Abstract
BMS-354825 (dasatinib) and AMN107 (nilotinib) are potent alternate Abl inhibitors with activity against many imatinib mesylate–resistant BCR-ABL kinase domain (KD) mutants, except T315I. We used N-ethyl-N-nitrosourea (ENU)–exposed Ba/F3-p210BCR-ABL cells to compare incidence and types of KD mutants emerging in the presence of imatinib mesylate, dasatinib, and nilotinib, alone and in dual combinations. Although ENU is expected to induce mutations in multiple proteins, resistant clones were almost exclusively BCR-ABL KD mutant at relevant concentrations of nilotinib and dasatinib, consistent with a central role of KD mutations for resistance to these drugs. Twenty different mutations were identified with imatinib mesylate, 10 with nilotinib (including only 1 novel mutation, E292V) and 9 with dasatinib. At intermediate drug levels the spectrum narrowed to F317V and T315I for dasatinib and Y253H, E255V, and T315I for nilotinib. Thus, cross-resistance is limited to T315I, which is also the only mutant isolated at drug concentrations equivalent to maximal achievable plasma trough levels. With drug combinations maximal suppression of resistant clone outgrowth was achieved at lower concentrations compared with single agents, suggesting that such combinations may be equipotent to higher-dose single agents. However, sequencing uniformly revealed T315I, consistent with the need for a T315I inhibitor, to completely block resistance.
Introduction
Imatinib mesylate (Gleevec, STI571) has become the standard of care for the treatment of patients with chronic myeloid leukemia (CML).1,2 Although responses in chronic phase tend to be durable, relapse after an initial response is common in patients with more advanced disease.3,4 Point mutations within the kinase domain (KD) of BCR-ABL are the most common mechanism of acquired drug resistance, found in 50% to 90% of such patients.5-10 These mutations impair imatinib mesylate binding by steric hindrance, by disruption of critical interactions between imatinib mesylate and Abl, or by preventing the kinase from adopting the specific inactive conformation that is required for imatinib mesylate binding.11,12
The problem of acquired resistance to imatinib mesylate has prompted the development of novel ATP-competitive small molecule tyrosine kinase inhibitors with activity against clinically observed BCR-ABL mutants. The compounds most advanced in clinical development are AMN107 (nilotinib)13 and BMS-354825 (dasatinib),14 both of which are currently in phase 2 clinical trials in patients with imatinib mesylate resistance.15,16 Dasatinib, a 2-aminothiazole-5-carboxamide, is structurally distinct from imatinib mesylate, was originally developed as a Src kinase inhibitor, and binds both active and inactive conformations of Abl.17 Compared with imatinib mesylate, dasatinib exhibits increased potency but reduced selectivity.14 In contrast, nilotinib was designed based on the imatinib mesylate scaffold and substituting the N-methylpiperazine with a trifluoromethyl/imidazole-substituted phenyl group.13 Like imatinib mesylate, nilotinib binds to the inactive Abl kinase conformation. In vitro studies have shown that nilotinib is at least 20-fold and dasatinib at least 300-fold more potent than imatinib mesylate against unmutated Abl.18 In addition, they are active against many imatinib mesylate–resistant BCR-ABL mutants.13,14,18 As these compounds enter the clinical arena, it would be useful to predict the profile of mutations that confer drug resistance because this would allow for a rational approach to the design of combination strategies.
Two different methods have been used to generate profiles of resistance mutations. Azam et al19 used “saturation mutagenesis” based on expression of BCR-ABL in a DNA repair–deficient strain of Escherichia coli. The mutagenized plasmids were then used to rescue Ba/F3 cells from growth factor withdrawal in the presence of drug. Although resistance mutation patterns generated with this assay are comprehensive,20 only BCR-ABL mutations are recovered, precluding the detection of alternative resistance mechanisms. Von Bubnoff et al21 isolated individual resistant clones from Ba/F3-p185BCR-ABL cells grown in the presence of inhibitors in a cell-based screen. Although this assay is unbiased and capable of detecting both BCR-ABL–dependent and –independent resistance mechanisms, it has the disadvantage of a relatively low yield, with the highest frequency of resistant colonies being 3.9 per 106 cells. To address these limitations we have developed a rapid mutagenesis assay that is based on Ba/F3-p210BCR-ABL cells chemically mutagenized with N-ethyl-N-nitrosourea (ENU), a potent inducer of point mutations.22 We used this assay to compare the resistance mutation profile of imatinib mesylate, nilotinib, and dasatinib, alone and in combination.
Materials and methods
Inhibitors
Imatinib mesylate was purchased from the Oregon Health and Science University (OHSU) pharmacy and stored as a 10-mM stock solution in deionized water. Nilotinib was kindly provided by Novartis Pharma (Basel, Switzerland) and dasatinib by Bristol-Meyers Squibb (New York, NY). Both were stored at –20°C as 10-mM stock solutions in DMSO. Fresh dilutions in complete media were prepared prior to the experiments.
N-ethyl-N-nitrosourea (ENU) mutagenesis
ENU was dissolved in DMSO at 50 mg/mL and stored in aliquots at –80°C. Ba/F3 cells expressing p210BCR-ABL were generated as previously described23 and maintained in complete culture medium (RPMI 1640 media containing 10% fetal bovine serum and penicillin/streptomycin) at an exponential growth rate. ENU was added to Ba/F3-p210BCR-ABL cells (5 × 106 cells/mL) at a concentration of 50 μg/mL, followed by culture for 12 to 24 hours. In preliminary experiments this dose of ENU had been established as minimally cytotoxic (data not shown). The cells were then washed 3 times with RPMI, replated in complete medium, and allowed to expand over 1 week under exponential growth conditions.
Resistance screen
ENU-exposed Ba/F3-p210BCR-ABL cells were cultured in 96-well plates at 1 × 105 cells/well in 200 μL complete media supplemented with graded concentrations of the respective inhibitors. Inhibitor concentrations were as follows: imatinib mesylate, 2 μM, 4 μM, 8 μM, 16 μM; nilotinib, 10 nM, 50 nM, 500 nM, 2000 nM, 5000 nM; and dasatinib, 5 nM, 10 nM, 25 nM, 100 nM, 500 nM. In several experiments, nonmutagenized Ba/F3-p210BCR-ABL cells, incubated with 16 μM imatinib mesylate, 5000 nM nilotinib, or 500 nM dasatinib, were included as controls. Wells were observed for cell growth by visual inspection under an inverted microscope and media color change every 2 to 3 days for at least 4 weeks. When growth in a well occurred, cells were transferred to 24-well plates and expanded in the presence of the corresponding inhibitor concentration used in the screen. If growth was simultaneously observed in all 96 wells of a given condition, only 24 wells were expanded for further analysis.
PCR and sequencing
DNA was extracted from the cell pellets of resistant cells using DNeasy Tissue kit (QIAGEN, Valencia, CA). The BCR-ABL kinase domain was amplified using primers B2A (5′-TTCAGAAGCTTCTCCCTGACAT-3′ and ABL4065 5′-TGAGTTCATAGACCTTCTCTGG-3′). The polymerase chain reaction (PCR) products were sequenced by a commercial contractor (Agencourt Bioscience Corporation, Beverly, MA) using primers BCRF4 (5′-TGGTTCATCATCATTCAACGG-3′) and U396 (5′-AGACGTCGGACTTGATGG-3′), as described.24 The chromatograms were analyzed for mutations using Mutation Surveyor software (SoftGenetics, State College, PA). The sensitivity of mutation detection in our hands is approximately 10% to 15% of mutant allele in a wild-type background. These study protocols were approved by the Oregon Health and Science University (OHSU) Institutional Review Board.
Results
Accelerated cell-based mutagenesis screen
In 2 independent experiments, mutagenized Ba/F3-p210BCR-ABL cells were exposed to graded concentrations of imatinib mesylate (2-16 μM), nilotinib (50-2000 nM), and dasatinib (5-100 nM). In the second experiment, dasatinib was also used at 500 nM and nilotinib at 5000 nM. The frequency of wells with growth decreased with increased concentrations of all 3 inhibitors and growth tended to occur later, although this was not uniformly observed (Table S1, available at the Blood website; see the Supplemental Tables link at the top of the online article). Control cells without prior exposure to ENU were tested at the highest concentration of inhibitors used in each experiment. Growth was not regularly observed and occurred only in isolated wells and at late time points. Cells from a total of 763 wells with growth were expanded and sequenced. KD mutations were detected in 681 wells. In 636 wells (93.4%), only 1 mutation was seen, as a rule, in 100% of alleles. Forty-five (6.6%) contained 2 different mutations. Review of the sequence traces revealed that in all cases the estimated proportions of the mutant added up to approximately 100%. This is consistent with the presence of 2 independent clones in the same well but not with 1 clone harboring 2 mutations. Thus, the 2 mutations were considered as independently derived, and the total number of KD mutant clones is 726.
Resistance to higher doses of imatinib mesylate, nilotinib, and dasatinib is exclusively the result of BCR-ABL kinase domain mutations
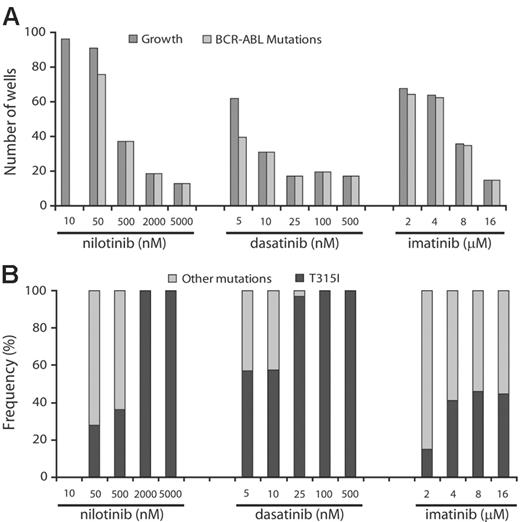
To determine whether the frequency of resistant clones was dose dependent, we cultured ENU-exposed Ba/F3-p210BCR-ABL cells in the presence of increasing concentrations of the 3 inhibitors. Greater than 50% of wells showed growth in the presence of 2 and 4 μM imatinib mesylate, 10 and 50 nM nilotinib, and 5 nM dasatinib. At higher inhibitor concentrations, growth was observed in progressively fewer wells (Figure 1). At levels equivalent to achievable plasma trough concentrations (100 nM for dasatinib, 2000 nM for nilotinib, 4 μM for imatinib mesylate at 800 mg daily dosing),15,25,26 fewer than 20 wells with growth were recorded for dasatinib and nilotinib but greater than 60 for imatinib mesylate. This suggests that resistance is less likely to emerge at clinically achievable concentrations of nilotinib and dasatinib.
Because ENU causes point mutations in multiple proteins, we hypothesized that in our assay, resistance to Abl inhibitors may be multifactorial. However, sequencing of BCR-ABL revealed KD mutations in the overwhelming majority of resistant clones, with the exception of the lowest concentration of nilotinib (10 nM), whereby sequencing was wild type in all 24 clones analyzed. Because the IC50 for inhibition of unmutated BCR-ABL by nilotinib is 13 nM,18 it is likely that this inhibitor concentration failed to generate sufficient selection pressure. In contrast, with nilotinib concentrations at 500 nM or higher, dasatinib at 10 nM or higher, and imatinib mesylate at 16 μM, only clones with KD mutations were recovered.
Prevalence of KD mutations in resistant cell clones. ENU-treated Ba/F3-p210BCR-ABL cells were cultured in the presence of graded concentrations of nilotinib, dasatinib, or imatinib mesylate. (A) At higher drug concentrations, only KD mutant clones were detected. (B) T315I is the single mutation that persists in highest concentrations of nilotinib and dasatinib.
Prevalence of KD mutations in resistant cell clones. ENU-treated Ba/F3-p210BCR-ABL cells were cultured in the presence of graded concentrations of nilotinib, dasatinib, or imatinib mesylate. (A) At higher drug concentrations, only KD mutant clones were detected. (B) T315I is the single mutation that persists in highest concentrations of nilotinib and dasatinib.
ENU-based mutagenesis faithfully reproduces the spectrum of mutations recovered from patients with imatinib mesylate resistance
To establish whether the ENU-based assay described here would reproduce the spectrum of mutations detected in patients with clinical resistance, we compared the types of mutations that emerged in the presence of imatinib mesylate with mutations described in patients with clinical resistance as compiled from 20 pertinent publications (Figure 2).27 Mutations were reported in 30 residues in clinical samples, 25 (83%) of which were reproduced in our assay. In 16 of these (64%) we detected the identical amino acid mutation. Taking into account the relative frequency of mutations reported in patients, the overall coverage of our assay was 92% for detecting residues and 82% for detecting the precise mutations. It is conceivable that our failure to recover all clinically detected mutations is because we used a minimal dose of 2 μM imatinib mesylate, which is likely to suppress the outgrowth of mutants conferring only low-level resistance. Conversely, the “false” discovery rate in our assay was low, with only 3 novel mutations that, to the best of our knowledge, have not been described in patients with imatinib mesylate resistance, including L248R, F311I, and L348M (Figure 2). Overall, these data indicate that the ENU-based assay faithfully reproduces the spectrum of mutations seen in patients with clinical resistance to imatinib mesylate.
Spectrum of point mutations in patients with imatinib mesylate resistance compared with mutations recovered from in vitro assay in the presence of imatinib mesylate. Ba/F3-p210BCR-ABL exposed to ENU and cultured in the presence of imatinib mesylate at graded concentrations. Bars represent the frequencies of mutations that occur in patients with imatinib mesylate resistance, as summarized by Hughes et al.27 Asterisks denote mutations that appeared in the screen but that have not been described clinically.
Spectrum of point mutations in patients with imatinib mesylate resistance compared with mutations recovered from in vitro assay in the presence of imatinib mesylate. Ba/F3-p210BCR-ABL exposed to ENU and cultured in the presence of imatinib mesylate at graded concentrations. Bars represent the frequencies of mutations that occur in patients with imatinib mesylate resistance, as summarized by Hughes et al.27 Asterisks denote mutations that appeared in the screen but that have not been described clinically.
Spectrum of mutations arising in the presence of nilotinib and dasatinib is narrow compared with imatinib mesylate, and only T315I is recovered at high drug levels
We identified a total of 726 mutants representing 26 different exchanges of 19 amino acid residues. Overall, Y253H, T315I, and E255K were the most frequent mutations in our screen (Table 1). For nilotinib and dasatinib there were 10 and 9 different mutations, respectively, compared with 20 different mutations recovered in the presence of imatinib mesylate. Y253H and T315I were the most prominent mutations recovered with nilotinib, whereas T315I and mutations of F317 (F317C/I/L/V) were predominant in the presence of dasatinib. With both nilotinib and dasatinib, the spectrum of kinase domain mutations narrowed at higher concentrations, and T315I emerged as the sole mutation (Figure 1). Mutants recovered in the presence of 500 nM nilotinib included Y253H, T315I, and E255V (Table 1). The concentrations at which these mutants were recovered correspond well with reported IC50s from cell proliferation assays.18 Thus, nilotinib inhibits Y253H and E255V with IC50s of approximately 450 nM, consistent with these mutants being recovered at nilotinib concentrations of 500 nM or less, but not at 2000 nM and greater. In contrast, the IC50 for T315I is greater than 2000 nM for nilotinib, consistent with this mutation being recovered at all concentrations. With intermediate levels of dasatinib (25 nM), the only mutants observed were T315I and F317V. F317V was also recovered from an E coli–based mutagenesis screen in the presence of 50 nM dasatinib and has 40-fold reduced sensitivity to dasatinib compared with unmutated BCR-ABL.28
Spectrum and frequency of BCR-ABL kinase domain mutations recovered in the presence of imatinib mesylate, nilotinib, and dasatinib
. | Imatinib mesylate . | . | . | . | . | Nilotinib* . | . | . | . | . | Dasatinib . | . | . | . | . | . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Incidence (%) . | 2 μM . | 4 μM . | 8 μM . | 16 μM . | Incidence (%) . | 50 nM . | 500 nM . | 2000 nM . | 5000 nM . | Incidence (%) . | 5 nM . | 10 nM . | 25 nM . | 100 nM . | 500 nM . | |||||||||||||
| M244V | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L248R | 2 (0.54) | 1 | 0 | 1 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L248V | 3 (0.81) | 3 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 3 (1.66) | 3 | 0 | 0 | 0 | 0 | |||||||||||||
| G250E | 4 (1.08) | 3 | 1 | 0 | 0 | 5 (2.89) | 5 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| Q252H | 5 (1.34) | 0 | 4 | 0 | 1 | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.55) | 0 | 1 | 0 | 0 | 0 | |||||||||||||
| Y253H | 168 (45.16) | 56 | 65 | 31 | 16 | 64 (36.99) | 14 | 50 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E255K | 22 (5.91) | 12 | 8 | 1 | 1 | 5 (2.89) | 5 | 0 | 0 | 0 | 2 (1.1) | 2 | 0 | 0 | 0 | 0 | |||||||||||||
| E255V | 1 (0.27) | 0 | 1 | 0 | 0 | 1 (0.58) | 0 | 1 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| D276G | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E292V† | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| V299L | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 3 (1.66) | 0 | 3 | 0 | 0 | 0 | |||||||||||||
| F311I | 4 (1.08) | 4 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F311V | 3 (0.81) | 3 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| T315I | 139 (37.37) | 39 | 56 | 29 | 15 | 88 (50.87) | 13 | 29 | 36 | 10 | 141 (77.9) | 20 | 33 | 32 | 39 | 17 | |||||||||||||
| F317C | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 7 (3.87) | 1 | 6 | 0 | 0 | 0 | |||||||||||||
| F317I | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 12 (6.63) | 2 | 10 | 0 | 0 | 0 | |||||||||||||
| F317L | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 10 (5.52) | 6 | 4 | 0 | 0 | 0 | |||||||||||||
| F317V | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 2 (1.1) | 1 | 0 | 1 | 0 | 0 | |||||||||||||
| M351T | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E355G | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F359C | 6 (1.61) | 6 | 0 | 0 | 0 | 6 (3.47) | 6 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F359V | 3 (0.81) | 3 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| V379I | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L384M | 1 (0.27) | 1 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L387F | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| H396R | 2 (0.54) | 1 | 1 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
. | Imatinib mesylate . | . | . | . | . | Nilotinib* . | . | . | . | . | Dasatinib . | . | . | . | . | . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Incidence (%) . | 2 μM . | 4 μM . | 8 μM . | 16 μM . | Incidence (%) . | 50 nM . | 500 nM . | 2000 nM . | 5000 nM . | Incidence (%) . | 5 nM . | 10 nM . | 25 nM . | 100 nM . | 500 nM . | |||||||||||||
| M244V | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L248R | 2 (0.54) | 1 | 0 | 1 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L248V | 3 (0.81) | 3 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 3 (1.66) | 3 | 0 | 0 | 0 | 0 | |||||||||||||
| G250E | 4 (1.08) | 3 | 1 | 0 | 0 | 5 (2.89) | 5 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| Q252H | 5 (1.34) | 0 | 4 | 0 | 1 | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.55) | 0 | 1 | 0 | 0 | 0 | |||||||||||||
| Y253H | 168 (45.16) | 56 | 65 | 31 | 16 | 64 (36.99) | 14 | 50 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E255K | 22 (5.91) | 12 | 8 | 1 | 1 | 5 (2.89) | 5 | 0 | 0 | 0 | 2 (1.1) | 2 | 0 | 0 | 0 | 0 | |||||||||||||
| E255V | 1 (0.27) | 0 | 1 | 0 | 0 | 1 (0.58) | 0 | 1 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| D276G | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E292V† | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| V299L | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 3 (1.66) | 0 | 3 | 0 | 0 | 0 | |||||||||||||
| F311I | 4 (1.08) | 4 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F311V | 3 (0.81) | 3 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| T315I | 139 (37.37) | 39 | 56 | 29 | 15 | 88 (50.87) | 13 | 29 | 36 | 10 | 141 (77.9) | 20 | 33 | 32 | 39 | 17 | |||||||||||||
| F317C | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 7 (3.87) | 1 | 6 | 0 | 0 | 0 | |||||||||||||
| F317I | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 12 (6.63) | 2 | 10 | 0 | 0 | 0 | |||||||||||||
| F317L | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 10 (5.52) | 6 | 4 | 0 | 0 | 0 | |||||||||||||
| F317V | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 2 (1.1) | 1 | 0 | 1 | 0 | 0 | |||||||||||||
| M351T | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| E355G | 1 (0.27) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F359C | 6 (1.61) | 6 | 0 | 0 | 0 | 6 (3.47) | 6 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| F359V | 3 (0.81) | 3 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| V379I | 2 (0.54) | 2 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L384M | 1 (0.27) | 1 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| L387F | 0 (0) | 0 | 0 | 0 | 0 | 1 (0.58) | 1 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
| H396R | 2 (0.54) | 1 | 1 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 (0) | 0 | 0 | 0 | 0 | 0 | |||||||||||||
For imatinib mesylate, N = 372; for nilotinib, N = 173; for dasatinib, N = 181.
No mutations were observed at nilotinib 10 nM.
Residue not previously implicated either clinically or in vitro.
Combinations of imatinib mesylate, nilotinib, and dasatinib minimize the outgrowth of resistant clones and narrow the spectrum of mutations to T315I
To test the efficacy of drug combinations at suppressing the outgrowth of resistant clones, we performed combination studies. In a pilot experiment, mutagenized Ba/F3-p210BCR-ABL cells were exposed to pairwise combinations of imatinib mesylate, nilotinib, and dasatinib (Table S2). In all cases, a more profound suppression of growth was observed with the combinations as compared with single agents. Sequencing revealed only T315I in resistant clones, with the exception of the lowest concentrations of imatinib mesylate (400 nM) and nilotinib (50 nM), whereby F359C and Y253H were recovered in addition to T315I (Table S2).
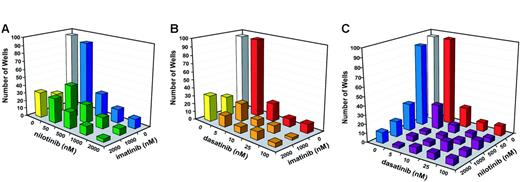
We then performed a comprehensive drug combination experiment using a broader range of concentrations for dasatinib and nilotinib. Compared with single agents, each of the 3 combinations (imatinib mesylate + nilotinib, imatinib mesylate + dasatinib, and nilotinib + dasatinib) was more effective at reducing the outgrowth of resistant cell clones (Figure 3). Consistent with the results of the pilot experiments, none of the 3 combinations completely suppressed growth, even at the highest concentrations of inhibitors. Interestingly, even at the highest concentrations of dasatinib and nilotinib, the addition of imatinib mesylate still appeared to further reduce the frequency of resistant cell clones (Figure 3). In contrast, combinations between nilotinib and dasatinib produced a maximal effect at low concentrations. For example, the addition of 50 nM nilotinib to 10 nM dasatinib reduced the number of resistant clones from 22 to 9. However, even with 2000 nM nilotinib in the combination, we still recovered 7 resistant clones. Similarly, when 500 nM nilotinib was combined with 5 nM dasatinib, resistant clones were reduced from 30 to 4. However, no further reduction was observed at higher levels of dasatinib (Figure 3).
Outgrowth of resistant cells in the presence of inhibitor combinations. (A) Number of wells with resistant growth in the presence of imatinib mesylate 1 μM and 2 μM combined with graded concentrations of nilotinib (50 nM, 500 nM, 1000 nM, and 2000 nM). (B) Number of wells with resistant growth in the presence of imatinib mesylate 1 μM and 2 μM combined with graded concentrations of dasatinib (5 nM, 10 nM, 25 nM, and 100 nM). (C) Number of wells with resistant growth in the presence of graded concentrations of nilotinib (50 nM, 500 nM, 1000 nM, and 2000 nM) and dasatinib (5 nM, 10 nM, 25 nM, and 100 nM).
Outgrowth of resistant cells in the presence of inhibitor combinations. (A) Number of wells with resistant growth in the presence of imatinib mesylate 1 μM and 2 μM combined with graded concentrations of nilotinib (50 nM, 500 nM, 1000 nM, and 2000 nM). (B) Number of wells with resistant growth in the presence of imatinib mesylate 1 μM and 2 μM combined with graded concentrations of dasatinib (5 nM, 10 nM, 25 nM, and 100 nM). (C) Number of wells with resistant growth in the presence of graded concentrations of nilotinib (50 nM, 500 nM, 1000 nM, and 2000 nM) and dasatinib (5 nM, 10 nM, 25 nM, and 100 nM).
Discussion
Alternative Abl kinase inhibitors are emerging as the most promising strategy to overcome drug resistance induced by BCR-ABL mutations. The compounds that have advanced furthest are nilotinib, an imatinib mesylate derivative,13 and dasatinib, a combined Abl/Src kinase inhibitor.14 Other agents such as SKI60629 and NS-18730 have only recently or not yet entered clinical testing. Although the use of these compounds is currently limited to imatinib mesylate–resistant disease, it is obvious that they may have potential in earlier stages of CML, either as single agents or perhaps in simultaneous or sequential drug combinations. Although both dasatinib and nilotinib are much more potent than imatinib mesylate and have activity against most kinase domain mutants, it is conceivable that their specific modes of binding to Abl may lead to new “vulnerable” sites that could confer drug resistance. Identification of such sites a priori would open the possibility to rationally design combination therapy strategies.
Here, we compared imatinib mesylate, nilotinib, and dasatinib using a cell-based assay that uses ENU-based mutagenesis prior to selection of resistant clones in the presence of drugs. We found that at concentrations approximately 2-fold higher than the IC90 against unmutated BCR-ABL in cell proliferation assays (2 μM imatinib mesylate, 50 nM nilotinib, 5 nM dasatinib),18 the overwhelming majority of resistant cell clones carried BCR-ABL KD mutations. The majority of mutations detected in patients treated with imatinib mesylate were observed (Table S3), and few novel mutations were detected, indicating that this assay faithfully reproduces the spectrum of clinically relevant mutations.
In contrast to imatinib mesylate, mutations recovered in the presence of 50 nM nilotinib were limited to L248V, p-loop (G250E, Y253H, E255K), T315I, F359C, L384M, and L387F. One novel mutation (E292V) was seen in a single clone at 50 nM. At 500 nM, only Y253H, E255V, and T315I were detected, in line with their reported level of resistance in cell proliferation assays.18 At concentrations of at least 2000 nM, only T315I was recovered. The fact that only one novel mutation occurred at a low concentration and the lack of novel, previously undescribed mutations at intermediate inhibitor concentrations (500 nM) suggests that the structural modifications of nilotinib compared with imatinib mesylate do not generate clinically relevant novel vulnerable sites. Because the plasma trough concentrations at the maximum tolerated dose (MTD) of nilotinib (400 mg twice daily) are in the range of 2000 nM, the prediction is that only T315I will emerge in patients treated with the MTD.25
At 5 nM dasatinib mutations of L248, Q252, E255, V299, T315, and F317 were recovered (Table 1), similar to the results of Burgess et al,28 whereas at intermediate drug levels (25 nM) only F317V was found in addition to T315I. Overall, F317 was the most frequently involved residue other than T315, and 4 different mutations were identified (F317I/V/L/C), implicating F317 as a potential vulnerable site for dasatinib. Consistent with this, F317C/L/V were also detected in Ba/F3-p185BCR-ABL cells grown in the presence of PD166326, a pyridopyrimidine type Abl/Src inhibitor.21 Crystal structure analysis of the Abl KD in complex with dasatinib has revealed that the compound binds an active conformation of Abl, with F317 as a contact residue.17 Additional mutants detected at 5 nM and 10 nM dasatinib include V299L (also a contact point14 ) and Q252H in the p-loop. Both mutants are relatively resistant to dasatinib in cell proliferation assays.18,28 At dasatinib concentrations of at least 100 nM only T315I was recovered, suggesting that only T315I may be capable of growing out at MTD plasma levels.15 In line with this, patients with T315I do not respond to dasatinib, and this mutation was identified in several patients at the time of relapse.31,32 Interestingly, F317I was also detected in a patient with acquired resistance to dasatinib.33 Whether this is a more common phenomenon remains to be seen as more data become available.
Dasatinib and nilotinib may eventually be used in combination with imatinib mesylate or each other. The value of such combinations could be either to minimize resistance by combining maximal doses of 2 inhibitors or to minimize side effects if less than maximal doses of each drug could be given. To test these possibilities we conducted a series of combination experiments (Figure 3). Although, as expected, fewer resistant clones were recovered with drug combinations than with the respective single agents, the observed patterns were variable between the various combinations. In combinations containing imatinib mesylate the frequency of resistant clones tended to follow a dose response along the imatinib mesylate gradient, most evidently for combinations of imatinib mesylate with nilotinib, whereby imatinib mesylate seemed to enhance suppression of resistant clones even at high levels of nilotinib (Figure 3A). Because nilotinib binds to the same Abl conformation, is more than 20-fold more potent than imatinib mesylate, and does not appear to have additional vulnerable sites, one would not have expected to see increased efficacy from adding comparatively low concentrations of imatinib mesylate, and the reason for this observation is not clear. In contrast, in combinations of nilotinib and dasatinib, maximal suppression was seen with 10 nM dasatinib + 50 nM nilotinib or 5 nM dasatinib + 500 nM nilotinib, suggesting that the combination leads to mutual elimination of mutants with intermediate resistance, consistent with predictions based on the single-agent experiments (Table 2). Thus, low doses of dasatinib combined with low doses of nilotinib may effectively suppress the emergence of mutations other than T315I. Because the nonhematologic side effects of nilotinib and dasatinib are not identical,15,25 patients with intolerance to either agent could be managed with combinations at low doses, avoiding toxicity while maintaining full antileukemic activity. Clearly, with T315I emerging as the “default” mutation, an inhibitor of T315I will ultimately be required to close the remaining gap.34
Cellular proliferation IC50 values of BCR-ABL point mutants recovered in the presence of nilotinib and dasatinib
. | Nilotinib . | . | Dasatinib . | . | Imatinib mesylate . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mutation . | IC50, nM . | Fold change . | IC50, nM . | Fold change . | IC50, nM . | Fold change . | |||
| L248V | NR* | NR* | NR | NR | NR | NR | |||
| G250E | 48* | 4* | 1.8 | 2 | 1350 | 5 | |||
| Q252H | 70 | 4 | 3.4† | 2† | 1325 | 5 | |||
| Y253H | 450† | 35† | 1.3 | 2 | > 6400 | > 25 | |||
| E255K | 200* | 14* | 5.6 | 7 | 5200 | 20 | |||
| E255V | 430† | 33† | 11 | 14 | > 6400 | > 25 | |||
| E292V | NR* | NR* | NR | NR | NR | NR | |||
| V299L | NR | NR | 18* | 13.4* | 540 | 1.7 | |||
| T3151 | > 2000‡ | > 154‡ | > 200‡ | > 250‡ | > 6400 | > 25 | |||
| F317C | NR | NR | NR* | NR* | 1200 | 3 | |||
| F317L | 50 | 4 | 7.4* | 9* | 1050 | 4 | |||
| F3171 | NR | NR | NR* | NR* | NR | NR | |||
| F317V | 350 | 1.1 | 53† | 40† | 500 | 1.3 | |||
| F359C | NR | NR | NR* | NR* | 1200 | 3 | |||
. | Nilotinib . | . | Dasatinib . | . | Imatinib mesylate . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mutation . | IC50, nM . | Fold change . | IC50, nM . | Fold change . | IC50, nM . | Fold change . | |||
| L248V | NR* | NR* | NR | NR | NR | NR | |||
| G250E | 48* | 4* | 1.8 | 2 | 1350 | 5 | |||
| Q252H | 70 | 4 | 3.4† | 2† | 1325 | 5 | |||
| Y253H | 450† | 35† | 1.3 | 2 | > 6400 | > 25 | |||
| E255K | 200* | 14* | 5.6 | 7 | 5200 | 20 | |||
| E255V | 430† | 33† | 11 | 14 | > 6400 | > 25 | |||
| E292V | NR* | NR* | NR | NR | NR | NR | |||
| V299L | NR | NR | 18* | 13.4* | 540 | 1.7 | |||
| T3151 | > 2000‡ | > 154‡ | > 200‡ | > 250‡ | > 6400 | > 25 | |||
| F317C | NR | NR | NR* | NR* | 1200 | 3 | |||
| F317L | 50 | 4 | 7.4* | 9* | 1050 | 4 | |||
| F3171 | NR | NR | NR* | NR* | NR | NR | |||
| F317V | 350 | 1.1 | 53† | 40† | 500 | 1.3 | |||
| F359C | NR | NR | NR* | NR* | 1200 | 3 | |||
Imatinib mesylate values are provided for comparison.
NR indicates not reported.
Mutants recovered at lower drug concentrations.
Mutations recovered at intermediate drug concentrations.
Prepublished online as Blood First Edition Paper, June 13, 2006; DOI 10.1182/blood-2006-02-004580.
Supported in part by the National Heart, Lung, and Blood Institute (grant HL082978-01) (M.W.D.); the Doris Duke Charitable Foundation (B.J.D.); and the Leukemia and Lymphoma Society (B.J.D. and M.W.D.). M.W.D. is a Junior Faculty Scholar of the American Society of Hematology.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Note added in proof. While this manuscript was under review, von Bubnoff et al have obtained results of a cell-based resistance screen investigating BCR-ABL mutations that arise in the presence of nilotinib, confirming the selection of T315I in higher drug concentrations.35
We thank Taiping Jia, OHSU, for help with experiments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal