline upon expression of torso (arrow in lane 6), although the baseline levels of phospho-STAT5 are not reduced in the presence of XFD.
Expression of an activated FGF receptor results in increased phosphorylation of STAT5 in Xenopus embryos. Shown are representative Western blotting experiments on samples that had been immunoprecipitated with antibodies specific for the myc epitope (M) or equivalent samples that were processed similarly using an isotype-matched control antibody (C). Lysates were derived from pools of 20 stage 12 embryos that had been injected with RNA encoding LacZ as a control (lanes 1, 2), myc-tagged xSTAT5 (myc STAT5, lanes 3, 4), the constitutively activate isoform of the FGFR1 (torso) + myc-STAT5 (myc S5, lanes 5, 6), eFGF + myc STAT5 (lanes 7, 8), myc-STAT5 + the dominant-negative FGFR1 (XFD, lanes 9, 10), or the constitutively active isoform of the FGFR1 (torso) alone (lanes 11, 12). Blotted samples were probed sequentially using antibodies for total STAT5 protein (top panel), STAT5 phosphorylated specifically at tyrosine 695 (P-STAT5, middle panel), or total myc-tagged proteins (myc, bottom panel). The phospho-STAT5 levels are increased over baseline upon expression of torso (arrow in lane 6), although the baseline levels of phospho-STAT5 are not reduced in the presence of XFD.
Discussion
STATs 1, 3, and 5 provide antiapoptotic signals in erythroid progenitors downstream of the erythropoietin receptor.39 Mammalian cells express 2 isoforms of STAT5, encoded by separate genes named STAT5A and STAT5B.40,41 Both the single and double STAT5 knockout mice survive to adulthood, although a proportion of the double-null mice die after 2 days of an unknown cause.42 None of the mutants, including the double knockouts, display an overt steady-state hematopoietic phenotype,42-44 perhaps due to compensation by STAT1 or STAT3. However, the double-knockout mice do show defects in fetal hematopoiesis and in the ability of bone marrow–derived hematopoietic stem cells to respond to hematopoietic growth factors.42,45-49 Since the null embryos survive, it has not been considered whether STAT5 regulates yolk sac primitive hematopoiesis. However, it should be pointed out that the individual gene knockout strategies may generate hypomorphic alleles, since a different strategy that targets deletion of both genes together from the germ line causes perinatal lethality, which might be explained by severe anemia.50
We tested a role for STAT5 in embryonic erythropoiesis using the Xenopus system, because of the relative ease in evaluating the analogous program, the ventral blood island. In Xenopus, only 3 members of the STAT family are described, xSTAT1, 3, and 5.25,51,52 The cytokine receptor gp130, acting through xSTAT3, has potent ventralizing activity,51,52 but only xSTAT5 is expressed in the VBI.25 Therefore, STAT5 is expressed at the right time and place to influence VBI development, and the Xenopus STAT system may be less compensated by family members compared to mouse. Depletion of STAT5 does cause a delay in primitive erythropoiesis, although this appears to reflect regulation at a relatively early stage, and the embryos eventually recover. Furthermore, rather than stimulating or inducing ectopic erythropoiesis, we find that expression of an activator isoform of xSTAT5 results in a potent block to primitive erythropoiesis. The results reveal several unanticipated aspects of STAT signaling during early embryogenesis.
First, at least in Xenopus, STAT5 does function to influence primitive blood-island development. Based on the morpholino experiments, this function could be either during gastrulation (when STAT5 is expressed ubiquitously) or during early stages of primitive erythropoiesis. STAT5 is not highly expressed around tailbud stage 25, when we note the delay in globin expression with STAT5 depletion. However, we did confirm by RT-PCR experiments that the gene is expressed at this time in ventral tissues, and so STAT5 might be involved in the initial stages of primitive erythropoiesis. Regardless if the function is during gastrulation or initiation of VBI development (or both), endogenous STAT5 seems to be either unnecessary or compensated by other factors during later stages, since in most embryos primitive erythropoiesis does recover. This interpretation assumes that the morpholino remains potent at blocking STAT5 translation in later stages. While this interpretation would also be consistent with the mouse mutant phenotype (in which case the delay might not have been documented), it is tempered by the experimental approach, since the morpholinos could also be less effective at later stages, as STAT5 transcript levels rise in the VBI.
Second, the data are consistent with the function of STAT5 during primitive erythropoiesis as a repressor rather than an activator of transcription. The overexpression of either STAT5 itself or the isoform created by fusion to a strong repressor domain (EnR) has little obvious effect on development, while expression of the isoform created by fusion to a strong activator domain (VP16) causes a block to blood-island development. This suggests that STAT5 functions as a repressor to turn off genes that normally inhibit blood-island development. If STAT5 is normally a repressor, it is predicted that reduction of STAT5 repressor activity should resemble the phenotype caused by overexpression of the activated isoform. This is what we observe in the morpholino-injected embryos. The morphant phenotype is not as strong as the block seen with the STAT5-VP16–injected embryos, but the morpholinos may generate a partial loss of STAT5, while the VP16-fused isoform is expected to potently activate the normally repressed STAT5 target genes. Although STATs were originally characterized as cytokine-dependent activators of transcription, there is precedence in the literature for considering repressor activity. For example, STAT5 is known to interact with the silencing mediator of retinoid and thyroid receptors (SMRT) corepressor,53 and this could mediate repression of target genes during development. The Dictyostelium STATa protein is a repressor during development, and mutations in this gene cause hypersensitivity to induction of differentiation caused by the dorsal-related immunity factor (DIF) morphogen.54
Finally, the results suggest that STAT5 functions normally to antagonize a subset of target genes relevant to VBI development that are activated indirectly by FGF signaling. Excessive FGF signaling blocks blood-island development, and expression of a dominant-negative FGF receptor is sufficient to expand the VBI.12 In our experiments, overexpression of the potent repressor isoform (fused to EnR) is sufficient to block this aspect of the phenotype caused by excessive FGF signaling. In general, the effects of manipulating STAT signaling are restricted to the VBI, indicating that STAT5 modulates only a subset of FGF target genes, or only indirect target genes. The xSTAT5 pathway appears to function in this respect distinct from the eFGF–mitogen-activated protein kinase (MAPK) pathway that is required for gastrulation movements and mesoderm induction, based on 2 observations. First, the dominant-negative FGF receptor (XFD) is known to block MAPK signaling, yet embryos expressing XFD have an expanded VBI,12,38,55,56 which is reduced by coexpression of xSTAT5-VP16. Second, embryos overexpressing eFGF exhibit major body axis defects,57 due to hyperactivation of MAPK signaling, whereas embryos expressing xSTAT5-VP16 do not display gross body axis defects.
Although the interaction of STAT5 with FGF appears to be indirect, there is strong precedence for STATs, including STAT5, in mediating FGF signaling, for example, with respect to chondrocyte development and bone disease.58-63 It is intriguing that hyperactive FGF signaling leads to phosphorylation of STAT5. This is presumably an indirect effect since the basal levels of STAT5 phosphorylation were not depleted by overexpression of XFD. Thus, FGF signaling may activate a secondary signaling pathway, leading to phosphorylation and activation of STAT5 repressor activity. We speculate that this could be a negative-feedback mechanism that regulates the initiation of erythropoiesis. While FGF inhibits erythropoiesis, the subsequent activation of the STAT5 repressor activity could represent one step leading to activation of the early hematopoietic program.
Interpreting experiments using forced expression of STAT5 fusion proteins must be done with caution, since overexpression could lead to misregulation of unintended target genes, for example, those regulated by related STAT proteins. However, the loss-of-function experiments support the repressor model for STAT5, since these embryos generate a phenotype that resembles that caused by the VP16 fusion protein and not the EnR fusion protein. Forced expression of xSTAT3-EnR causes axis duplications,51 which most likely does not reflect the normal function of STAT3, since axis duplication is not observed in STAT3 morphant embryos, which instead show defects in cell movements during gastrulation.64 A very recent report used an overexpression approach to analyze the function of mutant isoforms of STAT5 that are constitutively active.65 In this case some hematopoietic phenotypes were described that would be consistent with increased proliferation of hematopoietic progenitors, and in all cases the results were not different from overexpression of wild-type STAT5. We note that in some embryos we did observe what appeared to be enhanced levels of erythropoiesis in embryos expressing the STAT5-EnR isoform and so the “constitutively active” isoform may be functioning similar to an EnR isoform.
Our results manipulating STAT5 are very different from those obtained in experiments that analyzed overexpression of STAT5 in embryonic stem cells or hematopoietic stem cells, which leads to enhanced proliferation, differentiation, and survival of hematopoietic progenitors.66,67 However, we are targeting a much earlier developmental stage, not specific to hematopoietic progenitors, but rather throughout the embryo. While the loss-of-function phenotype is not reported in mice, this could reflect increased compensation in mammals. On the other hand, most of the morphant Xenopus embryos eventually recover globin-expressing cells and so compromised primitive erythropoiesis in mouse might not be embryonic lethal and might need to be evaluated quantitatively. In summary, although the mechanisms are entirely distinct, STAT5 has positive regulatory functions during the 2 distinct waves of hematopoiesis. As a repressor, we hypothesize that the protein facilitates the development of primitive erythroid cells by modulating indirectly the inhibitory effects of FGF-regulated signaling. Later, it functions as an activator to regulate (directly) the proliferation and survival of definitive erythrocytes downstream of EpoR. It will be of interest to determine the mechanisms by which distinct sets of genes are targeted by STAT5 in these separate developmental contexts.
Prepublished online as Blood First Edition Paper, July 11, 2006; DOI 10.1182/blood-2006-05-022137.
Supported by grants from the National Institutes of Health (NIH) (HL056182) and the Irma T. Hirschl Trust (T.E.), from the Centre National de la Receherche Scientifique (M.U.), and an NIH predoctoral training grant GM07491 (M.S.).
M.S. helped design the study and performed research and assisted in the writing; I.T. performed research; A.P. provided essential experimental assistance by generating most of the constructs used in the study; M.U. provided essential intellectual input and study design, and provided essential reagents for carrying out the experiments; T.E. helped design the study, provided intellectual input, and wrote the manuscript.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Jim Smith, Chris Wright, Doug Melton, Richard Harland, Enrique Amaya, Paul Krieg, and Christof Niehrs for providing reagents. We also thank Brian Zafonte for advice on the quantitative RT-PCR analysis, and John Wallingford's laboratory for assistance during the revision.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal