Abstract
Previous studies showed that most cases of ALK+ anaplastic large-cell lymphoma (ALK+ALCL) do not express SHP1, a tyrosine phosphatase and an important negative regulator for cellular signaling pathways such as that of JAK/STAT. To fully assess the biologic significance of loss of SHP1 in ALK+ALCL, we transfected SHP1 plasmids into 2 SHP1-, ALK+ALCL cell lines, Karpas 299 and SU-DHL-1. After 24 hours of transfection, pJAK3 and pSTAT3 were decreased, and these changes correlated with down-regulation of STAT3 downstream targets including cyclin D3, mcl-1, and bcl-2. Expression of SHP1 in these 2 cell lines also resulted in marked decreases in the protein levels of JAK3 and NPM-ALK, and these effects were reversible by proteosome inhibitor MG132. Conversely, when SHP1 expression in SUP-M2 (a SHP1+ ALK+ALCL cell line) was inhibited using siRNA, pSTAT3, pJAK3, JAK3, and NPM-ALK were all up-regulated. Coimmunoprecipitation studies showed that SHP1 was physically associated with JAK3 and NPM-ALK. SHP1 expression in Karpas 299 and SU-DHL-1 led to significant G1 cell cycle arrest but not apoptosis. To conclude, loss of SHP1 contributes to the pathogenesis of ALK+ALCL by 2 mechanisms: (1) it leaves the tyrosine phosphorylation and activation of JAK3/STAT3 unchecked and (2) it decreases proteosome degradation of JAK3 and NPM-ALK.
Introduction
SHP1 is a non-transmembrane protein tyrosine phosphatase expressed most abundantly in hematopoietic cells.1 It serves as an important negative regulator in cytokine-mediated signal transduction, including that of the JAK/STAT signaling pathway.2-7 Gene methylation and silencing of SHP1 are relatively common in hematologic neoplasms, and a significant proportion of lymphoid malignancies of both B- and T-cell lineages are negative for SHP1 protein expression.8-11 In view of its normal function, loss of SHP1 has been hypothesized to deregulate various signaling pathways and promote lymphomagenesis. Nevertheless, relatively few studies have been performed to directly test this hypothesis and delineate the mechanism by which loss of SHP1 contributes to lymphoma formation. One previous study has shown that transfection of SHP1 in a myeloid leukemia cell line leads to cell cycle arrest, but the biochemical basis of these SHP1-induced changes was not fully examined.12 Using a DNA methyltransferase inhibitor, Chim et al13 reported that SHP1 expression in myeloma cell lines correlated with a reduction in the tyrosine phosphorylation of STAT3. However, DNA methyltransferase inhibitors are not gene specific,14 and the relationship between SHP1 expression and down-regulation of STAT3 activation cannot be definitely established. In addition to its function as a tyrosine phosphatase, SHP1 also has been implicated in the regulation of the rate of proteosome degradation of certain cellular proteins,15,16 but the biologic significance of this relatively novel function of SHP1 has not been extensively studied in cancers.
ALK+ anaplastic large-cell lymphoma (ALK+ALCL) is a distinct type of non-Hodgkin lymphoma, characterized by CD30 expression, a sinusoidal infiltrative pattern, T/null-cell immunophenotype, and the expression of the oncogenic fusion chimeric protein, NPM-ALK.17,18 NPM-ALK is the result of the chromosomal translocation, t(2;5)(p23;q35), that juxtaposes the nucleophosmin (NPM) gene at 5q35 with the anaplastic lymphoma kinase (ALK) gene at 2p23.19 Recent studies have shown that NPM-ALK directly contributes to the pathogenesis of ALK+ALCL, by exerting its tyrosine kinase activity embodied in the ALK portion of the fusion protein and thereby deregulating multiple signaling pathways.20-24 One of these signaling pathways is the STAT3 pathway.25-27 STAT3 has been shown to be oncogenic in a wide variety of human cancers,28-33 and ample evidence indicates that STAT3 activation is crucial to the pathogenesis of ALK+ALCL.25,27,34 Although it is likely that NPM-ALK mediates oncogenesis via STAT3 activation, previous data also suggest that activation of STAT3 in ALK+ALCL is multifactorial. For instance, we previously showed that JAK3 (a physiologic activator of STAT3) is frequently activated in ALK+ALCL,35 and JAK3 contributes to STAT3 activation in ALK+ALCL.36 Zhang et al26 provided evidence to support the model of “multi-level dysregulation of STAT3 activation” in ALK+ALCL. With regard to SHP1, one of our previous studies identified gene methylation of SHP1 and loss of SHP1 expression in the vast majority of ALK+ALCL tumors.37 Because SHP1 has been implicated in JAK/STAT signaling, it is possible that loss of SHP1 potentiates JAK3/STAT3 signaling in ALK+ALCL cells and contributes to the lymphomagenesis in this cell type. This hypothesis, however, has not been directly tested. The less understood role of SHP1 as a modulator of proteosome degradation also has not been examined in ALK+ALCL.
To directly assess the biologic importance of loss of SHP1 in ALK+ALCL, we transfected 2 ALK+ALCL cell lines, Karpas 299 and SU-DHL-1, with specific SHP1 expression vectors. In view of the central pathogenetic roles of NPM-ALK, JAK3, and STAT3 in these tumors, we focused our analysis on these proteins with regard to their expression or tyrosine phosphorylation status. Whether restoration of SHP1 expression in ALK+ALCL cells can modulate degradation of these proteins was evaluated. These changes were then correlated with changes in cell proliferation and apoptosis.
Materials and methods
Cell lines, tissue culture, and reagents
Three ALK+ALCL cell lines, Karpas 299, SU-DHL-1, and SUP-M2, were included in this study. These cell lines were maintained at 37°C in RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum, 10 000 U/mL penicillin (Sigma, St Louis, MO), 10 mg/mL streptomycin (Sigma), and 200 mM l-glutamine (Life Technologies) in a humidified atmosphere containing 5% CO2 and 95% O2. MG132, purchased from Calbiochem (EMD Biosciences, San Diego, CA), was initially dissolved in DMSO and further diluted with sterile, deionized water to a final concentration of 1 mM. The aliquoted MG132 was then stored at -20°C. During the experiment, MG132 was thawed and diluted with tissue culture media to a final concentration of 10 μM.
Plasmid constructs and cell transfection
Two different SHP1 plasmids were constructed and used in this study. Full-length SHP1 cDNA was initially synthesized using a primer set including the 5′ end of SHP1 (Xho1)-F: 5′-AAGCCTCGAGGATGGTGAGGT-3′ and the 3′ end of SHP1 (EcoR1)-R: 5′-GCGGAATTCTTCCACAGGGTCA-3′ (Qiagen, Valencia, CA). Subsequently, the full-length SHP1 cDNA was cloned into pCI (Promega, Madison, WI) and pIRES2-EGFP (Clontech, BD Bioscience, Mountain View, CA). After amplification using a kit from Qiagen, the sequence and orientation of the SHP1 insert were confirmed by DNA sequencing using the ABI 3100 gene sequencer (ABI, San Jose, CA). The pIRES2-EGFP plasmid contains an internal ribosomal entry site that allows translation of SHP1 and GFP independently from a single bicistronic mRNA. Cell transfection was performed using a commercially available transfection instrument (Amaxa, Koeln, Germany), following the manufacturer's recommended protocol. Specifically, 2 × 106 SU-DHL-1 cells were transfected with 5 μ vector (SHP1 or empty) in 100 μL Nucleofector V-solution (Amaxa). Transfection for Karpas 299 cells was done similarly, except that only 3 μg vector was used to achieve the most optimal level of transfection.
siRNA to block SHP1 expression
siRNA specific for SHP1 was purchased from Qiagen. SUP-M2, an ALK+ALCL cell line confirmed to be SHP1+ by reverse transcriptionpolymerase chain reaction (RT-PCR) and Western blot in our study, was transfected with siRNA using the Amaxa instrument following the same transfection protocol described (see “Plasmid constructs and cell transfection”). Transfected cells were cultured for 30 hours, and cell lysates were prepared and subjected to Western blot analysis.
Flow cytometry and cell sorting
The pIRES2-EGFP- and pIRES2-EGFP-SHP1-transfected ALK+ALCL cells were subjected to cell cycle analysis as well as cell sorting. For cell cycle analysis, transfected cells were resuspended in a PBS-EDTA buffer, stained with Draq 5 (Biostatus, Shepshed Leicestershire, United Kingdom) according to the manufacturer's protocol, and analyzed using the fluorescence-activated cell sorting cytometer (FACSort; Becton Dickinson, San Jose, CA). The results were then analyzed using the Cell Quest Software (Verity Software House, Topsham, ME). For cell sorting, SHP1-transfected SU-DHL-1 cells were washed twice, high-speed sorted using the EPICS ALTRA (Beckman-Coulter, Mississauga, ON, Canada), and collected into fetal bovine serum. Purities of more than 98% were achieved on analysis of the sorted cell population. The cells were washed with PBS and centrifuged at 201g for 10 minutes. The supernatant was aspirated and the cell pellet was frozen at -80°C. All of the pIRES2-EGFP- or pIRES2-EGFP-SHP1-transfected SU-DHL-1 cells used throughout this study were cell sorted and purified.
Western blot analysis
Western blot analysis was performed using standard techniques. Briefly, the cells were washed in PBS (pH 7.5) and lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 1 mM phenylmethyl sulfonyl fluoride, 5 μg/mL aprotinin (5 μg/mL), sodium vanadate (1 mM), and leupeptin (5 μg/mL). After incubation on ice for 30 minutes, the lysates were subjected to centrifugation at 1608g for 10 minutes at 4°C. The supernatants were collected after the centrifugation. Protein concentration was determined with a protein assay kit (Bio-Rad, Hercules, CA). Each lane of a 6% or 12% polyacrylamide gel was loaded with 80 μg protein. After electrophoresis and transfer to nitrocellulose membranes (Bio-Rad) by electroblotting, blots were probed with specific primary and secondary antibodies and the enhanced chemiluminescence detection system (Amersham, Arlington Heights, IL) according to the manufacturer's protocol. The antibodies used in this study were those reactive with SHP1, STAT3, phospho-STAT3 (pSTAT3), JAK3, phospho-JAK3 (pJAK3), ALK1, cyclin D3, mcl-1, bcl-2, and PARP, all of which were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at a dilution of 1:500. Antibodies reactive with β-actin (Sigma), survivin (Sigma), active caspase-3 (Abcam, Cambridge, MA), and NPM1 (NeoMarkers, Fremont, CA) were also used at a dilution of 1:500.
Quantitative PCR assay to detect SHP1 in ALK+ALCL cell lines
Total cellular RNA was extracted using the RNeasy MINI Handbook Kit (Qiagen). We performed quantitative RT-PCR using a method adapted from a previous report.38 One-step RT-PCR was performed using the TaqMan Gold RT-PCR kit according to the manufacturer's protocol. Total RNA (50 ng) from Karpas 299, SUP-M2, and SU-DHL-1 cells was used as template. The SHP1 primer and probe sequences were as follows: forward: GGAGTCGGAGTACGGGAACAT; reverse: ATCCTCCTTGTGTTTGGACGA; probe: CCCCAGCCATGAAGAATGCCCA. The probe was labeled with FAM reporter dye on the 5′ end and measured at 518 nm wavelength, and the reporter dye was quenched by TAMPRA on the 3′ end. The TaqMan GAPDH control reagents were used. The thermal cycler parameters for SHP1 included 48°C for 30 minutes, 95°C for 10 minutes, and 40 cycles of denaturation (95°C for 15 seconds) and annealing/extension (54°C for 1 minute). The thermal cycler parameters for GAPDH were identical to those for SHP1, except that the annealing/extension condition was 60°C for 1 minute.
ALK+ALCL tumor samples and immunohistochemistry
A total of 17 cases of ALK+ALCL were retrieved randomly from the file at the Department of Laboratory Medicine and Pathology, Cross-Cancer Institute (Edmonton, AB, Canada). The use of these tissues was approved by our institutional ethics committee. All cases were routinely fixed in formalin and embedded in paraffin. Immunohistochemistry was performed and details of the method have been described elsewhere.33 Antibodies used included anti-mcl-1 (sc-819), anti-pJAK3 (sc-16567), and anti-SHP1 (sc-7289), all of which were purchased from Santa Cruz Biotechnology.
Results
SHP1 expression in ALK+ALCL cell lines
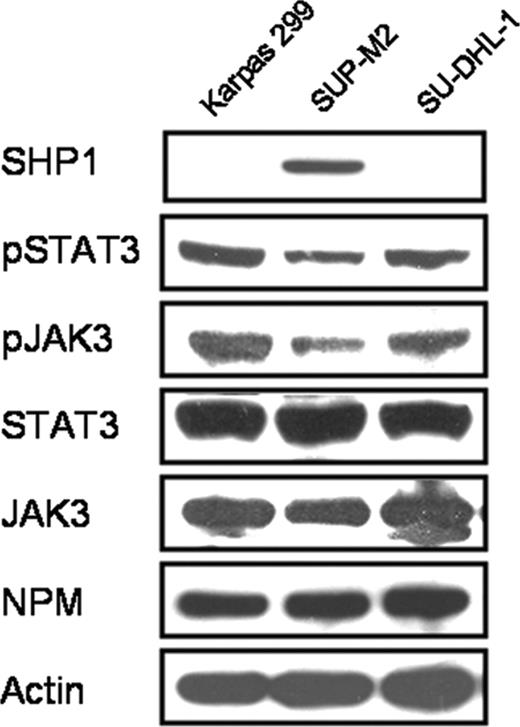
We first confirmed the status of SHP1 expression in 3 ALK+ALCL cell lines including Karpas 299, SU-DHL-1, and SUP-M2. As shown in Figure 1, Karpas 299 and SU-DHL-1 cells showed no detectable SHP1 protein expression on Western blots, whereas SUP-M2 cells expressed SHP1 relatively strongly. Of note, SUP-M2, the SHP1+ ALK+ALCL cell line, had a relatively lower expression of pSTAT3, pJAK3, and JAK3 compared with the 2 SHP1- ALK+ALCL cell lines. No appreciable differences were identified in the protein levels of STAT3 and NPM-ALK among these 3 cell lines. We also performed quantitative RT-PCR assay to measure SHP1 mRNA and the results correlated with those of Western blots, with the average threshold cycle for SUP-M2 being 29, and those for Karpas 299 and SU-DHL-1 being more than 40 (undetectable range).
SHP1 expression resulted in down-regulation of the JAK3/STAT3 pathway
To directly assess the biologic roles of SHP1 in ALK+ALCL cells, SU-DHL-1 cells were transfected with pIRES2-EGFP or pIRES2-EGFP-SHP1. Based on the expression of GFP analyzed by flow cytometry, the transfection efficiency was between 40% and 65%, with a median of 50% for both vectors (based on 5 independent experiments). Negative controls, which were SU-DHL-1 cells transfected with a pCI empty vector, showed no evidence of GFP-expressing cells.
Based on the GFP expression, transfected SU-DHL-1 cells were sorted using flow cytometry, as described in “Materials and methods,” and the purity of GFP-expressing cells was confirmed to be more than 98%. Western blot analysis was performed comparing sorted cells transfected with pIRES2-EGFP and those transfected with pIRES2-EGFP-SHP1. As shown in Figure 2A (left panel), SHP1 was highly expressed in cells transfected with pIRES2-EGFP-SHP1 but absent in those transfected with pIRES2-EGFP. Expression of SHP1 was associated with substantial decreases in the protein levels of pJAK3, JAK3, and pSTAT3. Triplicate experiments were performed and results from a representative experiment are illustrated.
Expression of SHP1, NPM-ALK, JAK3, and STAT3 in 3 ALK+ALCL cell lines, Karpas 299, SUP-M2, and SU-DHL-1. Only SUP-M2 expressed SHP1, and this cell line also had relatively lower levels of pSTAT3, pJAK3, and JAK3 compared with the other 2 cell lines.
Expression of SHP1, NPM-ALK, JAK3, and STAT3 in 3 ALK+ALCL cell lines, Karpas 299, SUP-M2, and SU-DHL-1. Only SUP-M2 expressed SHP1, and this cell line also had relatively lower levels of pSTAT3, pJAK3, and JAK3 compared with the other 2 cell lines.
Although Karpas 299 cells were also transducible with the pIRES2-EGFP-SHP1 plasmid, the transfection efficiency, based on the expression of GFP analyzed by flow cytometry, was only 10% of the cells. Nevertheless, we found that a relatively high transfection efficiency could be obtained if pCI-SHP1 was used instead of pIRES2-EGFP-SHP1. Western blot analysis was performed to compare Karpas 299 cells transfected with pCI-SHP1 and those transfected with the empty vector pCI. Triplicate experiments were performed and results from a representative experiment are illustrated in Figure 2A (right panel); the expression levels of pSTAT3, pJAK3, and JAK3 were decreased in pCI-SHP1-transfected cells compared to the empty pCI-transfected cells. The decrease in pJAK3 was more obvious than that of JAK3. The pattern of changes in the expression of JAK3, pJAK3, and pSTAT3 was thus similar to that seen in SHP1-transfected SU-DHL-1, although the changes were not as prominent in Karpas 299 cells, which can be explained by the fact that cell sorting was used for SU-DHL-1 cells but not for Karpas 299 cells. Compared with that of SU-DHL-1 cells, the short form of pSTAT3 was expressed at a relatively low level in Karpas 299 cells, and this band became undetectable after SHP1 transfection in these cells.
We then determined whether these alterations of the JAK3/STAT3 pathway led to any significant changes in the expression of STAT3 downstream targets, previously shown to be decreased when the STAT3 activity was inhibited by a STAT3 dominant-negative construct.27 These targets often are involved in either cell cycle progression or apoptosis, including cyclin D3, mcl-1, bcl-2 and survivin. As shown in Figure 2B, compared to the negative controls, SU-DHL-1 and Karpas 299 cells transfected with pIRES2-EGFP-SHP1 or pCI-SHP1, respectively, showed down-regulation of mcl-1, bcl-2, and cyclin D3. Relatively small decreases in survivin were also detected in both cell lines. The pattern of change was again similar between SU-DHL-1 and Karpas 299, with more prominent changes seen in SU-DHL-1 cells, again likely due to the fact that cell sorting was done with SU-DHL-1 but not Karpas 299 cells.
Although NPM-ALK is not known to be a STAT3 downstream target, we examined whether SHP1 expression modulates the expression of this oncogenic protein, which has a central pathogenetic role in ALK+ALCL. As shown in Figure 2B, NPM-ALK was substantially down-regulated in both cell lines after SHP1 transfection, compared to their negative controls.
To further confirm the role of SHP1 in down-regulating JAK3/STAT3 signaling and the expression of NPM-ALK, we blocked SHP1 expression in SUP-M2, an ALK+ALCL cell line that expresses a relatively high level of SHP1 at the steady state. As shown in Figure 2C, siRNA decreased SHP1 expression, which was associated with up-regulation of pSTAT3, JAK3, and pJAK3. NPM-ALK and the downstream effectors of STAT3 (such as cyclin D3) were also increased as the SHP1 protein expression was decreased.
SHP1 decreased expression of NPM-ALK and JAK3 via the proteosome pathway
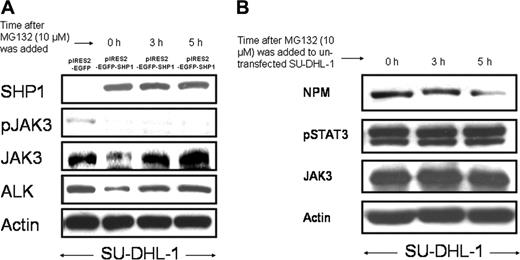
We then questioned whether the SHP1-mediated down-regulation of NPM-ALK and JAK3 is related to the proteosome pathway. SU-DHL-1 cells were treated with 10 μM MG132 at 24 hours after gene transfection with pIRES2-EGFP-SHP1 and cell sorting. As shown in Figure 3A, the initial down-regulation of both JAK3 and NPM-ALK was reversed at 3 to 5 hours after the addition of MG132. However, MG132 did not induce any changes in the pJAK3 level in the same experiment.
SHP1 expression and down-regulation of the JAK3/STAT3 pathway. (A) SHP1 expression induced down-regulation of the JAK3/STAT3 signaling pathway in SU-DHL-1 and Karpas 299 cells. SU-DHL1 cells transfected with pIRES2-EGFP or pIRES2-EGFP-SHP1 were sorted using flow cytometry and subjected to Western blot analysis 24 hours after gene transfection. The results showed substantial decreases in the protein expression of pSTAT3, pJAK3, and JAK3. In contrast, there were relatively few changes in the protein level of STAT3 between the 2 samples. Karpas 299 cells were transfected with pCI (empty vector) and pCI-SHP1, and subjected to Western blot analysis 24 hours after gene transfection. Similar to the SU-DHL-1 cells, Karpas 299 cells showed down-regulation of the JAK3/STAT3 signaling. (B) Modulation of STAT3 downstream targets as well as NPM-ALK in Karpas 299 and SU-DHL-1 cells after SHP1 gene transfection. SHP1 expression in Karpas 299 and SU-DHL-1 cells induced similar changes in the STAT3 downstream targets including bcl-2, mcl-1, and cyclin D3. Survivin was only slightly down-regulated. The protein level of NPM-ALK was also decreased. Cell lysates were prepared 24 hours after gene transfection. Cells transfected with pCI empty vector and pIRES2-EGFP served as negative controls for Karpas 299 and SU-DHL-1 cells, respectively. (C) Blockade of SHP1 expression using siRNA in SUP-M2. Inhibition of SHP1 in SU-DHL-1 cells using siRNA induced down-regulation of the expression of SHP1, with 200 pM more effective than 100 pM. There were increases in the expression of pSTAT3, pJAK3, JAK3, and NPM-ALK. One of the STAT3 downstream targets, cyclin D3, was also up-regulated. SUP-M2 cells transfected with the sense SHP1 siRNA served as negative controls.
SHP1 expression and down-regulation of the JAK3/STAT3 pathway. (A) SHP1 expression induced down-regulation of the JAK3/STAT3 signaling pathway in SU-DHL-1 and Karpas 299 cells. SU-DHL1 cells transfected with pIRES2-EGFP or pIRES2-EGFP-SHP1 were sorted using flow cytometry and subjected to Western blot analysis 24 hours after gene transfection. The results showed substantial decreases in the protein expression of pSTAT3, pJAK3, and JAK3. In contrast, there were relatively few changes in the protein level of STAT3 between the 2 samples. Karpas 299 cells were transfected with pCI (empty vector) and pCI-SHP1, and subjected to Western blot analysis 24 hours after gene transfection. Similar to the SU-DHL-1 cells, Karpas 299 cells showed down-regulation of the JAK3/STAT3 signaling. (B) Modulation of STAT3 downstream targets as well as NPM-ALK in Karpas 299 and SU-DHL-1 cells after SHP1 gene transfection. SHP1 expression in Karpas 299 and SU-DHL-1 cells induced similar changes in the STAT3 downstream targets including bcl-2, mcl-1, and cyclin D3. Survivin was only slightly down-regulated. The protein level of NPM-ALK was also decreased. Cell lysates were prepared 24 hours after gene transfection. Cells transfected with pCI empty vector and pIRES2-EGFP served as negative controls for Karpas 299 and SU-DHL-1 cells, respectively. (C) Blockade of SHP1 expression using siRNA in SUP-M2. Inhibition of SHP1 in SU-DHL-1 cells using siRNA induced down-regulation of the expression of SHP1, with 200 pM more effective than 100 pM. There were increases in the expression of pSTAT3, pJAK3, JAK3, and NPM-ALK. One of the STAT3 downstream targets, cyclin D3, was also up-regulated. SUP-M2 cells transfected with the sense SHP1 siRNA served as negative controls.
To support that the effects of MG132 in our experiments are not due to nonspecific toxicity of this pharmacologic agent, we added MG132 to nontransfected SU-DHL-1 cells. As illustrated in Figure 3B, in the absence of SHP1, MG132 induced no increase in the level of NPM-ALK. Thus, the MG132-mediated up-regulation of JAK3 and NPM-ALK in SHP1-transfected SU-DHL-1 cells is unlikely a nonspecific drug effect. No detectable changes in pSTAT3 and JAK3 were observed. There was a slight decrease in NPM, for which the mechanism is unclear.
SHP1-mediated down-regulation of JAK3 was mediated through tyrosine dephosphorylation and increased proteosome degradation
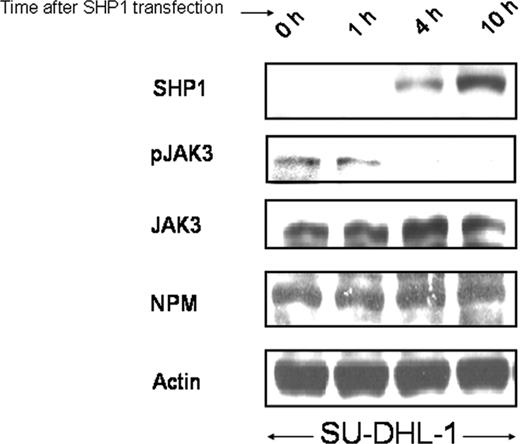
To provide additional evidence that the SHP1-mediated decrease in pJAK3 is not dependent on that of JAK3, we performed a time-course study using SU-DHL-1 cells transfected with pIRES-EGFP-SHP1, and the results are illustrated in Figure 4. The decrease in pJAK3 was detectable as early as 4 hours, at which time the protein expression of the transfected SHP1 became first detectable. Neither JAK3 nor NPM-ALK changed their protein levels within the first 10 hours after SHP1 gene transfection was performed.
Coimmunoprecipitation studies
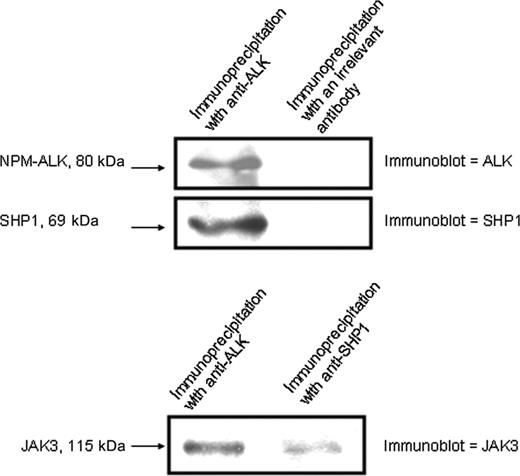
To further substantiate the functional interaction among SHP1 and NPM-ALK, we performed coimmunoprecipitation experiments. We found that transfected SHP1 in SU-DHL-1 coimmunoprecipitated with NPM-ALK (Figure 5). After immunoprecipitation with an anti-ALK antibody, the membrane was first immunoblotted with anti-ALK (upper panel). The same membrane was then immunoblotted with anti-SHP1. These results showed that SHP1 physically interacts with NPM-ALK. We then examined the relationship between JAK3 and NPM-ALK and between JAK3 and SHP1. As shown in Figure 5 (lower panel), JAK3 coimmunoprecipitated with NPM-ALK and SHP1. The overall results indicate that SHP, JAK3, and NPM-ALK physically interact with each other.
Expression of SHP1 induced cell cycle arrest but not apoptosis
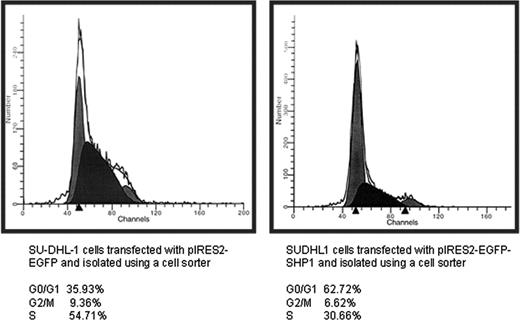
To determine the biologic effects of SHP1 in ALK+ALCL cells, cell cycle analysis using flow cytometry was performed. As shown in Figure 6, SU-DHL-1 cells transfected with pIRES2-EGFP-SHP1 showed evidence of G1 cell cycle arrest. The proportion of cells in the G0/1 phase was 63%, as opposed to 36% in cells transfected with pIRES2-EGFP. Triplicate experiments were performed and the difference was statistically significant (P < .05, Student t test). No significant changes in the sub-G0 phase were identified, indicating that there was no substantial increase in apoptosis.
Effect of MG-132. (A) MG-132 reversed the decrease of NPM-ALK and pJAK3 induced by SHP1. SU-DHL1 cells were transfected with pIRES2-EGFP-SHP1 and sorted based on GFP expression by flow cytometry. MG132 was then added 24 hours after gene transfection. The reduction in the levels of JAK3 and NPM-ALK induced by SHP1 was completely reversed by MG132 at 3 to 5 hours after the addition of MG132 to the cell culture. Cells transfected with pIRES2-EGFP (sorted based on GFP expression) served as negative controls. (B) MG132 did not induce up-regulation of JAK3, pSTAT3, and NPM-ALK in nontransfected SU-DHL cells. In contrast with the SHP1-transfected SU-DHL-1 cells, nontransfected SU-DHL-1 cells treated with MG132 for 3 to 5 hours showed no increase in the protein levels of NPM-ALK, pSTAT3, and JAK3.
Effect of MG-132. (A) MG-132 reversed the decrease of NPM-ALK and pJAK3 induced by SHP1. SU-DHL1 cells were transfected with pIRES2-EGFP-SHP1 and sorted based on GFP expression by flow cytometry. MG132 was then added 24 hours after gene transfection. The reduction in the levels of JAK3 and NPM-ALK induced by SHP1 was completely reversed by MG132 at 3 to 5 hours after the addition of MG132 to the cell culture. Cells transfected with pIRES2-EGFP (sorted based on GFP expression) served as negative controls. (B) MG132 did not induce up-regulation of JAK3, pSTAT3, and NPM-ALK in nontransfected SU-DHL cells. In contrast with the SHP1-transfected SU-DHL-1 cells, nontransfected SU-DHL-1 cells treated with MG132 for 3 to 5 hours showed no increase in the protein levels of NPM-ALK, pSTAT3, and JAK3.
A time-course experiment illustrating that SHP1 dephosphorylated JAK3 independent of down-regulation of the total JAK3 protein. SHP1 slowly increased its protein levels within the first 10 hours after SHP1 gene transfection into SU-DHL-1 cells. A decrease in pJAK3 was detectable at 4 hours, at which time no significant changes in the expression of JAK3 and NPM-ALK were observed.
A time-course experiment illustrating that SHP1 dephosphorylated JAK3 independent of down-regulation of the total JAK3 protein. SHP1 slowly increased its protein levels within the first 10 hours after SHP1 gene transfection into SU-DHL-1 cells. A decrease in pJAK3 was detectable at 4 hours, at which time no significant changes in the expression of JAK3 and NPM-ALK were observed.
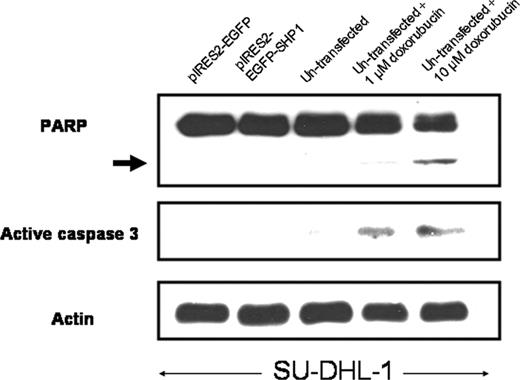
To further assess whether the expression of SHP1 induces apoptosis in both cell lines, we performed Western blots using antibodies reactive with PARP and active caspase-3; no evidence of PARP-cleaved products or increased active caspase-3 was detected in Karpas 299 and SU-DHL-1 cells transfected with SHP1 (illustrated in Figure 7). Nevertheless, SHP1 gene transfection into SU-DHL-1 cells (sorted based on GFP expression) induced a significant decrease in the number of viable cells (assessed by trypan blue exclusion test), with 33% reduction in the number of viable cells 24 hours after gene transfection, and 45% reduction in the number of viable cells 48 hours after gene transfection.
Immunohistochemical studies to compare pSTAT3 and pJAK3 expression in SHP1+ and SHP1- ALK+ALCL tumors
Previous studies have demonstrated that the vast majority of ALK+ALCL tumors are SHP1-; we found that only 14% of ALK+ALCL tumors express the SHP1 protein.37 Based on our in vitro data presented here, one would predict that SHP1-expressing tumors may have a lower level of activation in JAK3/STAT3 signaling compared to the SHP1- tumors. We performed immunohistochemical studies, in which we compared the expression of pSTAT3, pJAK3, and 2 of the STAT3 downstream targets (bcl-2 and mcl-1) in 17 ALK+ALCL tumors (2 SHP1+ and 15 SHP1-). Using more than 10% pSTAT3+ cells as the cut-off, we found that 1 of 2 (50%) SHP1+ tumors was pSTAT3-, whereas only 1 of 15 (7%) SHP1- tumors was pSTAT3-. As for pJAK3, 1 of 2 (50%) SHP1+ tumors was pJAK3+, as opposed to 9 of 11 (82%) SHP1- tumors assessed. As for bcl-2 and mcl-1, both of which are STAT3 downstream targets that are relatively sensitive to the STAT3 activity in vitro,27 no significant differences were detected.
Discussion
SHP1, a tyrosine phosphatase expressed most abundantly in hematopoietic cells, is important in regulating various cellular signaling pathways in lymphocytes.6 The biologic importance of SHP1 is highlighted by the moth-eaten (me) mice, in which SHP1 is not expressed (me/me phenotype) or dramatically decreased (meviable/meviable phenotype). Although me/me mice die shortly after birth, the homozygous meviable mice have patchy hair loss (thus the name `moth-eaten') as a result of sterile dermal abscesses, abnormal myeloid cell function and development, and an increased propensity of developing CD5+ lymphomas.39 In humans, gene silencing/methylation of SHP1 has been demonstrated in many hematologic cancers. In view of the normal biologic functions of SHP1, loss of SHP1 likely contributes to the pathogenesis of these cancers, although mechanistic studies to define the importance of this biochemical defect are relatively lacking. SHP1 has been previously shown to regulate the JAK/STAT pathway.5,40,41 Because loss of SHP1 is frequently found in ALK+ALCL,37 and constitutive activation of JAK3/STAT3 signaling plays an important role in the pathogenesis of ALK+ALCL, we believe that ALK+ALCL is an excellent experimental model to examine the importance of loss of SHP1 in promoting aberrant JAK3/STAT3 activation and lymphomagenesis.
In this study, we first confirmed that both ALK+ALCL cell lines (Karpas 299 and SU-DHL-1) do not have detectable SHP1 protein expression, and these findings correlated with those of the quantitative RT-PCR assay. The absence of SHP1 expression in SU-DHL-1 is in keeping with that of one in a previously published study,8 and in keeping with our previous findings that both of these 2 cell lines have gene methylation of the SHP1 promoter region.37 In the same experiment, we also documented that SUP-M2, another ALK+ALCL cell line, was SHP1 expressing. This cell line was used for siRNA silencing of SHP1 expression.
Using gene transfection with SHP1-specific vectors, we successfully transduced SHP1 expression in both Karpas 299 and SU-DHL-1 cells at a relatively high efficiency. Both of these 2 cell lines are known to be hard to transfect. As far as we know, this is the first study in which relatively high levels of gene transduction using expression vectors were achievable in these cells. We also documented a relatively high rate of siRNA transfection efficiency for SUP-M2 using a similar protocol. The availability of these techniques will likely improve our ability to examine the pathogenesis of ALK+ALCL.
In this study, we used 2 different SHP1 expression vectors for SU-DHL-1 and Karpas 299 because the transfection efficiency appeared to be dependent on the specific cell lines as well as plasmids. Although we were able to achieve relatively high levels of SHP1 expression in both cell lines, only the pIRES2-EGFP-SHP1-transfected SU-DHL-1 cells can be sorted by a flow cytometer and purified for analysis. Despite these experimental differences, both cell lines showed similar patterns of results; enforced expression of SHP1 completely abrogated the pJAK3 level in the sorted, SHP1-expressing SU-DHL-1 cells and dramatically decreased the pJAK3 level in the unsorted, SHP1-expressing Karpas 299 cells. These findings are in keeping with the concept that SHP1 serves as a tyrosine phosphatase for JAK3,1,2,40 thereby negatively regulating JAK3 function. The decrease in pJAK3 was more prominent in SU-DHL-1 cells than in Karpas 299, which can be explained by the fact that the SHP1-transfected Karpas 299 cells were not sorted and thus contaminated with nontransfected cells. Interestingly, the total JAK3 level was also substantially decreased after SHP1 expression in both cell lines. The extent of reduction in JAK3 was relatively small compared to that of pJAK3 in both cell lines, indicating that the decrease in pJAK3 is independent of that of JAK3. To support this concept, results from the time-course studies showed that pJAK3 was down-regulated much sooner than JAK3 after SHP1 transfection. Thus, down-regulation of pJAK3 cannot be explained by the decrease in the JAK3 total protein. Taken together, our results led us to conclude that down-regulation of pJAK3 was due to both SHP1-mediated tyrosine dephosphorylation and SHP1-induced down-regulation of the total JAK3 protein.
SHP1 coimmunoprecipitated with NPM-ALK and JAK3. Coimmunoprecipitation studies indicated that SHP1 physically interacts with NPM-ALK and JAK3.
SHP1 coimmunoprecipitated with NPM-ALK and JAK3. Coimmunoprecipitation studies indicated that SHP1 physically interacts with NPM-ALK and JAK3.
SHP1 induced G1cell cycle arrest and a decrease in the number of viable cells. Cell cycle analysis for SU-DHL1 cells transfected with pIRES2-EGFP and pIRES2-EGFP-SHP1. Cells were harvested 24 hours after gene transfection and subjected for cell sorting based on GFP expression. All GFP+ cells were then subjected to cell cycle analysis. SHP1 induced a significant increase in the G0/1 population, indicating cell cycle arrest.
SHP1 induced G1cell cycle arrest and a decrease in the number of viable cells. Cell cycle analysis for SU-DHL1 cells transfected with pIRES2-EGFP and pIRES2-EGFP-SHP1. Cells were harvested 24 hours after gene transfection and subjected for cell sorting based on GFP expression. All GFP+ cells were then subjected to cell cycle analysis. SHP1 induced a significant increase in the G0/1 population, indicating cell cycle arrest.
In addition to JAK3 and pJAK3, SHP1 also effectively down-regulated STAT3 activation in both cell lines. These changes can be partly attributed to the decrease in the protein level and activation of JAK3, a physiologic activator of STAT3. Using pharmacologic inhibitors for JAK3, we have previously demonstrated that JAK3 potentiates STAT3 activation in ALK+ALCL cells.36 Importantly, NPM-ALK, a potent activator of STAT3,25 was also decreased by SHP1. Thus, there are at least 2 mechanisms by which SHP1 can inhibit STAT3 activation: (1) tyrosine dephosphorylation of JAK3 and (2) down-regulation of the protein levels of JAK3 and NPM-ALK. Other mechanisms may be involved. A recent study by Honorat et al also showed that NPM-ALK is a substrate of SHP1, and SHP1 mediates dephosphorylation of NPM-ALK.42 We also do not exclude the possibility that SHP1 may directly inactivate STAT3 because it has been reported that these 2 proteins can physically interact with each other in some cell types.43
One of the most important observations of this study is that of SHP1-mediated down-regulation of JAK3 and NPM-ALK in ALK+ALCL via the proteosome degradation pathway. Using MG132, a proteosome inhibitor, we found evidence that SHP1 promotes proteosome-mediated proteolysis of both of these 2 proteins. We also found that SHP1-induced down-regulation of pJAK3 was not reversed by MG132 after the restoration of JAK3 expression, further supporting the concept that tyrosine dephosphorylation of JAK3 by SHP1 can be dissociated with the regulation of the JAK3 protein level. Nevertheless, because MG132 is a nonspecific pharmacologic agent, further studies are needed to substantiate these findings and to delineate how SHP1 may regulate the rate of proteosome degradation of JAK3 and NPM-ALK. Our coimmunoprecipitation study showed that SHP1 indeed physically interacts with both JAK3 and NPM-ALK, probably forming a complex. The physical interaction between SHP1 and NPM-ALK also has been shown in the recent study by Honorat et al.42 In view of the tyrosine phosphatase property of SHP1, it is possible that SHP1 may modulate the interaction between NPM-ALK/JAK3 with proteins in the proteosome pathway by regulating the tyrosine phosphorylation status of these proteins. Of note, a previous study that examined NPM-ALK-binding partners using mass spectrometry did not identify SHP1,44 and we believe that this finding is related to the fact that Karpas 299 is a SHP1- cell line.
We also examined whether there were any changes to the STAT3 downstream targets in ALK+ALCL revealed in our previous study. With only a partial down-regulation of pSTAT3 after SHP1 expression, it is not surprising to observe that only a subset of the examined STAT3 downstream targets was down-regulated. There were substantial decreases in mcl-1, bcl-2, and cyclin D3, and, to a lesser extent, survivin. This pattern of changes suggests that the expression of various STAT3 downstream targets is dependent on different STAT3 activation levels. Thus, a relatively low level of STAT3 activation is sufficient to activate survivin but not mcl-1 and cyclin D3. Down-regulation of cyclin D3 correlates with the G1 cell cycle arrest. In view of the importance of survivin in antiapoptosis,45,46 it is possible that the lack of apoptosis in our experimental models is due to the relatively high level of survivin that persists in the presence of SHP1.
SHP1 induced no significant apoptosis. In addition to the lack of an increase in the sub-G0/1 cell population, SHP1 expression in SU-DHL-1 cells also did not lead to detectable cleaved PARP product (arrow) and active caspase-3. Nontransfected SU-DHL-1 cells treated with different doses of doxorubicin served as positive controls.
SHP1 induced no significant apoptosis. In addition to the lack of an increase in the sub-G0/1 cell population, SHP1 expression in SU-DHL-1 cells also did not lead to detectable cleaved PARP product (arrow) and active caspase-3. Nontransfected SU-DHL-1 cells treated with different doses of doxorubicin served as positive controls.
Overall, our in vitro findings support the hypothesis that loss of SHP1 significantly contributes to the aberrant activation of JAK3/STAT3 and lymphomagenesis of ALK+ALCL. At the time when this manuscript was revised, Honorat et al reported that NPM-ALK is a substrate of SHP1 and that SHP1-mediated tyrosine dephosphorylation of NPM-ALK leads to decreased cell growth and tumorigenecity.42 Thus, data from Hororat et al are in parallel with those of our study, supporting that loss of SHP1 plays important pathogenetic roles in ALK+ALCL.
Because SHP1 expression in ALK+ALCL tumors is heterogeneous, the biologic importance of SHP1 in these tumors can be further validated, by comparing both clinical and biologic parameters in SHP1-expressing and SHP1- ALK+ALCL tumors. Our findings from the pSTAT3 and pJAK3 immunostaining in 17 cases of ALK+ALCL tumors are in keeping with the concept that loss of SHP1 may be associated with a higher percentage of pSTAT3+/pJAK3+ cells. Nevertheless, due to the relatively small sample size in this study, future studies including a large number of cases are needed to draw definitive conclusions. With regard to the relationship between STAT3 and SHP1, a recent study suggests that STAT3 plays direct roles in contributing to gene methylation and silencing of SHP1 in ALK+ALCL.47 Thus, STAT3 activation and loss of SHP1 may, in fact, form a vicious positive feedback loop, in which STAT3 activation promotes gene silencing of SHP1, and the loss of SHP1 potentiates STAT3 activation. In view of the fact that constitutive activation of STAT3 and loss of SHP1 are relatively common in lymphoid malignancies other than ALK+ALCL, disruption of the STAT3/SHP1 interaction may be a potentially useful therapeutic approach in treating various types of hematologic malignancies.
Prepublished online as Blood First Edition Paper, July 6, 2006; DOI 10.1182/blood-2006-04-017434.
Supported by research grants from the Alberta Cancer Board and the University of Alberta Hospital Research Foundation (R.L.). H.A. is a recipient of the University of Texas M. D. Anderson Physician Scientist Program Award.
Y.H., H.A., and R.L. designed the experiments; Y.H., B.F., X.S., and C.F. performed all the experimental procedures; Y.H., H.A., and R.L. analyzed the data; H.A. and R.L. reviewed the immunostaining results; and Y.H. and R.L. wrote the manuscript.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal