Abstract
Ectodermal dysplasia with immune deficiency (EDI) is caused by alterations in NEMO (nuclear factor [NF]–κB essential modulator). Most genetic mutations are located in exon 10 and affect the C-terminal zinc finger domain. However, the biochemical mechanism by which they cause immune dysfunction remains undetermined. In this report, we investigated the effect of a cysteine-to-arginine mutation (C417R) found in the NEMO zinc finger domain on dendritic cell (DC) function. Following CD40 stimulation of DCs prepared from 2 unrelated patients with the NEMO C417R mutation, we found NEMO ubiquitination was absent, and this was associated with preserved RelA but absent c-Rel activity. As a consequence, CD40 stimulated EDI DCs failed to synthesize the c-Rel–dependent cytokine interleukin-12, had impaired up-regulation of costimulatory molecules, and failed to support allogeneic lymphocyte proliferation in vitro. In contrast, EDI DCs stimulated with the TLR4 ligand lipopolysaccharide (LPS) showed normal downstream NF-κB activity, DC maturation, and NEMO ubiquitination. These findings show for the first time how mutations in the zinc finger domain of NEMO can lead to pathway specific defects in NEMO ubiquitination and thus immune deficiency.
Introduction
Dendritic cells (DCs) recognize, capture, and process foreign antigens in order to activate T cells through antigen presentation, expression of surface costimulatory molecules, and secretion of effector cytokines.1 In the presence of a cognate antigen, DC-primed T lymphocytes express CD40 ligand (CD40L), which interacts with CD40 on the surface of DCs to promote their further maturation.2 CD40L elevates expression of the major histocompatibility complex (MHC) and costimulatory molecules as well as the secretion of cytokines such as interleukin (IL)–12 to drive the differentiation and expansion of T cells.3,4 Impaired CD40-CD40L interaction in both humans and mice leads to impaired T-helper 1 (Th1) T-cell differentiation and predisposes the host to opportunistic infections.5,6
Signaling induced by CD40L, pathogen-derived molecules that bind the Toll-like receptors (TLRs), or proinflammatory mediators secreted in the local microenvironment, including tumor necrosis factor-α (TNF-α) and IL-1β converge on the IκB kinase complex (IKK) to activate the transcription factor nuclear factor-κB (NF-κB).1,7 The IKK signalosome consists of 2 related proteins with intrinsic kinase activity, IKKα and IKKβ, and a regulatory noncatalytic subunit NEMO (NF-κB essential modulator, also known as IKKγ). After cellular stimulation, upstream mediators promote activation of the IKK complex and the phosphorylation and subsequent degradation of the cytosolic inhibitors IκBs, facilitating the nucleus translocation of NF-κB dimers.8-10 In the IKK-dependent NF-κB signaling pathway, the NF-κB transcription factors RelA (p65) and c-Rel heterodimerize with p50 to regulate the transcription of genes responsible for development and activation of DCs.11
Hypomorphic mutations in the gene encoding NEMO give rise to a X-linked disease known as ectodermal dysplasia with immunodeficiency (EDI). This disorder is characterized by severe immunologic impairment as well as abnormal development of ectoderm-derived structures leading to misshaped or absent teeth, lack of eccrine sweat glands, and sparse scalp hair.12-14 We have shown previously that B cells from EDI patients with a C417R missense mutation have a profound defect in the activation of NF-κB in response to CD40 ligation.15 Their peripheral blood B cells invariably express surface immunoglobulin M (IgM) and IgD, are devoid of somatic mutations in the Ig variable region and fail to undergo class-switch recombination in response to CD40 agonists in vitro.
In the present study, we sought to define the biochemical mechanism underlying impaired CD40L responses using patient DCs. Our data show that CD40 and TLR4 use separate pathways to ubiquitinate NEMO, and defective NEMO ubiquitination impairs DC function. Interestingly, interferon-γ (IFN-γ) costimulation can enhance CD40-induced RelA activation and some DC functions, but it does not rescue c-Rel activity. Using microarray analysis of patient DCs, we identified downstream effects of impaired NF-κB activation as well as candidate factors that may be necessary for DC function and cell-mediated immunity.
Patients, materials, and methods
Patients and protocols
Patients were studied at the Clinical Center, NIAID, NIH (protocol 89-1-0158). The diagnosis of EDI was established by medical history and by sequencing the gene encoding NEMO by methods previously described.15 Unaffected family members or other immunologically healthy volunteers served as controls. The Institutional Review Board of NIAID approved the open protocol and informed consent was obtained from all patients or their parents before enrollment in the study.
Cells and reagents
Immature monocyte-derived dendritic cells (MoDCs) were prepared by culturing elutriated monocytes in RPMI 1640 supplemented with IL-4 (R&D Systems, Minneapolis, MN) and granulocyte-macrophage colony-stimulating factor (GM-CSF; Berlex Laboratories, Richmond, VA) as previously described.16 The following reagents were used for stimulation: CD40L (2.5 μg/mL; Amgen, Seattle, WA), IFN-γ (2000 U/mL; Genzyme, Brisbane, CA) and LPS (2.5 μg/mL; Sigma, St Louis, MO).
Flow cytometry analysis
After stimulation, MoDCs were immunostained with monoclonal antibodies, including anti-CD40 (5C3), anti-CD54 (HA58), anti-CD80 (L307.4), anti-CD83 (HB15e), and anti-CD86 (2331). After washing, 10 000 events were collected on a FACScalibur (BD Biosciences, Palo Alto, CA) flow cytometer. Acquired events were analyzed using FlowJo software (Treestar, Ashland, OR).
Morphologic studies
MoDCs were incubated in culture chamber slides (Nunc, Rochester, NY), and differential interference contrast (DIC) images were collected on a Leica DMIRBE (Leica Microsystems, Exton, PA) using a 40 ×/1.25 NA oil-immersion objective lens. Images were processed using Leica TCS-NT/SP software and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
Mixed-lymphocyte reaction
Allogeneic CD3+ T lymphocytes were purified from peripheral blood mononuclear cells (PBMCs) by positive selection. Briefly, cells were incubated with magnetic-activated cell-sorter (MACS) CD3 microbeads (Miltenyi Biotec, Auburn, CA) for 15 minutes at 4°C. After washing, cell pellets were resuspended and applied onto a MACS column (Miltenyi Biotec) in a magnetic field. Positively selected cells were collected and the purity assessed by flow cytometry (> 95%). Patient and normal MoDCs were added to allogeneic CD3+ T cells at different ratios and stimulated with LPS (2.5 μg/mL) or CD40L (2.5 μg/mL) alone or CD40L supplemented with IFN-γ (2000 U/mL) or IL-12p70 (10 ng/mL; R&D Systems) in a round-bottom 96-well plate. After 7 days of coculture, cells were pulsed with 1 μCi (37 KBq) tritiated thymidine for an additional 18 hours at 37°C. Cells were harvested and thymidine incorporation measured using a β scintillation counter (Topcount; Perkin-Elmer, Wellesley, MA).
Microarray analysis
RNA was prepared from MoDCs using Qiagen RNeasy reagents (Valencia, CA) according to the manufacturer's recommendations. RNA (5 μg total) from individual samples was labeled according to standard protocols (Affymetrix, Santa Clara, CA) and hybridized to individual U95A arrays (Affymetrix) consisting of approximately 12 000 human genes from Unigene build 95. Hybridized chips were processed as recommended and scanned images were analyzed using Resolver software (Rosetta Biosoftware, Kirkland, WA) and Spotfire Decision Site (Spotfire, Somerville, CA). Hierarchic clustering was performed using the Eisen Cluster program and Cluster Trees produced with Java Treeview (Alok Saldanha, Stanford University, Stanford, CA).
Expression profiles were validated by flow cytometry (Figure 3A) and by quantitative real-time PCR for the following genes using methods described by the manufacturer (CD54, Hs00164932_m1; CD44, Hs00174139_m1; Rab11a, Hs00366449_g1; CD86, Hs00199349_m1; and Bcl6, Hs00153368_m1; Applied Biosystems, Foster City, CA).
Electrophoretic mobility shift assay
Cellular lysates were prepared by resuspending cell pellets in 100 mM NaCl, 50 mM Tris-HCl at pH 8.0, 0.5% NP-40, 50 mM NaF, 30 mM Na4PPi, 1 mM Na3VO4, 0.6% diisopropyl fluorophosphate (Sigma), and 1 complete protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN). Protein concentrations were determined using a Micro BCA protein assay (Pierce, Rockford, IL), and c-Rel and p65 binding to a 32P-labeled oligonucleotide probe (5′-AGCTTGGGGTATTTCCAGCCG-3′) was performed using a c-Rel Gelshift kit (Active Motif, Carlsbad, CA) following the manufacturer's instructions. For c-Rel supershift, cell extracts were first incubated with a human c-Rel–specific rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 20 minutes. The gels were dried and exposed to phosphoimaging screens (Kodak, Eastman, NY), and signal intensities of specific DNA-protein complexes were analyzed using a Scion Imager (Scion, Frederick, MD).
Immunoprecipitations and immunoblotting
To study NEMO ubiquitination, cellular lysates were prepared with immunoprecipitation (IP) lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 1% Triton X-100, 10% glycerol, 3 mM EGTA, 3 mM EDTA, 0.1 mM Na3VO4, 10 μM N-ethylmaleimide, and 1 complete protease inhibitor mixture (Roche Diagnostics). The supernatants prepared from the lysates were next precleared with protein A/G Plus-Agarose (Santa Cruz Biotechnology) for 1 hour at 4°C and then incubated with anti-NEMO mouse monoclonal antibody (BD Biosciences) for 4 hours at 4°C. Protein A/G Plus-Agarose was then added for 1 hour at 4°C. Immunoprecipitates were washed 3 times with IP lysis buffer and heat-denatured at 70°C for 10 minutes in 4 × diluted NuPage LDS sample buffer supplemented with 2.5% 2-ME. Samples were resolved on a 4% to 12% polyacrylamide gel (Invitrogen, Carlsbad, CA), and proteins were transferred onto nitrocellulose membrane and probed with anti–ubiquitin rabbit polyclonal antibodies (Imgenex [San Diego, CA] and Santa Cruz Biotechnology). Detection was performed using an horseradish peroxidase (HRP)–conjugated donkey anti–rabbit IgG (Amersham Biosciences, Arlington Heights, IL) and developed by the chemiluminescence method (Amersham Biosciences).
Results
Impaired NF-κB heterodimers activation in EDI MoDCs
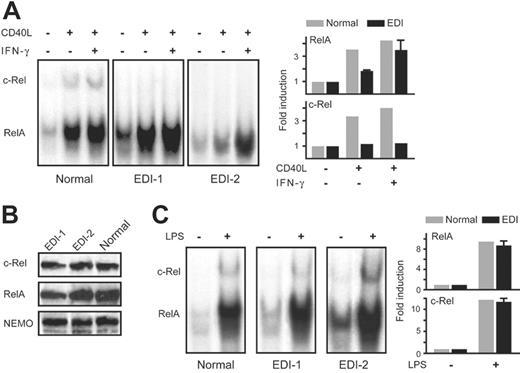
In initial studies, we extended our previous findings concerning NF-κB activation in B cells of patients with EDI to MoDCs. Monocytes from 2 unrelated EDI patients with a missense mutation affecting the cysteine residue at position 417 of NEMO were cultured in the presence of IL-4 and GM-CSF for 1 week to induce their differentiation into immature MoDCs.16 Notably, no morphologic differences were observed between normal and patient immature MoDCs, indicating that the NEMO C417R mutation does not affect this phase of DC development. To characterize the signaling defect in EDI MoDCs we assessed the NF-κB function by measuring DNA-binding activity of both RelA and c-Rel in an electrophoretic mobility shift assay (EMSA). Compared with healthy controls, RelA binding was modestly reduced in EDI MoDCs after stimulation with CD40L. In contrast, c-Rel binding was notably absent in patient MoDCs as determined by a c-Rel–specific antibody (Figure 1A). The level of NEMO, c-Rel, and RelA protein in EDI MoDCs were comparable to that in healthy controls (Figure 1B); thus, the defects in RelA and c-Rel activation were not due to aberrant protein synthesis of these NF-κB signaling constituents. These data highlight the importance of the NEMO zinc finger domain for downstream NF-κB signaling.
Recognizing that IFN-γ costimulation enhances CD40L activation of MoDCs, we next assessed whether addition of IFN-γ improved RelA and c-Rel induction in CD40L-stimulated MoDCs. While IFN-γ alone did not modify NF-κB activity (data not shown), IFN-γ costimulation with CD40L enhanced RelA activation in both normal and patient MoDCs as well as c-Rel induction in control MoDCs (Figure 1A). Nevertheless, c-Rel activity remained absent in IFN-γ–costimulated EDI MoDCs. The mechanism by which IFN-γ enhances NF-κB signaling awaits further investigation. When we repeated our costimulation experiments in presence of cycloheximide, an inhibitor of new protein synthesis, IFN-γ no longer improved CD40L-induced NF-κB activation (data not shown). This suggests that the proximal IFN-γ signaling effectors do not act directly on the NF-κB pathway, and new protein synthesis is required for IFN-γ–mediated enhancement of NF-κB activity.
NEMO C417R mutation impairs RelA activation and abolishes c-Rel induction in MoDCs stimulated with CD40L but not with LPS. (A) Left panel shows DNA binding activity of NF-κB dimers in MoDCs stimulated with CD40L (2.5 μg/mL) or CD40L plus IFN-γ (2000 U/mL) for 4 hours. Anti–c-Rel antibody was added prior to incubation with the labeled NF-κB DNA probe to supershift c-Rel containing dimers. Right panel shows quantification of RelA and c-Rel activity represented as means ± SEM of fold change compared with unstimulated cells (set at 1.0). (B) NEMO, c-Rel, and RelA protein levels in MoDCs. Cell lysates from 2 healthy volunteers and 1 patient with EDI were subjected to immunoblotting. Blots were probed with anti–c-Rel, anti-RelA, or anti-NEMO antibody. (C) Left panel shows DNA-binding activity of NF-κB dimers in MoDcs stimulated with LPS (2.5 μg/mL) for 4 hours and processed as in panel A. Right panel shows quantification of RelA and c-Rel activity represented as means ± SEM of fold change compared with unstimulated cells (set at 1.0).
NEMO C417R mutation impairs RelA activation and abolishes c-Rel induction in MoDCs stimulated with CD40L but not with LPS. (A) Left panel shows DNA binding activity of NF-κB dimers in MoDCs stimulated with CD40L (2.5 μg/mL) or CD40L plus IFN-γ (2000 U/mL) for 4 hours. Anti–c-Rel antibody was added prior to incubation with the labeled NF-κB DNA probe to supershift c-Rel containing dimers. Right panel shows quantification of RelA and c-Rel activity represented as means ± SEM of fold change compared with unstimulated cells (set at 1.0). (B) NEMO, c-Rel, and RelA protein levels in MoDCs. Cell lysates from 2 healthy volunteers and 1 patient with EDI were subjected to immunoblotting. Blots were probed with anti–c-Rel, anti-RelA, or anti-NEMO antibody. (C) Left panel shows DNA-binding activity of NF-κB dimers in MoDcs stimulated with LPS (2.5 μg/mL) for 4 hours and processed as in panel A. Right panel shows quantification of RelA and c-Rel activity represented as means ± SEM of fold change compared with unstimulated cells (set at 1.0).
CD40L and TLR ligand signaling pathways activate the IKK complex. We therefore investigated whether EDI MoDCs stimulated with LPS, a TLR4 agonist, also displayed differential RelA and c-Rel induction. Interestingly, LPS stimulation results in both RelA and c-Rel activation in the EDI C417R MoDCs, and the level of this activation was comparable with what was observed in healthy controls (Figure 1C). These results imply that the NEMO C417R zinc finger domain mutation impairs CD40-mediated, but not TLR4-mediated, NF-κB signaling pathway.
NEMO ubiquitination is impaired in CD40L-stimulated EDI patients
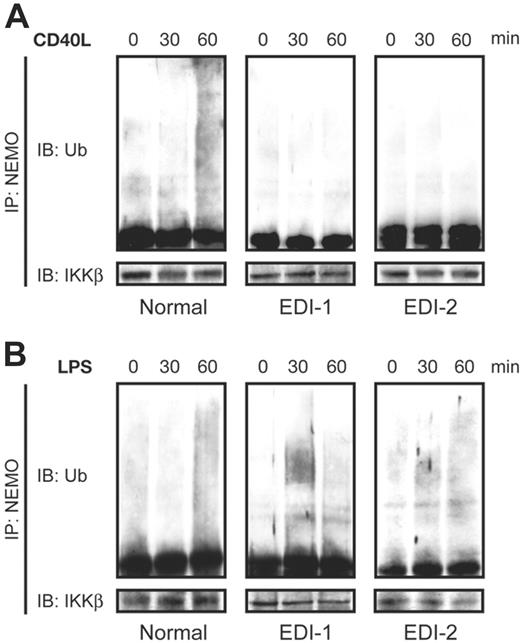
Ubiquitination is a posttranslational modification in which the 76–amino acid ubiquitin protein is reversibly conjugated to a lysine residue on a target protein. The type of ubiquitin linkage determines the fate of cellular proteins. Polyubiquitin (poly-Ub) chains linked through lysine 48 (K48) target proteins for degradation by the proteosome.8 However, several recent studies have reported that K63-linked poly-Ub chains can activate NF-κB through a degradation-independent mechanism.17,18 NEMO has been shown to be K63 polyubiquitinated at K399, which lies within its zinc finger motif.11,19 To determine whether the NEMO C417R mutation affects its ubiquitination, we stimulated EDI antigen-presenting cells with CD40L and examined NEMO ubiquitination with immunoprecipitation using an anti-NEMO antibody followed by immunoblotting with an antiubiquitin antibody. A smear of slower-migrating forms of NEMO, corresponding to the polyubiquinated protein, was maximally observed at 60 minutes in control cells stimulated with CD40L, but the absence of NEMO ubiquitination was observed in the CD40L-stimulated EDI cells (Figure 2A).
In parallel, we examined the ubiquitination of NEMO in cells stimulated with LPS. Consistent with our EMSA findings, the NEMO C417R mutation does not alter the TLR4 signaling pathway, as NEMO is normally ubiquitinated in patients with EDI in response to LPS (Figure 2B). Taken together, our findings suggest that CD40L and LPS signaling pathways use separate cofactors to catalyze NEMO ubiquitination.
Impaired maturation of MoDCs in vitro
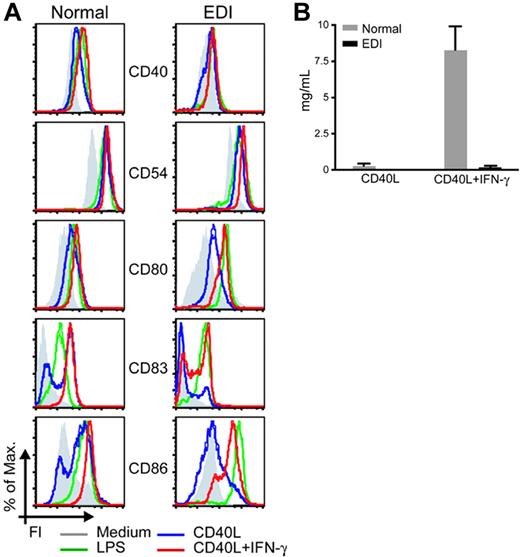
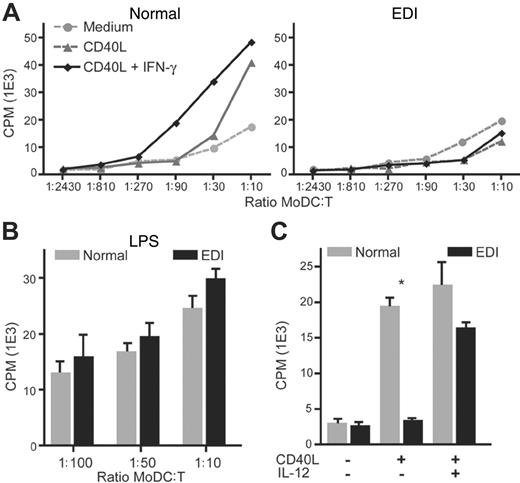
In the next series of experiments, we studied the impact of the defective CD40 signaling on MoDC maturation and functions. While EDI MoDC maturation was unaffected in response to TLR4 stimulation, they failed to up-regulate surface maturation markers such as the B7 family members (CD80 and CD86), CD40, CD54, and CD83 in response to CD40L stimulation (Figure 3A). As IFN-γ can enhance DC activation,20 the maturation profile of MoDCs was assessed after stimulation. IFN-γ alone did not affect the expression of surface molecules, while IFN-γ with CD40L costimulation partly rescued the expression of all maturation markers tested (Figure 3A). In agreement with previous findings on patient PBMCs, EDI MoDCs failed to secrete the heterodimeric cytokine IL-12p70 in response to CD40L, and this deficit was not restored by the addition of IFN-γ (Figure 3B). These findings show that CD40-mediated MoDC maturation is impaired in cells with a NEMO C417R mutation, and that some but not all of these defects can be complemented by IFN-γ costimulation.
K-63 polyubiquitination of C417R mutated NEMO is impaired in EDI MoDCs stimulated through CD40 but not through TLR4. (A) Polyubiquination of NEMO in response to CD40L (2.5 μg/mL) for the indicated time periods in normal and EDI MoDCs. (B) Polyubiquitination of NEMO when stimulated with LPS (2.5 μg/mL) for the indicated time periods in normal and EDI MoDCs. Cell lysates were immunoprecipitated (IP) with anti-NEMO monoclonal antibody and then analyzed with an anti-Ub antibody by immunoblotting (IB). The blot was reprobed with an anti-IKKβ antibody that served as a loading control.
K-63 polyubiquitination of C417R mutated NEMO is impaired in EDI MoDCs stimulated through CD40 but not through TLR4. (A) Polyubiquination of NEMO in response to CD40L (2.5 μg/mL) for the indicated time periods in normal and EDI MoDCs. (B) Polyubiquitination of NEMO when stimulated with LPS (2.5 μg/mL) for the indicated time periods in normal and EDI MoDCs. Cell lysates were immunoprecipitated (IP) with anti-NEMO monoclonal antibody and then analyzed with an anti-Ub antibody by immunoblotting (IB). The blot was reprobed with an anti-IKKβ antibody that served as a loading control.
EDI MoDCs fail to up-regulate surface markers and secrete IL-12 in response to CD40L stimulation. (A) MoDCs from 1 healthy control and 1 patient with EDI (EDI-1) were incubated with medium (gray histogram), CD40L (2.5 μg/mL; blue histogram), CD40L plus IFN-γ (2000 U/mL; red histogram) or LPS (2.5 μg/mL; green histogram) for 24 hours. Cells were immunostained with specific antibodies for surface markers and 10 000 events were collected. Each diagram represents the fluorescence intensity as measured by using a flow cytometer. One representative experiment of 3 is shown. (B) MoDCs from 3 healthy controls and 2 EDI patients were incubated 24 hours with CD40L (2.5 μg/mL) or CD40L plus IFN-γ (2000 U/mL). IL-12p70 secretion was measured in collected supernatants by Luminex (Austin, TX). Data are shown as means ± SEM.
EDI MoDCs fail to up-regulate surface markers and secrete IL-12 in response to CD40L stimulation. (A) MoDCs from 1 healthy control and 1 patient with EDI (EDI-1) were incubated with medium (gray histogram), CD40L (2.5 μg/mL; blue histogram), CD40L plus IFN-γ (2000 U/mL; red histogram) or LPS (2.5 μg/mL; green histogram) for 24 hours. Cells were immunostained with specific antibodies for surface markers and 10 000 events were collected. Each diagram represents the fluorescence intensity as measured by using a flow cytometer. One representative experiment of 3 is shown. (B) MoDCs from 3 healthy controls and 2 EDI patients were incubated 24 hours with CD40L (2.5 μg/mL) or CD40L plus IFN-γ (2000 U/mL). IL-12p70 secretion was measured in collected supernatants by Luminex (Austin, TX). Data are shown as means ± SEM.
Impaired clustering of CD40L-stimulated EDI MoDCs
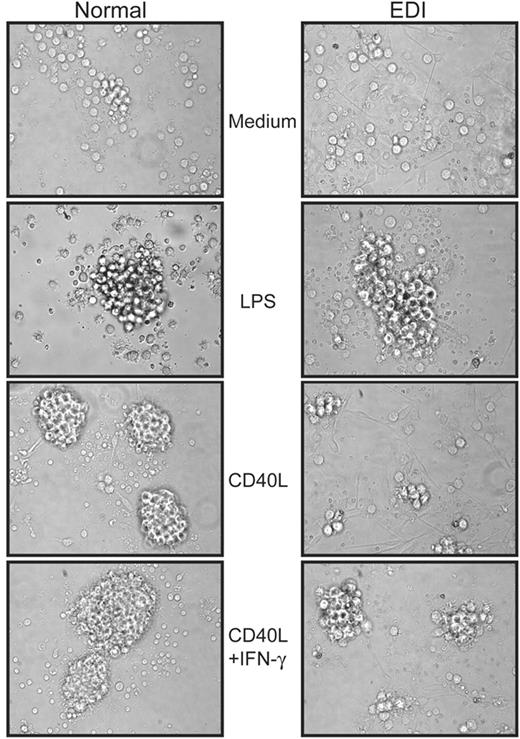
Upon activation DCs undergo a reorganization of their actin cytoskeleton and extend membrane protrusions, called dendrites, to facilitate interaction with potential antigen-restricted T cells.21 Furthermore, clustering of mature DCs supports efficient activation and expansion of lymphocytes. We therefore analyzed the clustering ability and the dendrite extension of CD40L-stimulated MoDCs by microscopy. Tight and dense aggregates were observed when normal MoDCs were incubated for 24 hours with CD40L, and in EDI DCs stimulated with LPS (Figure 4). In contrast, EDI MoDCs failed to form clusters and dendrite formation was greatly diminished in the presence of CD40L. Whereas the addition of IFN-γ improved the clustering of normal MoDCs, only a weak enhancement was detected for EDI MoDCs, suggesting morphogenic responses associated with CD40 signaling is impaired in EDI MoDCs.
CD40L-stimulated EDI MoDCs fail to trigger allogeneic T-cell proliferation
As EDI MoDCs exhibit several activation defects, we next assessed whether patient MoDCs can support the proliferation of allogeneic T cells in a mixed leukocyte culture. Isolated T lymphocytes from a healthy control were cocultured in the presence of MoDCs prepared from an unrelated healthy individual or a patient with EDI. Next, lymphoproliferation was measured by thymidine incorporation. CD40L stimulation greatly enhanced allogeneic T-cell expansion when cultured with normal MoDCs (Figure 5A). In contrast, T lymphocyte proliferation remained weak when cultured with CD40L-stimulated EDI MoDCs. Because the addition of IFN-γ partly rescues the maturation of patient MoDCs, we measured T-cell proliferation in the presence of CD40L and IFN-γ. Whereas increased T-cell proliferation is observed with normal MoDCs, CD40L plus IFN-γ–stimulated EDI MoDCs failed to induce T-cell expansion, indicating that T-cell activation requires the expression of additional factors that are lacking in EDI MoDCs. A likely candidate is the proinflammatory cytokine IL-12, which is not secreted by EDI MoDCs in response to stimulation with CD40L and IFN-γ (Figure 3B). In agreement with this hypothesis, the addition of human recombinant IL-12 with CD40L induced the allogeneic T-cell proliferation when cultured with EDI MoDCs. These findings confirm that EDI MoDCs are impaired in the expression of c-Rel regulated cytokines that are normally synthesized in response to CD40L and are critical for T-cell activation.
EDI MoDCs fail to form tight and dense clusters when stimulated with CD40L. After 24 hours of stimulation with CD40L (2.5 μg/mL), CD40L plus IFN-γ (2000 U/mL), or LPS (2.5 μg/mL), healthy control (left panel) and EDI patient (right panel) MoDCs were subjected to differential interference contrast (DIC) microscopy, and images were taken at 40 × magnification. One experiment of 2 is shown.
EDI MoDCs fail to form tight and dense clusters when stimulated with CD40L. After 24 hours of stimulation with CD40L (2.5 μg/mL), CD40L plus IFN-γ (2000 U/mL), or LPS (2.5 μg/mL), healthy control (left panel) and EDI patient (right panel) MoDCs were subjected to differential interference contrast (DIC) microscopy, and images were taken at 40 × magnification. One experiment of 2 is shown.
CD40L-stimulated EDI MoDCs fail to support the proliferation of allogeneic T cells. (A) Positively selected CD3+ cells (purity > 99%) from a healthy donor were mixed with increasing number of unrelated MoDCs from a healthy control (left panel) or patient with EDI (right panel) for 6 days in the presence of medium alone, CD40L (2.5 μg/mL), CD40L plus IFN-γ (2000 U/mL), or LPS (2.5 μg/mL). (B) CD3+ cell proliferation was evaluated after incorporation of triated thymidine by using a β scintillation counter. Data are shown as means ± SEM of counts per minute (cpm). (C) Addition of recombinant human IL-12 (10 ng/mL) rescues CD3+ lymphoproliferation in the presence of CD40L-stimulated EDI MoDCs. Data are shown as means ± SEM of one representative DC/T-cell ratio (1:20). *P < .05.
CD40L-stimulated EDI MoDCs fail to support the proliferation of allogeneic T cells. (A) Positively selected CD3+ cells (purity > 99%) from a healthy donor were mixed with increasing number of unrelated MoDCs from a healthy control (left panel) or patient with EDI (right panel) for 6 days in the presence of medium alone, CD40L (2.5 μg/mL), CD40L plus IFN-γ (2000 U/mL), or LPS (2.5 μg/mL). (B) CD3+ cell proliferation was evaluated after incorporation of triated thymidine by using a β scintillation counter. Data are shown as means ± SEM of counts per minute (cpm). (C) Addition of recombinant human IL-12 (10 ng/mL) rescues CD3+ lymphoproliferation in the presence of CD40L-stimulated EDI MoDCs. Data are shown as means ± SEM of one representative DC/T-cell ratio (1:20). *P < .05.
We also examined allogeneic T-cell proliferation when EDI MoDCs are stimulated with LPS. Consistent with our EMSA findings and NEMO ubiquitination studies, the NEMO C417R mutation does not alter EDI MoDC activation following TLR4 stimulation, as allogeneic T cells proliferate normally (Figure 5C). Taken together, our findings indicate that CD40L and LPS signaling use separate and distinct cofactors to activate the IKK complex and DC activation.
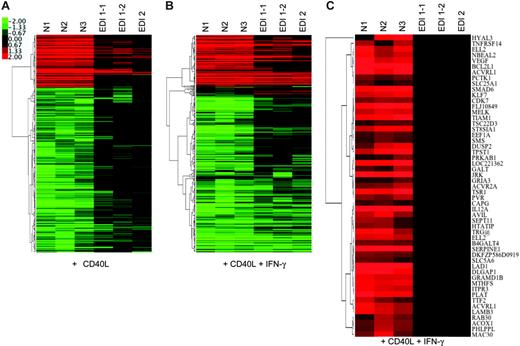
Microarray analysis of EDI MoDCs stimulated with CD40L and CD40L plus IFN-γ. MoDCs from healthy (N1-N3) and EDI (EDI patient 1; EDI1-1 and EDI1-2 from 2 independent cultures: EDI patient 2; EDI2) subjects were stimulated with CD40L alone or CD40L plus IFN-γ for 24 hours. Each row represents the ratio of expression in stimulated versus unstimulated cells for each gene, and each column represents the data from an independent stimulation experiment. The color bar shows the magnitude of gene expression changes on a log scale: significantly induced (red), unchanged (black), or repressed (green). (A) Genes that show consistent regulation in normal MoDCs in response to CD40L but abnormal regulation in EDI MoDCs. A hierarchic cluster map of 400 probe sets that show consistent regulation in normal MoDCs (> 2-fold; P < .01) but significantly different regulation in EDI MoDCs (ANOVA at P < .01). (B) Addition of IFN-γ restores a portion of the CD40L expression signature in EDI MoDCs. A hierarchic cluster map of the expression patterns for the 400 probe sets from panel A in response to CD40L plus IFN-γ. Note that many of the probe sets show similar expression in EDI and normal samples, illustrating the effects of IFN-γ on complementing the CD40L expression signature in EDI MoDCs. (C) EDI MoDCs fail to up-regulate a subset of CD40L-induced genes even in the presence of IFN-γ. A number of probe sets (53) are shown that passed the following filter criteria: significantly regulated in all 3 normal CD40L plus IFN-γ cultures (P < .01); and no significant regulation in any of the 3 EDI CD40L plus IFN-γ cultures. Note that for this analysis any ratio values with P > .01 were set to log (ratio) = 0.
Microarray analysis of EDI MoDCs stimulated with CD40L and CD40L plus IFN-γ. MoDCs from healthy (N1-N3) and EDI (EDI patient 1; EDI1-1 and EDI1-2 from 2 independent cultures: EDI patient 2; EDI2) subjects were stimulated with CD40L alone or CD40L plus IFN-γ for 24 hours. Each row represents the ratio of expression in stimulated versus unstimulated cells for each gene, and each column represents the data from an independent stimulation experiment. The color bar shows the magnitude of gene expression changes on a log scale: significantly induced (red), unchanged (black), or repressed (green). (A) Genes that show consistent regulation in normal MoDCs in response to CD40L but abnormal regulation in EDI MoDCs. A hierarchic cluster map of 400 probe sets that show consistent regulation in normal MoDCs (> 2-fold; P < .01) but significantly different regulation in EDI MoDCs (ANOVA at P < .01). (B) Addition of IFN-γ restores a portion of the CD40L expression signature in EDI MoDCs. A hierarchic cluster map of the expression patterns for the 400 probe sets from panel A in response to CD40L plus IFN-γ. Note that many of the probe sets show similar expression in EDI and normal samples, illustrating the effects of IFN-γ on complementing the CD40L expression signature in EDI MoDCs. (C) EDI MoDCs fail to up-regulate a subset of CD40L-induced genes even in the presence of IFN-γ. A number of probe sets (53) are shown that passed the following filter criteria: significantly regulated in all 3 normal CD40L plus IFN-γ cultures (P < .01); and no significant regulation in any of the 3 EDI CD40L plus IFN-γ cultures. Note that for this analysis any ratio values with P > .01 were set to log (ratio) = 0.
Putative c-Rel transcriptional targets defined by microarray analysis
The specific defect in c-Rel activation in EDI MoDCs offers a unique opportunity to define putative downstream c-Rel target genes. For this purpose, we used gene expression arrays. Initially, we profiled the CD40L-induced expression signature in MoDCs derived from 3 independent healthy donors. A CD40L-induced expression signature was generated by filtering for genes showing a 2-fold or greater change in expression (P < .01) upon stimulation in all 3 normal MoDC cultures. This analysis was designed to stringently select for genes that show consistent and significant expression changes and resulted in a signature of 764 probe sets. A 1-way analysis of variance (ANOVA) was then used to select for genes from this signature that showed significant differences between the healthy and EDI groups. This yielded 400 probe sets at P < .01 (Figure 6A). Next, we assessed the effect of adding IFN-γ to CD40L on the expression of these 400 probe sets. As can be seen in Figure 6B, IFN-γ is able to enhance the regulation induced by CD40L for a large number of genes, consistent with our observations of the effects of IFN-γ on CD40L-induced NF-κB binding in EDI MoDCs.
Because EDI MoDCs fail to induce c-Rel DNA binding in response to CD40L plus IFN-γ (Figure 1A), we reasoned that we should be able to identify c-Rel genomic targets by looking for genes that show consistent up-regulation in normal MoDCs but not in EDI MoCs when stimulated with these mediators. Analysis of the 400 probe sets that we defined as demonstrating significant differences in regulation between the normal and EDI MoDC cultures enabled us to identify 53 probe sets (corresponding to 50 different genes) that show significant and consistent up-regulation in normal MoDCs in response to CD40L plus IFN-γ, but no significant regulation in any of the 3 EDI MoDC cultures. A hierarchic cluster of these 53 probe sets is shown in Figure 6C, and a summary table of the corresponding 50 genes is listed in Table 1. The validity of this gene set as c-Rel transcriptional targets is supported by the appearance of IL12A in the gene list, a locus previously shown to be regulated by c-Rel in DCs.22 Strikingly, the putative function of many of these genes correlates well with the known defects we have demonstrated in EDI MoDCs. These include genes involved in adhesion and the extracellular matrix as well as cytoskeletal components that may be responsible for the morphologic and clustering defects we have described.
Putative c-Rel targets derived from expression profiling of EDI MoDCs
Sequence name . | Sequence code . | Accession no. . | Sequence description . | Function . |
|---|---|---|---|---|
| DKFZP586D0919 | 39986_at | AL050100 | Hepatocellularcarcinoma-associated antigen HCA557a | Unknown |
| GRAMD1B | 41559_at | AA434319 | GRAM domain containing 1B | Unknown |
| HYAL3 | 35540_at | AF040710 | Hyaluronidase (LUCA-3) mRNA | Unknown |
| JRK | 37873_g_at | AF072468 | Jerky homolog (mouse) | Unknown |
| LOC221362 | 37538_at | AL049354 | Hypothetical protein LOC221362 | Unknown |
| MAC30 | 32791_at | L19183 | Hypothetical protein MAC30 | Unknown |
| NBEAL2 | 35763_at | AB011112 | Neurobeachin-like 2 | Unknown |
| PHLPPL | 32735_at | AB023148 | PH domain and leucine rich repeat protein phosphatase-like | Unknown |
| LAD1 | 33282_at | U42408 | Ladinin 1 | Adhesion and extracellular matrix |
| LAMB3 | 36929_at | U17760 | Laminin, beta 3 | Adhesion and extracellular matrix |
| PLAT | 33452_at | M15518 | Plasminogen activator, tissue | Adhesion and extracellular matrix |
| SERPINE1 | 38125_at | M14083 | Serine proteinase inhibitor, clade E (plasminogen activator inhibitor type 1), member 1 | Adhesion and extracellular matrix |
| ST8SIA1 | 40678_at | X77922 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | Adhesion and extracellular matrix |
| TPST1 | 32718_at | AF038009 | Tyrosylprotein sulfotransferase 1 | Adhesion and extracellular matrix |
| BCL2L1 | 34742_at | Z23115 | BCL2-like 1 | Apoptosis |
| HTATIP | 465_at | U74667 | HIV-1 Tat interactive protein, 60 kDa | Apoptosis |
| AVIL | 41582_at | AF041449 | Advillin | Cytoskeleton |
| CAPG | 38391_at | M94345 | Capping protein (actin filament), gelsolin-like | Cytoskeleton |
| DLGAP1 | 40388_at | AB000277 | Discs, large (Drosophila) homolog-associated protein 1 | Cytoskeleton |
| SEPT11 | 33172_at | T75292 | Septin 11 | Cytoskeleton |
| TIAM1 | 37460_at | U16296 | T-cell lymphoma invasion and metastasis 1 | Cytoskeleton |
| ACVR2A | 35162_s_at | D31770 | Activin A receptor, type II | Growth and differentiation |
| ACVRL1 | 32714_s_at | L17075 | Activin A receptor type II-like 1 | Growth and differentiation |
| CDK7 | 1969_s_at | X77743 | Cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | Growth and differentiation |
| IL12A | 33955_at | M65291 | Interleukin 12A (cytotoxic lymphocyte maturation factor 1, p35) | Growth and differentiation |
| MELK | 38847_at | D79997 | Maternal embryonic leucine zipper kinase | Growth and differentiation |
| VEGF | 36101_s_at | M63978 | Vascular endothelial growth factor | Growth and differentiation |
| ACOX1 | 40460_s_at | X71440 | Acyl-Coenzyme A oxidase 1, palmitoyl | Metabolism |
| B4GALT4 | 39432_at | AF038662 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | Metabolism |
| GALT | 36664_at | M60091 | Galactose-1-phosphate uridylyltransferase | Metabolism |
| MTHFS | 39064_at | L38928 | 5,10-methenyltetrahydrofolate synthetase | Metabolism |
| PRKAB1 | 35348_at | AF022116 | Protein kinase, AMP-activated, beta 1 noncatalytic subunit | Metabolism |
| SLC25A1 | 38998_g_at | X96924 | Solute carrier family 25 (citrate transporter), member 1 | Metabolism |
| SLC5A6 | 35256_at | AL096737 | Solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | Metabolism |
| SMS | 38792_at | AD001528 | Spermine synthase | Metabolism |
| DUSP2 | 1292_at | L11329 | Dual specificity phosphatase 2 | Signaling |
| GRIA3 | 35050_at | X82068 | Glutamate receptor, ionotrophic, AMPA 3 | Signaling |
| ITPR3 | 37343_at | U01062 | Inositol 1,4,5-triphosphate receptor, type 3 | Signaling |
| PCTK1 | 1224_at | X66363 | PCTAIRE protein kinase 1 | Signaling |
| PVR | 32698_at | X64116 | PVR gene for poliovirus receptor | Signaling |
| RAB30 | 807_at | U57092 | RAB30, member RAS oncogene family | Signaling |
| TNFRSF14 | 39424_at | U70321 | TNF receptor superfamily, member 14 | Signaling |
| TRG@ | 41468_at | M30894 | T-cell receptor gamma locus | Signaling |
| ELL2 | 40606_at | U88629 | RNA polymerase II elongation factor | Transcription |
| KLF7 | 34217_at | AB015132 | Kruppel-like factor 7 (ubiquitous) | Transcription |
| SMAD6 | 1955_s_at | AF035528 | SMAD, mothers against DPP homolog 6 (Drosophila) | Transcription |
| TSC22D3 | 36629_at | AI635895 | TSC22 domain family, member 3 | Transcription |
| TTF2 | 37870_at | AF073771 | Transcription termination factor, RNA polymerase II | Transcription |
| EEF1A1 | 1288_s_at | J04617 | Eurkaryotic translation elongation factor 1 alpha 1 | Translation |
| TSR1 | 40982_at | AA926957 | TSR1, 20S rRNA accumualtion, homolog (yeast) | Translation |
Sequence name . | Sequence code . | Accession no. . | Sequence description . | Function . |
|---|---|---|---|---|
| DKFZP586D0919 | 39986_at | AL050100 | Hepatocellularcarcinoma-associated antigen HCA557a | Unknown |
| GRAMD1B | 41559_at | AA434319 | GRAM domain containing 1B | Unknown |
| HYAL3 | 35540_at | AF040710 | Hyaluronidase (LUCA-3) mRNA | Unknown |
| JRK | 37873_g_at | AF072468 | Jerky homolog (mouse) | Unknown |
| LOC221362 | 37538_at | AL049354 | Hypothetical protein LOC221362 | Unknown |
| MAC30 | 32791_at | L19183 | Hypothetical protein MAC30 | Unknown |
| NBEAL2 | 35763_at | AB011112 | Neurobeachin-like 2 | Unknown |
| PHLPPL | 32735_at | AB023148 | PH domain and leucine rich repeat protein phosphatase-like | Unknown |
| LAD1 | 33282_at | U42408 | Ladinin 1 | Adhesion and extracellular matrix |
| LAMB3 | 36929_at | U17760 | Laminin, beta 3 | Adhesion and extracellular matrix |
| PLAT | 33452_at | M15518 | Plasminogen activator, tissue | Adhesion and extracellular matrix |
| SERPINE1 | 38125_at | M14083 | Serine proteinase inhibitor, clade E (plasminogen activator inhibitor type 1), member 1 | Adhesion and extracellular matrix |
| ST8SIA1 | 40678_at | X77922 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | Adhesion and extracellular matrix |
| TPST1 | 32718_at | AF038009 | Tyrosylprotein sulfotransferase 1 | Adhesion and extracellular matrix |
| BCL2L1 | 34742_at | Z23115 | BCL2-like 1 | Apoptosis |
| HTATIP | 465_at | U74667 | HIV-1 Tat interactive protein, 60 kDa | Apoptosis |
| AVIL | 41582_at | AF041449 | Advillin | Cytoskeleton |
| CAPG | 38391_at | M94345 | Capping protein (actin filament), gelsolin-like | Cytoskeleton |
| DLGAP1 | 40388_at | AB000277 | Discs, large (Drosophila) homolog-associated protein 1 | Cytoskeleton |
| SEPT11 | 33172_at | T75292 | Septin 11 | Cytoskeleton |
| TIAM1 | 37460_at | U16296 | T-cell lymphoma invasion and metastasis 1 | Cytoskeleton |
| ACVR2A | 35162_s_at | D31770 | Activin A receptor, type II | Growth and differentiation |
| ACVRL1 | 32714_s_at | L17075 | Activin A receptor type II-like 1 | Growth and differentiation |
| CDK7 | 1969_s_at | X77743 | Cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | Growth and differentiation |
| IL12A | 33955_at | M65291 | Interleukin 12A (cytotoxic lymphocyte maturation factor 1, p35) | Growth and differentiation |
| MELK | 38847_at | D79997 | Maternal embryonic leucine zipper kinase | Growth and differentiation |
| VEGF | 36101_s_at | M63978 | Vascular endothelial growth factor | Growth and differentiation |
| ACOX1 | 40460_s_at | X71440 | Acyl-Coenzyme A oxidase 1, palmitoyl | Metabolism |
| B4GALT4 | 39432_at | AF038662 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | Metabolism |
| GALT | 36664_at | M60091 | Galactose-1-phosphate uridylyltransferase | Metabolism |
| MTHFS | 39064_at | L38928 | 5,10-methenyltetrahydrofolate synthetase | Metabolism |
| PRKAB1 | 35348_at | AF022116 | Protein kinase, AMP-activated, beta 1 noncatalytic subunit | Metabolism |
| SLC25A1 | 38998_g_at | X96924 | Solute carrier family 25 (citrate transporter), member 1 | Metabolism |
| SLC5A6 | 35256_at | AL096737 | Solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | Metabolism |
| SMS | 38792_at | AD001528 | Spermine synthase | Metabolism |
| DUSP2 | 1292_at | L11329 | Dual specificity phosphatase 2 | Signaling |
| GRIA3 | 35050_at | X82068 | Glutamate receptor, ionotrophic, AMPA 3 | Signaling |
| ITPR3 | 37343_at | U01062 | Inositol 1,4,5-triphosphate receptor, type 3 | Signaling |
| PCTK1 | 1224_at | X66363 | PCTAIRE protein kinase 1 | Signaling |
| PVR | 32698_at | X64116 | PVR gene for poliovirus receptor | Signaling |
| RAB30 | 807_at | U57092 | RAB30, member RAS oncogene family | Signaling |
| TNFRSF14 | 39424_at | U70321 | TNF receptor superfamily, member 14 | Signaling |
| TRG@ | 41468_at | M30894 | T-cell receptor gamma locus | Signaling |
| ELL2 | 40606_at | U88629 | RNA polymerase II elongation factor | Transcription |
| KLF7 | 34217_at | AB015132 | Kruppel-like factor 7 (ubiquitous) | Transcription |
| SMAD6 | 1955_s_at | AF035528 | SMAD, mothers against DPP homolog 6 (Drosophila) | Transcription |
| TSC22D3 | 36629_at | AI635895 | TSC22 domain family, member 3 | Transcription |
| TTF2 | 37870_at | AF073771 | Transcription termination factor, RNA polymerase II | Transcription |
| EEF1A1 | 1288_s_at | J04617 | Eurkaryotic translation elongation factor 1 alpha 1 | Translation |
| TSR1 | 40982_at | AA926957 | TSR1, 20S rRNA accumualtion, homolog (yeast) | Translation |
Sequence names correspond to Human Genome Organization (HUGO) symbols retrieved from the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/database/). Sequence codes refer to Affymetrix U95A probe set identifiers. Functions were assigned through reference to Entrez annotations (NCBI) and PubMed literature searches.
Discussion
While the role of the IKK signalosome for NF-κB activation is well documented,23 the precise mechanism by which the complex integrates signals delivered by numerous upstream stimuli to differentially activate members of the NF-κB family is not fully understood. Insight into this mechanism can be obtained from the study of individuals with specific NEMO mutations who manifest defined immunologic defects. In the present study, we report the consequence of a mutation targeting a highly conserved cysteine residue at position 417 of NEMO that occurs in patients with EDI. We show that antigen-presenting cells in such patients exhibit defective NEMO ubiquitination upon CD40L but not LPS stimulation, and this leads to a specific impairment in CD40L-mediated c-Rel activation. We therefore establish how a particular defect in NEMO ubiquitination leads to immunologic deficiency.
Several recent studies have shown that NF-κB activation requires the posttranslational modification of NEMO upon cellular stimulation. Addition of K63-linked polyubiquitin chains, thought to strengthen protein-protein interactions, appears to be a key modification in intracellular signaling.9,10,24 Interestingly, different regulators, including Bcl10 and TRAF6, have been shown to mediate K63 ubiquitination of NEMO via the putative zinc finger motif where the point mutation in patients with EDI lies.19,25 Thus far, CYLD and A20 have been shown to turn off NF-κB–mediated transcriptional activity by targeting ubiquitinated NEMO.26 Our results indicate that CD40L fails to support the ubiquitination of mutated NEMO. In light of the EMSA findings, NEMO ubiquitination appears to be dispensable for RelA activation, whereas c-Rel induction is apparently dependent on an intact C-terminal region and, therefore, on a K63-ubiquinated NEMO. Taken together, our findings imply that a subtle variation, NEMO ubiquitination, in the shared NF-κB signaling cascade can dramatically influence the outcome of the cellular stimulation by mobilizing different NF-κB subunits.
In contrast to CD40L, LPS still induces RelA and c-Rel activation, as well as NEMO ubiquitination in EDI DCs. The LPS-specific surface receptor TLR4 recruits multiple downstream adaptor molecules in order to activate the NF-κB pathway.27 Among the cofactors shared with the CD40 pathway is TNF receptor-associated factor 6 (TRAF6).28 Interestingly, the DCs from TRAF6–/– knockout mice display a profound defect in the activation of NF-κB and in the up-regulation of CD86 in response to LPS stimulation, while such defects are less prominent in response to CD40 stimulation.29,30 Moreover, it has been shown that the TRAF2 binding site on CD40, but not the TRAF6 binding site, is required for Ig class-switching.31 This observation is consistent with the clinical features of patients with EDI who lack sera IgG and IgE. These results suggest that TRAF2 rather than TRAF6 is used by CD40 to ubiquitinate NEMO, and that the NEMO C417R mutation is permissive for LPS signaling because NEMO ubiquitination in response to TLR4 stimulation is dependent on TRAF6 and not TRAF2.
Both patients with EDI have a history of atypical mycobacterial infections. EDI-1 developed Mycobacterium avium complex bursitis and EDI-2 developed Mycobacterium abscessus infection of vertebrae. Functional studies have shown that cellular stimulation through CD40 is necessary for defense against atypical mycobacterial infection and for the efficient induction of cellular memory by bacterial epitopes with reduced immunogenicity.32-34 Among CD40-expressing cells, DCs compose a bridge between innate and adaptive immunity, initiating cellular immune responses upon their differentiation into mature cells.1 After stimulation with CD40L, the phenotype of EDI DCs remained largely immature. Specifically, they were impaired in their maturation-associated morphologic features, expression of costimulatory markers, production of proinflammatory cytokines including IL-12, and their capacity to support the allogeneic lymphoproliferation. We have linked these functional deficiencies of EDI DCs to a failure in NEMO ubiquitination after CD40L stimulation. Flow cytometry and geneexpression profiling analysis of EDI DCs indicates that IFN-γ costimulation can restore the expression of some genes normally regulated by CD40L signaling. However, c-Rel activity and the capacity to support allogeneic T-cell proliferation remain deficient. We reason that the failure of CD40-stimulated EDI DCs to activate T cells is largely a consequence of their inability to synthesize the c-Rel–regulated cytokine IL-12, and offer a compelling explanation for the high susceptibility of patients with EDI to recurrent bacterial infections.
Having established EDI MoDCs as a unique resource to identify genomic targets whose expression is absolutely dependent on the presence of activated c-Rel, we used expression profiling to compare the genes induced by CD40L in normal and EDI MoDCs. We identified 50 genes that are normally highly up-regulated by CD40L and IFN-γ, but their regulation is blocked in EDI MoDCs. We identified the known c-Rel target IL-12p35 (IL12A) in this gene set as well as a number of genes that are likely to be critical to the maturation and function of DCs. Among these, LAD1, LAMB3, and SERPINE1 have functions in adhesion or in the extracellular matrix and likely influence the clumping behavior of DCs; cytoskeletal components such as AVIL, CAPG, and SEPT11 may regulate dendrite formation. Further studies will be needed to associate the expression of other members of this gene set with specific DC functions.
In summary, our study indicates that the NEMO C417R mutations cause immune deficiency by affecting NEMO ubiquitination in response to CD40 signaling. It is noted that other missense mutations in exon 10 of NEMO have been associated with different phenotypes. Some of these patients have osteopetrosis and lymphedema, while others have a marked predisposition infection with Streptococcus pneumoniae or cytomegalovirus. The patients described here with the NEMO C417R mutation have normal bone density and do not have a medical history of limb swelling or severe viral or pyogenic infections. We anticipate that some of these other exon 10 mutations in NEMO affect its ubiquitination in response to stimulation by receptor-activator of NF-κB (RANK), vascular endothelial growth factor (VEGF), or other TLRs. Thus, these clinical differences, in light of accompanying differences in NEMO ubiquitination, suggest that the various cellular receptors that activate NF-κB use different cofactors to ubiquitinate NEMO. Further characterization of these stimuli-restricted proteins could allow for the development of rationally designed drugs that target different aspects of immunity.
Prepublished online as Blood First Edition Paper, June 22, 2006; DOI 10.1182/blood-2006-04-017210.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank R. Hornung for cytokine analysis; C. Russell, D. Prosser, K. Kerkof, M. Timour, and S. Dorje for DNA microarray support; O. M. Schwartz and J. Kabat for microscopy assistance; Warren Strober and Uli Siebenlist for helpful discussions; and Phil Murphy for careful review of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal