Abstract
uPA (urokinase-type plasminogen activator) activates plasminogen with high efficiency when bound to its cellular receptor uPAR, but only after a prolonged lag phase during which generated plasmin activates pro-uPA. How the activity of this proteolytic system might be rapidly initiated is unknown. We have now found that 2 monocytic cell lines display distinct patterns of plasminogen activation. U937 cells, but not THP-1 cells, displayed the expected lag phase, suggesting a constitutive initiation mechanism on the latter. This was shown to be due to the plasmin-independent activation of uPAR-bound pro-uPA by a cell surface-associated protease and to correlate with the expression of matriptase, a type II transmembrane serine protease that was highly expressed in THP-1 cells but undetectable in U937 cells. Kinetic analysis demonstrated that matriptase is a relatively poor activator of pro-uPA in solution, approximately 100-fold less efficient than plasmin (kcat/Km 1.16 × 105 M-1s-1 cf 1.21 × 107 M-1s-1). However, down-regulation of matriptase expression in THP-1 cells by siRNA reduced the activation of cell-associated pro-uPA and the subsequent rapid initiation of plasminogen activation by 76% to 93%. Matriptase was also found to be expressed by peripheral blood monocytes and may therefore be a specific mechanism for the rapid initiation and regulation of plasminogen activation by these cells.
Introduction
uPAR, the specific GPI (glycosyl-phosphatidylinositol)-anchored cellular receptor for uPA (urokinase-type plasminogen activator), is expressed by most types of leukocytes1 and exerts multiple effects on the adhesion and migration of these cells during their recruitment from the circulation to extravascular sites of inflammation.2 Proteolytic activity generated by binding of uPA to uPAR and the subsequent generation of plasmin contribute to this process, facilitating cell migration and degrading the fibrin associated with inflammatory sites.
Although fibrinolysis is only mildly impaired in uPAR-deficient mice,3,4 uPAR has been shown to be necessary for the proteolytic activity of the uPA in various model systems in vivo,5,6 confirming in vitro observations that the presence of uPAR on the cell surface greatly increases plasminogen activation.7,8 These studies, primarily using the monocytic cell line U937 as a model, show that uPAR-bound uPA generates plasmin with high efficiency, due to a large reduction in the apparent Km for cell-associated plasminogen.7,8 In common with other serine proteases, uPA is synthesized as a single-chain zymogen, with negligible activity.9,10 When pro-uPA is the uPAR ligand, the effect on plasmin generation is even more pronounced, as a consequence of the activation of pro-uPA by plasmin in a process of reciprocal zymogen activation.10-12 Plasmin generated by the action of uPA activates pro-uPA, and the catalytic efficiency of this reaction is also increased by uPAR on the cell surface.13 Under these conditions, plasmin generation is initially undetectable but, after a substantial lag phase, increases exponentially finally achieving a rate equivalent to that observed with tc-uPA (activated, 2-chain uPA). The initial proteolytic event leading to the initiation of plasmin generation on the cell surface is thought to come from the low intrinsic catalytic activity of pro-uPA, and a mechanistic model has shown that this activity can quantitatively account for the initiation of plasmin generation on the cell surface.14 This conclusion is supported by cellular studies performed in the presence of plasminogen activator inhibitors.15,16 However, although plasmin generation can be initiated in the absence of activated proteases by this mechanism, these observations do not exclude the existence of alternative mechanisms for the rapid initiation of the activity of this proteolytic system.
In contrast to most serine protease zymogens, pro-uPA is promiscuous with respect to its activation; as in purified systems, a wide range of both serine and nonserine proteases can catalyze this reaction (reviewed in Ellis17 ), including the catalytic domain of the TTSP (type II transmembrane serine protease) matriptase.18,19 Although this suggests that these proteases may contribute to the initiation of plasminogen activation, there is no evidence that plasmin-independent activation of pro-uPA has a role in plasminogen activation. For example, the uPA that accumulates in conditioned media of many cell types in the absence of plasmin(ogen) is almost exclusively in the single-chain form,9,20,21 and pro-uPA activation in an experimental model in knock-out mice has been shown to be dependent on the presence of both plasminogen and uPAR.5 Therefore, if other proteases do contribute to the initiation of plasminogen activation, it may be expected to occur only under very specific conditions, perhaps restricted to the activation of uPAR-bound pro-uPA at the cell surface.
We have observed that when a directly GPI-anchored form of uPA is expressed in the monocytic cell lines U937 and THP-1, the plasminogen activation behavior of the 2 cell lines is quite different.22 In U937 cells, the activity of the GPI-anchored uPA is indistinguishable from uPAR-bound pro-uPA, displaying an extended lag phase during which pro-uPA becomes activated by generated plasmin. By contrast, in THP-1 cells, the GPI-anchored uPA activates plasminogen with no detectable lag phase, plasmin generation being essentially linear from the start of the reaction and comparable with that observed with active tc-uPA. These observations suggest that THP-1 cells express a plasmin-independent proteolytic activator of pro-uPA. This putative activity appears to possess properties ideal for a proteolytic initiator of the cell surface plasminogen activation system, being efficient, active at the cell surface, and potentially constitutively active.
In the present study, we establish that THP-1 cells rapidly activate uPAR-bound pro-uPA in a plasmin-independent manner, leading to the constitutive initiation of plasminogen activation. This pro-uPA activation activity is membrane associated and correlates with the expression of matriptase. Kinetic analysis demonstrates that matriptase is an intrinsically poor activator of pro-uPA in comparison with plasmin. Nevertheless, matriptase is the protease responsible for the specific activation of pro-uPA on these cells as this activity was abolished by transfection with matriptase-specific siRNA. Therefore, our data demonstrate that THP-1 cells, but not U937 cells, possess a matriptase-dependent mechanism for the specific proteolytic initiation of pericellular plasminogen activation. We also demonstrate that matriptase is expressed by peripheral blood monocytes and B cells, and therefore this may be a mechanism by which plasminogen activation is regulated by these cells.
Materials and methods
Cells and cell culture
THP-1 and U937 cells (American Type Cell Culture [ATCC], Manassas, VA) were routinely grown in full RPMI-1640 medium containing l-glutamine and 10% fetal calf serum. All cell culture media were from Invitrogen (Paisley, United Kingdom).
Proteins and antibodies
Pro-uPA was expressed in Drosophila S2 cells and purified by immunoaffinity chromatography. Active tc-uPA was prepared by activation of pro-uPA with Sepharose-immobilized plasmin (Merck Biosciences, Nottingham, United Kingdom). Native Glu-plasminogen was purified as previously described.7 Soluble uPAR expressed in S2 cells was a gift of Dr Michael Ploug (Finsen Laboratory, Copenhagen, Denmark). The monoclonal antibody M32 recognizing the third LDL receptor domain of human matriptase was a gift of Dr Chen-Yong Lin (Georgetown University, Washington, DC), and the rabbit antimatriptase polyclonal antibody IM1014 was from Calbiochem (Nottingham, United Kingdom). The anti-uPAR monoclonal R4 was a gift of Dr Gunilla Høyer-Hansen (Finsen Laboratory).
Measurement of cell-associated plasminogen activation
Plasmin generation by uPAR-bound pro-uPA was determined essentially as described previously.7,8 Briefly, cells were washed in RPMI 1640 buffered in 25 mM HEPES (pH 7.4), and incubated for 5 minutes with 0.05 M glycine hydrochloride (pH 3.0), 0.1 M NaCl to dissociate endogenously bound uPA, followed by neutralization and further washing. The cells were resuspended at a density of 1 × 107/mL and incubated with either uPA or pro-uPA for 20 minutes at 37°C, followed by washing. Cells were incubated at a final density of 1 × 106/mL in 0.05 M Tris-HCl (pH 7.4), 0.1 M NaCl, 0.01% Tween-80 in the presence of 0.2 mM H-d-Val-Leu-Lys-AMC (7-amino-4-methyl coumarin; Bachem, St Helens, United Kingdom) and 50 nM plasminogen at 37°C. Plasmin generation was determined by recording AMC fluorescence intensity at 60-second intervals, using a PerkinElmer LS-5B luminescence spectrometer (PerkinElmer, Beaconsfield, United Kingdom).
Determination of plasmin generation and data fitting
Plasmin generation was quantified as change in fluorescence/min versus time. In experiments where pro-uPA was the activator initially bound to cellular uPAR, these data were fitted to the previously described kinetic model for plasminogen and pro-uPA reciprocal zymogen activation kinetics.14,23 The derived data were used to estimate the amount of activated uPA initially present prior to the addition of plasminogen.
Measurement of plasmin-independent pro-uPA activation
To determine the activation of pro-uPA on U937 and THP-1 cells in the absence of plasminogen, cells were incubated with uPA or pro-uPA and washed as described in “Measurement of cell-associated plasminogen activation,” then assayed directly for uPA activity using H-Glu-Gly-Arg-AMC (0.2 mM). Substrate hydrolysis at 37°C was monitored continuously using a PerkinElmer LS-5B luminescence spectrometer.
Subcellular fractionation
THP-1 or U937 cells (1 × 108 cells) were harvested, washed, and resuspended in 10 mM HEPES (pH 7.4), 0.34 M sorbitol in the absence of protease inhibitors. Cells were disrupted by sonication for 3 × 20-second bursts at 4°C. The postnuclear supernatant, recovered by centrifugation at 1000g for 10 minutes, was then layered onto a continuous density gradient of 0.5 to 1.8 M sucrose in 10 mM HEPES (pH 7.4). Fractions were collected after centrifugation at 40 000g for 2 hours at 4°C. The fractions were assayed for pro-uPA-activating activity by incubation with pro-uPA (5 nM) and the uPA-specific chromogenic pyroGlu-Gly-Arg-p-nitroaniline (0.2 mM, S-2444; Quadratech, Epsom, United Kingdom) in 0.05 M Tris-HCl (pH 7.8), 0.1 M NaCl, 0.01% Tween-80 at 37°C. Substrate hydrolysis was measured as the absorbance ratio 405:490 nm using a ThermoMax microplate reader (Molecular Devices, Sunnyvale, CA). Plasma membrane fractions were identified either by prior biotinylation of cell surface proteins using sulfo-NHS-biotin (Pierce Biotechnology, Rockford, IL) and detection by Western blotting using horseradish peroxidase-conjugated streptavidin (Amersham Biosciences, Chalfont St Giles, United Kingdom) or by assay of alkaline phosphatase activity using p-nitrophenylphosphate (Sigma-Aldrich, Poole, United Kingdom).
Expression and purification of matriptase protease domain
The catalytic domain of matriptase was polymerase chain reaction (PCR) amplified from T47D cDNA using the forward primer 5′-CGGGATCCTGCGACTGTGGGCTGCGGTCAT-3′ and the reverse primer 5′-AACTGCAGCTATACCCCAGTGTTCTCTTTGATCC-3′. The product was digested with BamH1 and Pst1, ligated into the pQE-30 vector (Qiagen, Crawley, United Kingdom), and transformed into Escherichia coli M15[pREP4] (Qiagen). IPTG-induced cells were harvested and resuspended in 0.2 M Tris-HCl (pH 8.0), 100 mM NaCl, 0.1 mg/mL DNase (Sigma). Inclusion bodies were solubilized in wash buffer (100 mM NaH2PO4 [pH 8.0], 10 mM Tris, 8 M urea). This solution was incubated with Ni-NTA agarose (Qiagen) at 4°C for 1 hour. The slurry was poured into a column and washed with 2 column volumes of wash buffer. The column was further washed using wash buffer at pH 5.9, and matriptase eluted in the same buffer at pH 4.5. Urea was removed by dialysis against 0.1 M Tris-HCl (pH 8.0), 0.2 M NaCl. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting analysis revealed the matriptase catalytic domain to be 95% pure and in the activated 2-chain form (data not shown).
Pro-uPA activation kinetics
Matriptase and plasmin were quantified by active-site titration with 4-methylumbelliferyl p-guanidobenzoate (Sigma). Activation of pro-uPA by plasmin and the catalytic domain of matriptase was determined essentially as described previously.23 Briefly, varying concentrations of pro-uPA (0.1-5 μM) were incubated with plasmin (10 pM) or matriptase (0.3 nM) in 0.05 M Tris-HCl (pH 7.8), 0.1 M NaCl, 0.01% Tween-80 containing H-Glu-Gly-Arg-AMC (0.2 mM) at 37°C. In experiments where the effect of soluble uPAR was assessed, this was included at a concentration equimolar with pro-uPA. Change in fluorescence was read continuously in a SpectraMax Gemini microplate reader (Molecular Devices). uPA generation was quantified as the rate of change of fluorescence by comparison with active site-titrated uPA. Kinetic parameters were calculated by nonlinear regression.
Western blot analysis of matriptase
Cells were harvested by centrifugation and pellets lysed in reporter lysis buffer (Promega, Madison, WI) containing complete protease inhibitor cocktail (Roche, Mannheim, Germany). Conditioned medium was concentrated 15-fold using Ultrafree-MC centrifugal filter devices (Millipore, Hertfordshire, United Kingdom). Protein concentration was determined by micro BCA protein assay (Pierce Biotechnology). Nonreduced samples containing 10 μg protein or 10 μL concentrated conditioned medium were subjected to SDS-PAGE on 10% gels and transferred to PVDF membranes (Bio-Rad, Hemel Hempstead, United Kingdom). After incubation in blocking buffer (0.05 M Tris-HCl [pH 7.4], 0.1 M NaCl, 0.01% Tween-20, 5% skimmed-milk powder), blots were incubated with 1:1500 dilution of the monoclonal antibody M32. Blots were washed in 0.05 M Tris-HCl (pH 7.4), 0.1 M NaCl, 0.01% Tween-20 followed by incubation with 1:1000 dilution of horseradish peroxidase-conjugated anti-mouse IgG (DAKO, Glostrup, Denmark) in blocking buffer. The blots were washed and chemiluminescence was detected using enhanced chemiluminescence (ECL) Plus (Amersham Biosciences).
qRT-PCR (quantitative real-time PCR) analysis
qRT-PCR was used to measure matriptase mRNA expression in THP-1 and U937 cells, and peripheral blood leukocytes. Total RNA was extracted using Trizol (Invitrogen). RNA (1 μg) was reverse transcribed using 2 μg random hexamers (Amersham Biosciences) and Superscript II reverse transcriptase (Invitrogen). The fluorogenic probe, forward primer, and reverse primer were 5′-CAAAACAGTACAGAGGACCCAGGACAACAGC-3′, 5′-CACCTCAGTGGTGGCTTTCC-3′, and 5′-GCGTGCAGGCCAAAGCT-3′, respectively. The 18S ribosomal RNA gene was used as an endogenous control. PCR was performed using an ABI Prism 7700 (Applied Biosystems, Foster City, CA). Each reaction was performed in 25 μL and contained 5 ng reverse-transcribed RNA (1 ng RNA for the 18S analyses), 32% TaqMan 2 × PCR Master Mix (Applied Biosystems), 100 nM of each primer, and 200 nM probe. Conditions for the PCR were 2 minutes at 50°C, 10 minutes at 95°C, and then 40 cycles, each consisting of 15 seconds at 95°C, and 1 minute at 60°C. Standard curves and relative RNA levels were determined as described previously.24
Immunofluorescence microscopy
THP-1 cells were put on glass slides by cytocentrifugation using 0.5 × 106 cells per slide. Cells were fixed with 4% formaldehyde, blocked with 0.1 M glycine/PBS followed by 1% BSA/PBS. Incubations with primary antibodies were for 12 hours at 4°C using 10 μg/mL rabbit polyclonal anti-matriptase IM1014 and 20 μg/mL mouse monoclonal anti-uPAR R4 in 1% BSA/PBS, followed by washing in 0.1% Tween-20/PBS. Fluorescein-conjugated goat anti-mouse IgG and rhodamine red-X-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, PA) at 30 μg/mL were applied for 30 minutes at 37°C, and cells were washed again. Slides were mounted with Vectashield (Vector Laboratories). Cells were visualized using a Zeiss Plan-Apochromat 20 ×/0.8 numeric aperture lens on a Zeiss Axiovert 200M wide-field microscope (Carl Zeiss, Heidelberg, Germany), with a 100W mercury arc lamp light source and dichroic filter sets for FITC and Alexa 546. Images were acquired using a CoolSNAP charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) and were processed using Zeiss Axiovision 4 and Adobe Photoshop 7 (Adobe Systems, San Jose, CA) software.
Fluorescence-activated cell sorter (FACS) analysis
Human peripheral blood mononuclear cells (PBMCs) were isolated using standard Ficoll-Hypaque (Amersham Pharmacia Biotech, Uppsala, Sweden) density gradient centrifugation from venous blood of healthy consenting volunteers, in accordance with institutional guidelines. Matriptase expression was assessed by flow cytometry (FACScan; BD Biosciences, San Jose, CA) using rabbit anti-matriptase IM1014 (25 μg/mL), followed by PE-conjugated goat anti-rabbit IgG (100 ng/200 000 cells; Cedarlane, Hornby, ON). Multicolor flow cytometry was applied to evaluate matriptase expression on specific immune cell subsets by adding conjugated antibodies to markers of T cells (CD3), B cells (CD19), and monocytes (CD14), or corresponding isotype controls (all from BD-Pharmingen, San Diego, CA). A total of 10 000 events per sample was analyzed (CellQuest software; BD Biosciences) in each experiment.
Inhibition of matriptase expression using siRNA (small-interfering RNA duplexes)
siRNAs were generated against matriptase. Mat162 (5′-CAACGUCAAGAAGGUGGAAdTdT-3′) targeted nucleotides 126 to 144 downstream of the start codon, and mat1725 (5′-CGUCGUCACUUGUACCAAAdTdT-3′) targeted nucleotides 1689 to 1707. A nontargeting duplex matscram (5′-UAACGUCGAGAAGGCGGAAdTdT-3′) contained mutations in the mat162 sequence. siRNAs were synthesized by Qiagen as duplexes with a 3′ fluorescein label to enable visualization of transfection efficiency (data not shown). Transfection was performed using Oligofectamine (Invitrogen) in 6-well plates according to the manufacturer's instructions. Briefly, 24 hours prior to transfection, cells were plated at 5 × 105 cells/well in 2 mL RPMI-1640, 10% fetal calf serum. Oligofectamine (5 μL) was diluted in 10 μL serum-free OPTIMEM medium (Invitrogen), added to 10 μL of 20 μM siRNA, diluted in 165 μL OPTIMEM, incubated at room temperature for 25 minutes, and added to 2 mL of cells in full RPMI. In various experiments, siRNAs were used both singly and in combination. Measurements of cell-associated plasminogen activation or real-time PCR analysis were performed 24 hours after transfection.
Results
THP-1 and U937 cells display differential plasminogen activation
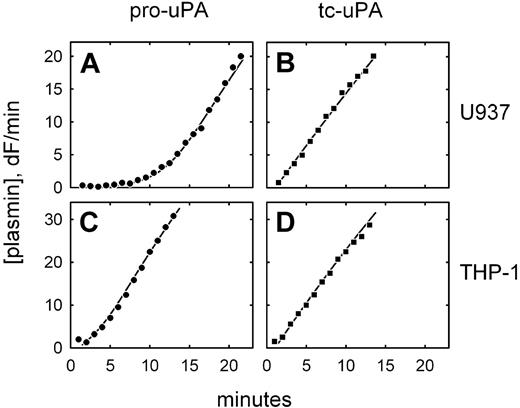
We have previously observed that 2 monocytic cell lines, U937 and THP-1, expressing a directly GPI-anchored uPA construct, each activate plasminogen with distinctly different characteristics.22 To determine whether this also occurs with uPAR-bound pro-uPA, similar experiments were performed on native, untransfected cells. Figure 1A shows that U937 cells preincubated with pro-uPA display an extended lag phase in plasmin generation, during which pro-uPA is activated by trace amounts of plasmin, followed by a rapid acceleration of plasmin generation (ie, a process of reciprocal zymogen activation).13,14 This mechanism is highlighted by the linear plasmin generation observed when active tc-uPA was initially bound to the cells (Figure 1B). By contrast, under identical conditions, no lag phase was detectable when THP-1 cells were preincubated with pro-uPA (Figure 1C), comparable with what was observed with active uPA (Figure 1D). This suggests that uPAR-bound pro-uPA has become proteolytically activated prior to the addition of plasminogen by a plasmin-independent mechanism.
Differential plasminogen activation on U937 and THP-1 cells. Plasmin generation is shown for U937 cells (A-B) and THP-1 cells (C-D) after incubation of the cells with either pro-uPA (A,C) or active, 2-chain uPA (B,D). The data are shown fitted to integrated rate equations describing the plasminogen activation system and using previously published kinetic constants. In the case of U937 cells, the data are consistent with 100% of the bound uPA being either in the active or pro form; in the case of THP-1 cells, the best fit of the data in the presence of pro-uPA is achieved by assuming that 50% of the bound uPA is initially present in the activated form. Plasminogen activation was not detected on cells preincubated with the blocking anti-uPAR monoclonal antibody R3.
Differential plasminogen activation on U937 and THP-1 cells. Plasmin generation is shown for U937 cells (A-B) and THP-1 cells (C-D) after incubation of the cells with either pro-uPA (A,C) or active, 2-chain uPA (B,D). The data are shown fitted to integrated rate equations describing the plasminogen activation system and using previously published kinetic constants. In the case of U937 cells, the data are consistent with 100% of the bound uPA being either in the active or pro form; in the case of THP-1 cells, the best fit of the data in the presence of pro-uPA is achieved by assuming that 50% of the bound uPA is initially present in the activated form. Plasminogen activation was not detected on cells preincubated with the blocking anti-uPAR monoclonal antibody R3.
Constitutive activation of pro-uPA on THP-1 cells
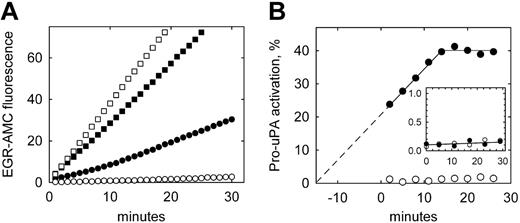
To confirm the presence of activated uPA on the surface of THP-1 cells subsequent to incubation with pro-uPA, and the plasmin independence of this activation, direct measurements of uPA activity were made using a uPA-specific fluorogenic peptide substrate. Figure 2A shows that U937 cells (open symbols) incubated with pro-uPA (circles) display very low uPA activity compared with cells incubated with active tc-uPA (squares). By contrast THP-1 cells (Figure 2A closed symbols) show significant uPA activity when preincubated with pro-uPA. Replotting these data (Figure 2B) reveals that activation of pro-uPA on THP-1 cells continues throughout the assay period, and that this increase in activity extrapolates back to the time point when the cells were first exposed to pro-uPA (ie, 15 minutes prior to assay for uPA activity). These data also show that the generation of uPA activity reaches an apparent plateau at approximately 40% pro-uPA activation. By comparison, substrate hydrolysis with U937 cells was at the lower limit of detection (0.05 δF/min), equating to maximum pro-uPA activation of less than 1.5% in the absence of plasminogen.
Plasmin-independent activation of pro-uPA bound to THP-1 cells. (A) Pro-uPA (○, •) or active tc-uPA (□, ▪) was incubated with either U937 (open symbols) or THP-1 (closed symbols) cells for 10 minutes, washed to remove unbound enzyme, and assayed directly for uPA activity using the substrate H-Glu-Gly-Arg-AMC (EGR-AMC). (B) The data for bound pro-uPA are converted to pro-uPA activation from the rate of substrate hydrolysis with time, and expressed as a percentage of the activity of bound tc-uPA. For THP-1 cells (•), uPA activity is extrapolated back to zero activation, which coincides with the start of the incubation with pro-uPA. The inset shows the activation of pro-uPA in the cell supernatant over the same time period, which is below 0.2% for both cell types.
Plasmin-independent activation of pro-uPA bound to THP-1 cells. (A) Pro-uPA (○, •) or active tc-uPA (□, ▪) was incubated with either U937 (open symbols) or THP-1 (closed symbols) cells for 10 minutes, washed to remove unbound enzyme, and assayed directly for uPA activity using the substrate H-Glu-Gly-Arg-AMC (EGR-AMC). (B) The data for bound pro-uPA are converted to pro-uPA activation from the rate of substrate hydrolysis with time, and expressed as a percentage of the activity of bound tc-uPA. For THP-1 cells (•), uPA activity is extrapolated back to zero activation, which coincides with the start of the incubation with pro-uPA. The inset shows the activation of pro-uPA in the cell supernatant over the same time period, which is below 0.2% for both cell types.
Pro-uPA activation on THP-1 cells is plasma-membrane associated
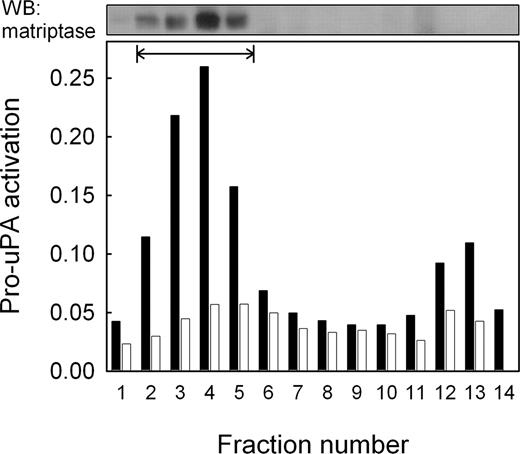
The activation of uPAR-associated pro-uPA observed on THP-1 cells suggests that it is activated by a membrane-associated protease. This assumption is supported by the observation that unbound pro-uPA present in the cell supernatant was not activated to any significant extent (Figure 2B inset). To further investigate membrane association, both THP-1 and U937 cells were subjected to subcellular fractionation on a sucrose density gradient and the fractions obtained assayed for their ability to activate pro-uPA. Figure 3 shows that THP-1 cells displayed a peak of pro-uPA-activating activity that was not present in U937 cells, and that this peak of activity corresponds to the plasma membrane fractions.
Matriptase is differentially expressed in monocytic cell lines
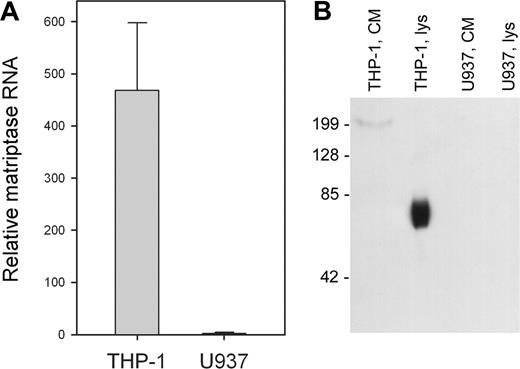
Candidate proteases for the activation of pro-uPA on THP-1 cells are numerous, although very few would be cell associated and possibly constitutively active. An exception is the TTSP matriptase, which has been reported to activate pro-uPA in solution.18,19 Therefore, we analyzed expression of matriptase mRNA in the 2 cell lines using qRT-PCR. Figure 4A shows that matriptase mRNA is highly expressed in THP-1 cells, but essentially undetectable in U937 cells. Matriptase protein expression was analyzed by Western blotting of cell lysates and was detected in THP-1 cell lysates (Figure 4B lane 2), but was absent from U937 cell lysates (lane 4). Matriptase was also absent from the concentrated conditioned media of both cell lines (Figure 4B lanes 1 and 3). Therefore, both the cellular expression of matriptase and its cellular location correlate with the observed activation of uPAR-bound pro-uPA on THP-1 cells.
Kinetics of pro-uPA activation by matriptase catalytic domain
We next sought to determine the efficiency of matriptase as an activator of pro-uPA. The catalytic domain of matriptase was expressed in E coli and activated autocatalytically, and its concentration was determined by active-site titration. The kinetics of pro-uPA activation by recombinant matriptase protease domain was compared with those of plasmin and is shown in Table 1. These data confirm that matriptase is an activator of pro-uPA, but also show that this reaction is unfavorable in comparison with plasmin-catalyzed activation, which is approximately 100-fold more efficient as assessed by kcat/Km.
Kinetic constants for pro-uPA activation
suPAR . | kcat, s-1 . | Km, μM . | Kcat/Km, M-1s-1 . |
|---|---|---|---|
| Plasmin | |||
| Without | 12.8 ± 1.3 | 1.06 ± 0.17 | 1.21 × 107 |
| With | 6.82 ± 2.23 | 0.79 ± 0.37 | 8.63 × 106 |
| Matriptase | |||
| Without | 0.466 ± 0.016 | 4.01 ± 0.17 | 1.16 × 105 |
| With | 0.129 ± 0.010 | 1.24 ± 0.15 | 1.04 × 105 |
suPAR . | kcat, s-1 . | Km, μM . | Kcat/Km, M-1s-1 . |
|---|---|---|---|
| Plasmin | |||
| Without | 12.8 ± 1.3 | 1.06 ± 0.17 | 1.21 × 107 |
| With | 6.82 ± 2.23 | 0.79 ± 0.37 | 8.63 × 106 |
| Matriptase | |||
| Without | 0.466 ± 0.016 | 4.01 ± 0.17 | 1.16 × 105 |
| With | 0.129 ± 0.010 | 1.24 ± 0.15 | 1.04 × 105 |
Data are shown for the activation of pro-uPA by plasmin and matriptase, both in the presence and absence of equimolar amounts of soluble uPAR. Values for kcat and Km are shown as mean ± standard error of the parameter estimated by nonlinear regression
Localization of pro-uPA-activating activity by subcellular fractionation. THP-1 and U937 cells were fractionated by sucrose density gradient centrifugation and the fractions assayed for pro-uPA-activating activity. Data are represented as absorbance due to hydrolysis of the uPA-specific chromogenic peptide substrate S-2444 for THP-1 cells (closed bars) and U937 cells (open bars). Plasma membrane fractions, identified by biotinylation of cell surface proteins (fractions 2 to 5), are shown by the horizontal bar. Matriptase was subsequently detected in these fractions by Western blotting (upper panel). The minor peak of activity in fractions 12 and 13 did not contain detectable levels of matriptase.
Localization of pro-uPA-activating activity by subcellular fractionation. THP-1 and U937 cells were fractionated by sucrose density gradient centrifugation and the fractions assayed for pro-uPA-activating activity. Data are represented as absorbance due to hydrolysis of the uPA-specific chromogenic peptide substrate S-2444 for THP-1 cells (closed bars) and U937 cells (open bars). Plasma membrane fractions, identified by biotinylation of cell surface proteins (fractions 2 to 5), are shown by the horizontal bar. Matriptase was subsequently detected in these fractions by Western blotting (upper panel). The minor peak of activity in fractions 12 and 13 did not contain detectable levels of matriptase.
Differential expression on matriptase by THP-1 and U937 cells. (A) RNA was isolated from THP-1 and U937 cells, reverse transcribed, and subjected to qRT-PCR. Values are corrected for RNA content by comparison with 18S ribosomal RNA and shown as mean ± SD (n = 3). (B) Cell lysates (lys, 10 μg protein) and concentrated conditioned medium (CM, 10 μL) were isolated from THP-1 and U937 cells in the presence of protease inhibitor cocktail, subject to 10% SDS-PAGE, and probed with an antibody to matriptase (M32). The mobility of matriptase is consistent with the molecular weight of the form proteolytically processed at Gly149,25 but this does not lead to shedding of the protease as it is not detectable in the cell-conditioned medium.
Differential expression on matriptase by THP-1 and U937 cells. (A) RNA was isolated from THP-1 and U937 cells, reverse transcribed, and subjected to qRT-PCR. Values are corrected for RNA content by comparison with 18S ribosomal RNA and shown as mean ± SD (n = 3). (B) Cell lysates (lys, 10 μg protein) and concentrated conditioned medium (CM, 10 μL) were isolated from THP-1 and U937 cells in the presence of protease inhibitor cocktail, subject to 10% SDS-PAGE, and probed with an antibody to matriptase (M32). The mobility of matriptase is consistent with the molecular weight of the form proteolytically processed at Gly149,25 but this does not lead to shedding of the protease as it is not detectable in the cell-conditioned medium.
To determine whether the efficiency of pro-uPA activation by matriptase might be increased when bound to uPAR, activation of pro-uPA in complex with recombinant soluble uPAR was determined. No increase in kcat/Km was observed in the presence of uPAR (Table 1). Although there were significant effects on the individual kinetic constants, overall these led to only a minor and negative effect on kcat/Km. Similar effects were observed with plasmin, as we have previously reported.23
Plasminogen activation is impaired in THP-1 cells with knock down of matriptase expression
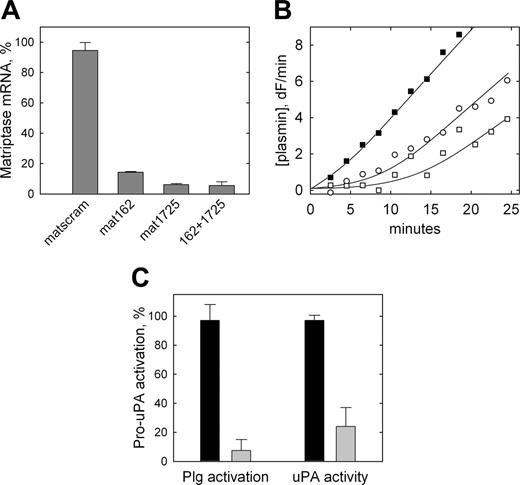
To directly determine whether matriptase is the protease responsible for the plasmin-independent activation of pro-uPA observed on THP-1 cells, siRNA was used to knock down matriptase expression in THP-1 cells. The matriptase-specific siRNA duplexes, mat162 and mat1725, led to a knock down of matriptase mRNA expression by 86% and 94%, respectively, compared with cells transfected with the control nontargeting siRNA (Figure 5A). The effect was not greater when both targeting siRNAs were cotransfected (94%).
Inhibition of pro-uPA activation on THP-1 cells by siRNA-mediated down-regulation of matriptase expression. (A) THP-1 cells were transiently transfected with 100 nM nontargeting siRNA (matscram), each matriptase specific siRNA (mat162 or mat1725), or both matriptase specific siRNAs in combination (162 + 1725). Twenty-four hours after transfection, RNA was isolated, reverse transcribed, and subjected to qRT-PCR. Values are corrected for 18S ribosomal RNA levels and expressed as a percentage of matriptase mRNA in nontransfected cells. Data are shown as mean ± SD (n = 3). None of the siRNAs used affected expression levels of uPA, uPAR, HAI-1, PAI-1, or PAI-2 as determined by qRT-PCR. (B) Representative cell surface plasminogen activation on control THP-1 cells transfected with nontargeting siRNA (▪) compared with cells transfected with mat162 + 1725 (○, □), representing data from 3 independent transfection experiments. The 2 plasmin generation curves shown for targeted cells (○, □) are fitted using 15% and 0% initial activation of pro-uPA, respectively. (C) Data from these plasminogen activation experiments and similar experiments in which pro-uPA activation by the cells was determined directly using H-Glu-Gly-Arg-AMC (as in Figure 2). Data are shown as mean ± SD (n = 3) for nontargeting (dark bars) and targeting (light bars) siRNA and are expressed as a percentage of the pro-uPA activation observed in nontransfected cells. Similar data were obtained when each of the targeting siRNAs was transfected individually.
Inhibition of pro-uPA activation on THP-1 cells by siRNA-mediated down-regulation of matriptase expression. (A) THP-1 cells were transiently transfected with 100 nM nontargeting siRNA (matscram), each matriptase specific siRNA (mat162 or mat1725), or both matriptase specific siRNAs in combination (162 + 1725). Twenty-four hours after transfection, RNA was isolated, reverse transcribed, and subjected to qRT-PCR. Values are corrected for 18S ribosomal RNA levels and expressed as a percentage of matriptase mRNA in nontransfected cells. Data are shown as mean ± SD (n = 3). None of the siRNAs used affected expression levels of uPA, uPAR, HAI-1, PAI-1, or PAI-2 as determined by qRT-PCR. (B) Representative cell surface plasminogen activation on control THP-1 cells transfected with nontargeting siRNA (▪) compared with cells transfected with mat162 + 1725 (○, □), representing data from 3 independent transfection experiments. The 2 plasmin generation curves shown for targeted cells (○, □) are fitted using 15% and 0% initial activation of pro-uPA, respectively. (C) Data from these plasminogen activation experiments and similar experiments in which pro-uPA activation by the cells was determined directly using H-Glu-Gly-Arg-AMC (as in Figure 2). Data are shown as mean ± SD (n = 3) for nontargeting (dark bars) and targeting (light bars) siRNA and are expressed as a percentage of the pro-uPA activation observed in nontransfected cells. Similar data were obtained when each of the targeting siRNAs was transfected individually.
Mat1725 was then used to knock down matriptase expression in THP-1 cells prior to quantification of pro-uPA activation, both indirectly via plasminogen activation or by direct hydrolysis of the uPA-specific fluorogenic peptide substrate. Cells transfected with nontargeting siRNA displayed a rapid, constant rate of plasmin generation (Figure 5B, ▪) consistent with the previous observations. By contrast, cells transfected with matriptase-targeting siRNA displayed an extended lag phase consistent with a greatly reduced level of constitutive pro-uPA zymogen activation on these cells (Figure 5B, open symbols), that is, as previously observed on U937 cells that lack matriptase expression. Estimation of pro-uPA activation on THP-1 cells by fitting of the plasmin generation curves to a model describing reciprocal zymogen activation indicates a mean inhibition of pro-uPA activation of 93% by the matriptase-targeting siRNA (Figure 5C), consistent with the observed reduction in matriptase expression. To confirm that this was a direct effect on pro-uPA activation, the generation of uPA activity was determined directly in the absence of plasminogen and found to be reduced by 76% (Figure 5C).
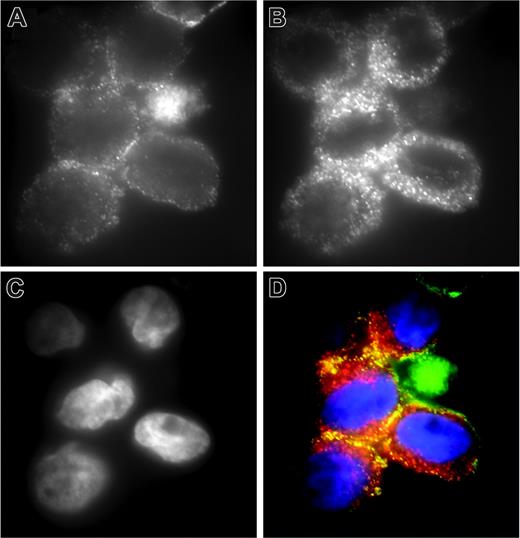
For matriptase to act as a cell-surface activator of uPAR-bound pro-uPA, the 2 proteins might be expected to be colocalized. Double immunofluorescence staining of THP-1 cells demonstrates that although uPAR (Figure 6A) is less widely distributed on the cell surface than matriptase (Figure 6B) and more associated with the periphery of the cells, there is significant colocalization of the 2 proteins (Figure 6D). Of interest, the data in Figure 2B indicates that not all uPAR-associated pro-uPA is activated on THP-1 cells (in the absence of plasmin), consistent with the partial colocalization of uPAR with matriptase.
Colocalization of matriptase and uPAR on THP-1 cells. THP-1 cells were fixed with formaldehyde and stained for (A) uPAR using monoclonal antibody R4 and (B) matriptase using polyclonal antibody IM1014. Detection of uPAR and matriptase was with fluorescein- and rhodamine-conjugated secondary antibodies, respectively. (C) Nuclear staining with DAPI. (D) The merged images with uPAR in green, matriptase in red, and DAPI in blue. The specificity of the observed colocalization was demonstrated in control experiments, with cells stained for matriptase and β1-integrin, in which no colocalization was observed in merged images (data not shown).
Colocalization of matriptase and uPAR on THP-1 cells. THP-1 cells were fixed with formaldehyde and stained for (A) uPAR using monoclonal antibody R4 and (B) matriptase using polyclonal antibody IM1014. Detection of uPAR and matriptase was with fluorescein- and rhodamine-conjugated secondary antibodies, respectively. (C) Nuclear staining with DAPI. (D) The merged images with uPAR in green, matriptase in red, and DAPI in blue. The specificity of the observed colocalization was demonstrated in control experiments, with cells stained for matriptase and β1-integrin, in which no colocalization was observed in merged images (data not shown).
Expression of matriptase by peripheral blood monocytes
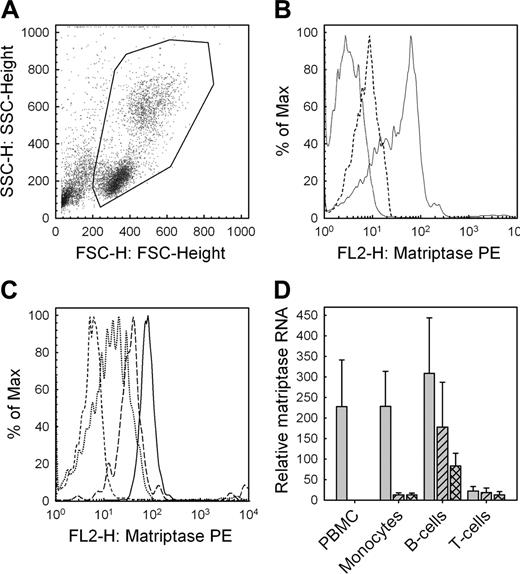
Given the biologic significance matriptase may have in pro-uPA activation in human monocytes, we investigated whether matriptase is expressed in human peripheral blood mononuclear cells. Using flow cytometry, we found that whole PBMCs obtained from healthy donors following Ficoll centrifugation (Figure 7A) express matriptase on their surface (Figure 7B). We then examined the relative surface expression of matriptase on immune cell subsets using multicolor flow cytometry and found that monocytes and B cells express significantly higher levels of matriptase compared with T cells (Figure 7C). Matriptase RNA expression in PBMCs and in isolated human immune cell subsets was assessed by quantitative real-time PCR. In keeping with the flow cytometry data, we found that matriptase is highly expressed in the ex vivo PBMCs and that this expression comes largely from the monocyte and B-cell subsets with significantly less expression observed in T cells (Figure 7D). Of interest, matriptase expression was highly down-regulated in monocytes when cultured ex vivo and this expression could not be recovered by stimulation with interferon-γ and lipopolysaccharide.
Discussion
The mechanism leading to the initiation of a proteolytic cascade is potentially its most important regulatory point, excellent examples being those involved in the blood coagulation and complement activation systems.26 However, it is often the most difficult step to identify experimentally. The mechanism that initiates the generation of the powerful multifunctional serine protease plasmin by uPA in the pericellular environment is not fully understood, although uPAR is known to have a key role and a variety of mechanisms have been proposed. We demonstrate here that on monocytes the TTSP matriptase has the potential to fulfill this role, acting as a specific activator of uPAR-bound pro-uPA at the cell surface. We observed a rapid initiation of plasminogen activation on THP-1 cells that correlated with the presence of a cell surface-associated protease and with the expression of matriptase, and that siRNA knock down of matriptase expression abolishes this rapid initiation. The activation by THP-1 cells was restricted to pro-uPA bound to uPAR on the cell surface and was catalyzed by cell-associated matriptase. By contrast, U937 cells, which were found not to express matriptase, did not display a rapid initiation of plasminogen activation.
The binding of uPA to uPAR on the cell surface facilitates the activation both of plasminogen and of pro-uPA by generated plasmin, and, therefore, the overall process of reciprocal zymogen activation is greatly increased by uPAR. Trace amounts of active plasmin or uPA will lead to rapid activation of the system, but under conditions where these are excluded, the low intrinsic activity of pro-uPA is thought to be sufficient to initiate plasmin generation.14 This mechanism of reciprocal zymogen activation is associated with a characteristic lag phase in the generation of plasmin activity, as is observed here in the experiments using U937 cells. The presence of plasminogen activator inhibitors (eg, plasminogen activator inhibitor-1) is thought to suppress initiation occurring via this mechanism.15,16 Therefore, activation of pro-uPA by proteases other than plasmin must also be considered as a potential initiation mechanism.
Protein and mRNA expression of matriptase by peripheral blood mononuclear cells. (A) Flow cytometry of PBMCs obtained following Ficoll centrifugation of venous blood from healthy consenting donors (B) exhibit matriptase surface expression (solid line indicates matriptase; dashed line, isotype control; and dotted line, intrinsic fluorescence). (C) Multicolor flow cytometry demonstrates significantly higher expression of matriptase on monocytes (solid line) and B cells (long dashes), compared with T cells (dotted). The control for intrinsic fluorescence control is shown (short dashes). (D) Matriptase expression quantified by qRT-PCR for the total mononuclear cell population immediately ex vivo (PBMC), and individual cell populations purified using magnetic activated cell sorting (MACS) magnetic beads to obtain CD3+ T cells, CD19+ B cells, and CD14+ monocytes. Data are shown for cells either immediately after purification (gray bars), after 8 hours in culture unstimulated (hatched bars), or after stimulation of monocytes, B cells, and T cells with IFN-γ and LPS, anti-IgG/IgM, or PMA and ionomycin, respectively (crosshatched bars). Values are corrected for RNA content by comparison to 18S ribosomal RNA. Data are shown as mean ± SD (n = 6). The observed reductions in matriptase expression by monocytes were statistically significant (paired t test, P < .05).
Protein and mRNA expression of matriptase by peripheral blood mononuclear cells. (A) Flow cytometry of PBMCs obtained following Ficoll centrifugation of venous blood from healthy consenting donors (B) exhibit matriptase surface expression (solid line indicates matriptase; dashed line, isotype control; and dotted line, intrinsic fluorescence). (C) Multicolor flow cytometry demonstrates significantly higher expression of matriptase on monocytes (solid line) and B cells (long dashes), compared with T cells (dotted). The control for intrinsic fluorescence control is shown (short dashes). (D) Matriptase expression quantified by qRT-PCR for the total mononuclear cell population immediately ex vivo (PBMC), and individual cell populations purified using magnetic activated cell sorting (MACS) magnetic beads to obtain CD3+ T cells, CD19+ B cells, and CD14+ monocytes. Data are shown for cells either immediately after purification (gray bars), after 8 hours in culture unstimulated (hatched bars), or after stimulation of monocytes, B cells, and T cells with IFN-γ and LPS, anti-IgG/IgM, or PMA and ionomycin, respectively (crosshatched bars). Values are corrected for RNA content by comparison to 18S ribosomal RNA. Data are shown as mean ± SD (n = 6). The observed reductions in matriptase expression by monocytes were statistically significant (paired t test, P < .05).
Although many proteases are capable of activating pro-uPA in purified systems,17 there is no evidence that any of these are of relevance to plasminogen activation in vivo. In contrast, the observations presented here suggest that matriptase could have an important biologic role as an activator of pro-uPA and that it has the characteristics of a true initiator of the plasminogen activation system. Together with its cell surface localization, the most relevant characteristic is that matriptase is capable of autocatalytic activation.19,27 This is a very unusual property among the serine proteases, and in this context eliminates the need for the presence of a further upstream protease or other mechanism as part of the initiation process.
The autocatalytic activation of both the bacterially expressed recombinant catalytic domain of matriptase and the wild-type protein has been demonstrated. The latter has also been shown to be dependent on the presence of the matriptase inhibitor HAI-1, by an as-yet-unresolved mechanism.28 However, THP-1 cells were found not to express HAI-1 as assessed by qRT-PCR, suggesting that the role of HAI-1 in the autoactivation mechanism may be cell-type specific. Furthermore, the activation of pro-uPA by matriptase on these cells was not inhibited by the addition of concentrations of recombinant HAI-1 that were effective in solution (data not shown). Another potentially cell type-specific property of matriptase is its shedding from the cell surface. Although the molecular weight of matriptase determined by SDS-PAGE is consistent with proteolytic processing at Gly149,25 this does not lead to the shedding of matriptase from the surface of THP-1 cells as it was not detectable in the cell-conditioned medium. This cleavage has also been linked to the activation of matriptase28 and appears to be a prerequisite for autocatalytic shedding of matriptase on epithelial cells.25,29 Therefore, the regulation of matriptase activity may differ substantially between leukocytes and the epithelial cells that have previously been studied.
Despite the apparently ideal characteristics of matriptase as an initiator of plasminogen activation, comparison of the catalytic efficiencies of pro-uPA activation by matriptase and plasmin in solution revealed matriptase to be a relatively poor activator. The presence of soluble uPAR, to mimic the substrate complex on the cell surface, did not enhance activation. Therefore, the relatively efficient activation of pro-uPA by matriptase observed on THP-1 cells suggests that the reaction is promoted on the cell surface. This either could be by nonspecific approximation effects, or the activation of pro-uPA by matriptase could be specifically facilitated on the cell surface. A possible mechanism here could be specific interactions between the N-terminal domains of matriptase and the uPA/uPAR complex. In support of this general mechanism, the N-terminal domains of other TTSPs have been shown to influence their interaction with their physiologic substrates. For example, the activation of trypsinogen by enteropeptidase30 and pro-ANP by corin31 is greatly reduced in the absence of these domains. In this respect, it is of interest that the modular structure of the TTSPs is dissimilar to that of the fibrinolytic and coagulation proteases, using a completely distinct range of protein modules.32
The expression of matriptase has previously been considered to be restricted to epithelial cells.33,34 We observe differential expression of matriptase by 2 monocytic cell lines, THP-1 and U937, both derived from acute myeloid leukemias, and also expression in ex vivo peripheral blood monocytes and B cells, both of which express matriptase at levels approaching those found in THP-1 cells (compare data in Figures 4A and 7D). Therefore matriptase is expressed by cells of both myeloid and lymphocytic lineages. The observation of differential expression in the 2 cell lines, and the rapid loss of matriptase expression in cultured peripheral blood monocytes, suggests that matriptase expression is highly regulated in these cells. Potential cell type-specific differences in the regulation of matriptase activity have been mentioned earlier, and this may extend to substrate recognition. Evidence has previously been presented that matriptase on the HRA ovarian cancer cell line can activate pro-uPA,35 although the effect of this on plasminogen activation was not determined. However, a general role for matriptase in the initiation of pericellular plasminogen activation in epithelial cells is perhaps unlikely given the distinct cellular localizations of matriptase and uPAR. Matriptase has been reported to be targeted basolaterally in polarized epithelial cells,36 whereas uPAR is targeted apically.37 By contrast, we find colocalization of matriptase and uPAR on THP-1 cells. Therefore, in epithelial cells, other targets for matriptase activity may be more relevant, for example hepatocyte growth factor/scatter factor,19 the receptor for which (c-Met) is also localized basolaterally.38
The data presented here demonstrate that matriptase is an efficient and specific activator of uPAR-bound pro-uPA on the surface of THP-1 cells and that this is sufficient for the rapid initiation of plasmin generation on these cells. Matriptase-deficient mice have been generated and are viable, but die postnatally from an epidermal barrier defect,39 due to a lack of matriptase expression in keratinocytes that is unrelated to the activation of pro-uPA.40 Therefore, it is not yet possible to assess the potential role of matriptase in plasminogen activation by leukocytes in vivo. However, the highly regulated expression of matriptase by leukocytes, together with the known importance of plasminogen activation in the biology of these cells, suggests that matriptase may be a key molecule in the physiologic and pathologic regulation of plasminogen activation by these cells.
Prepublished online as Blood First Edition Paper, June 22, 2006; DOI 10.1182/blood-2006-02-001073.
Supported by grants from the British Heart Foundation (PG/1998053, PG/04/037/16919). V.E. is a Senior Research Fellow of the British Heart Foundation (FS/1999073).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Chen-Yong Lin, Michael Ploug, and Gunilla Høyer-Hansen for supplying valuable reagents used in this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal