Abstract
The capacity of clopidogrel to inhibit ADP-induced platelet aggregation shows wide intersubject variability. To determine whether frequent functional variants of genes coding for candidate cytochrome P450 (CYP) isoenzymes involved in clopidogrel metabolic activation (CYP2C19*2, CYP2B6*5, CYP1A2*1F, and CYP3A5*3 variants) influence the platelet responsiveness to clopidogrel, we conducted a prospective pharmacogenetic study in 28 healthy white male volunteers treated for 7 days with clopidogrel 75 mg/d. We observed that pharmacodynamic response to clopidogrel was significantly associated with the CYP2C19 genotype. Twenty of the subjects were wild-type CYP2C19 (*1/*1) homozygotes, while the other 8 subjects were heterozygous for the loss-of-function polymorphism CYP2C19*2 (*1/*2). Baseline platelet activity was not influenced by the CYP2C19 genotype. In contrast, platelet aggregation in the presence of 10 μM ADP decreased gradually during treatment with clopidogrel 75 mg once daily in *1/*1 subjects, reaching 48.9% ± 14.9% on day 7 (P < .001 vs baseline), whereas it did not change in *1/*2 subjects (71.8% ± 14.6% on day 7, P = .22 vs baseline, and P < .003 vs *1/*1 subjects). Similar results were found with VASP phosphorylation. The CYP2C19*2 loss-of-function allele is associated with a marked decrease in platelet responsiveness to clopidogrel in young healthy male volunteers and may therefore be an important genetic contributor to clopidogrel resistance in the clinical setting.
Introduction
Clopidogrel, a thienopyridine derivative, inhibits platelet aggregation induced by adenosine diphosphate (ADP). Clopidogrel is a prodrug requiring several biotransformation steps, mediated mainly by cytochrome P-450 isoenzymes (CYP), in order to generate an active metabolite that binds irreversibly to the platelet ADP receptor P2Y12.1-3 Several large clinical trials4-6 have shown that clopidogrel effectively prevents thrombotic events in patients with atherosclerotic vascular disease. Clopidogrel, alone or combined with aspirin, is routinely used to treat patients with a variety of vascular disorders.7
The pharmacodynamic response to clopidogrel varies widely from subject to subject, and about 25% of patients treated with standard clopidogrel doses display low ex vivo inhibition of ADP-induced platelet aggregation.8,9 This poor response to clopidogrel is associated with an increased risk of recurrent ischemic events.8 The mechanisms underlying clopidogrel resistance are unclear.10,11
In addition to nonadherence to treatment, certain genetic factors may be involved in this phenomenon. A gain-of-function haplotype (H2) of the gene encoding the P2Y12 receptor has been described,12 but P2Y12 genetic variants are unlikely to have a marked effect on the response to clopidogrel.13,14 On the other hand, several functional polymorphisms have been found in genes encoding cytochrome P-450 isoforms involved in clopidogrel metabolic activation upstream of P2Y12 (eg, CYP 3A4/5, 2C19, 2B6, and 1A2),15,16 but their influence on the pharmacodynamic response to clopidogrel has not been systematically investigated.
The aim of this pharmacogenetic study was to determine whether the antiplatelet effect of standard-dose clopidogrel in healthy white subjects is influenced by functional variants in genes encoding CYP isoenzymes involved in clopidogrel metabolic activation. We show that the CYP2C19 genotype is a major determinant of the pharmacodynamic response to clopidogrel in healthy volunteers.
Patients, materials, and methods
Subjects and study protocol
This study was primarily designed to investigate the pharmacogenetics of clopidogrel in healthy young white men aged 18 to 35 years. Briefly, 29 such subjects were enrolled, based on the following criteria: nonsmokers, normal physical and laboratory results (including white and red blood cell count, liver enzymes, blood glucose, serum creatinin, prothrombin time, activated partial thromboplastin time, and plasma fibrinogen). The following inclusion criteria were also applied: normal (150-400 × 109/L) platelet count; normal (> 50%) maximal platelet aggregation in response to 5 and 10 μM ADP (Sigma Aldrich, St Louis, MO), 1 mM arachidonic acid (Helena Biosciences, Sunderland, United Kingdom), 1 μg/mL Horm collagen (Nycomed, Oslo, Norway), and 1 μM U46619 (Calbiochem, San Diego, CA); and a normal (80-160 seconds) PFA-100 assay (collagen-EPI cartridge; Dade Behring, Deerfield, IL).
The subjects were asked to come to the Clinical Investigations Center every day at 9 am, where they were given a standard dose of clopidogrel 75 mg once daily for 7 consecutive days in the presence of medical staff. It was checked that the tablets had been swallowed. Blood samples for pharmacodynamic studies were drawn just before the clopidogrel intakes on days 1, 2, 3, 4, 5, and 7. Blood samples were also drawn on days 9, 10, 11, and 14.
The study was approved by the Committee for the Protection of Human Subjects in Biomedical Research, which functions for the Cochin University Hospital (Paris, France), and all the subjects gave their written informed consent to participate, in accordance with the Declaration of Helsinki.
Laboratory methods
Genotyping. Genotyping failed in 1 of the 29 subjects, because of DNA degradation. We studied single nucleotide polymorphisms (SNPs) in genes encoding CYP2C19, 2B6, 1A2, and 3A4/5, based on the following criteria: (1) known functional consequences on enzyme activity and drug metabolism; and (2) a minor allele frequency higher than 5% in whites. The selection was based on publicly available databases (dbSNP, PharmGKB) and on Medline publications for selected SNPs. The primers and methods used for genotyping are reported in Table 1.
Genotyping methods
SNP (allele); dbSNP accession no. . | Primers used for PCR . | Genotyping method . |
|---|---|---|
| CYP2C19 | ||
| 681 G>A (*2); rs4244285 | Sense: 5′-CAGAGCTTGGCATATTGTATC-3′ | Restriction fragment length polymorphism (RFLP) with SmaI |
| Anti-sense: 5′-GTAAACACACAAAACTAGTCAATG-3′ | ||
| 636 G>A (*3); rs4986893 | Sense: 5′-AACATCAGGATTGTAAGCAC-3′ | RFLP with BamHI |
| Anti-sense: 5′-TCAGGGCTTGGTCAATATAG-3′ | ||
| 1 A>G(*4); rs28399504 | Sense: 5′-GCACACACACTTAATTAGCATGG-3′ | Direct sequencing |
| Anti-sense: 5′-TACCTTTTGCAAGCCACTGA-3′ | ||
| 1297 C>T (*5); — | Sense: 5′-TGCATATTCTGTCTGTGCCAGT-3′ | Direct sequencing |
| Anti-sense: 5′-AGCAGCCAGACCATCTGT-3′ | ||
| 395 G>A (*6); — | Sense: 5′-CCTGGGATCTCCCTCCTAGT-3′ | Direct sequencing |
| Anti-sense: 5′-AGGAGAGCAGTCCAGAAAGG-3′ | ||
| CYP3A5 | ||
| 6986 G>A (*3); rs776746 | Sense: 5′-GAGTGGCATAGGAGATACCCAC-3′ | Direct sequencing |
| Anti-sense: 5′-TCTAGTTCATTAGGGTGTGACACACA-3′ | ||
| CYP2B6 | ||
| 1459 C>T (*5); rs3211371 | Sense: 5′-TGAGAATCAGTGGAAGCCATAGA-3′ | RFLP with BglII |
| Anti-sense: 5′-TAATTTTCGATAATCTCACTCCTGC-3′ | ||
| CYP1A2 | ||
| -164 A>C (*1F); rs762551 | Sense: 5′-CCCAGAAGTGGAAACTGAGA-3′ | RFLP with ApaI |
| Anti-sense: 5′-GGGTTGAGATGGAGACATTC-3′ |
SNP (allele); dbSNP accession no. . | Primers used for PCR . | Genotyping method . |
|---|---|---|
| CYP2C19 | ||
| 681 G>A (*2); rs4244285 | Sense: 5′-CAGAGCTTGGCATATTGTATC-3′ | Restriction fragment length polymorphism (RFLP) with SmaI |
| Anti-sense: 5′-GTAAACACACAAAACTAGTCAATG-3′ | ||
| 636 G>A (*3); rs4986893 | Sense: 5′-AACATCAGGATTGTAAGCAC-3′ | RFLP with BamHI |
| Anti-sense: 5′-TCAGGGCTTGGTCAATATAG-3′ | ||
| 1 A>G(*4); rs28399504 | Sense: 5′-GCACACACACTTAATTAGCATGG-3′ | Direct sequencing |
| Anti-sense: 5′-TACCTTTTGCAAGCCACTGA-3′ | ||
| 1297 C>T (*5); — | Sense: 5′-TGCATATTCTGTCTGTGCCAGT-3′ | Direct sequencing |
| Anti-sense: 5′-AGCAGCCAGACCATCTGT-3′ | ||
| 395 G>A (*6); — | Sense: 5′-CCTGGGATCTCCCTCCTAGT-3′ | Direct sequencing |
| Anti-sense: 5′-AGGAGAGCAGTCCAGAAAGG-3′ | ||
| CYP3A5 | ||
| 6986 G>A (*3); rs776746 | Sense: 5′-GAGTGGCATAGGAGATACCCAC-3′ | Direct sequencing |
| Anti-sense: 5′-TCTAGTTCATTAGGGTGTGACACACA-3′ | ||
| CYP2B6 | ||
| 1459 C>T (*5); rs3211371 | Sense: 5′-TGAGAATCAGTGGAAGCCATAGA-3′ | RFLP with BglII |
| Anti-sense: 5′-TAATTTTCGATAATCTCACTCCTGC-3′ | ||
| CYP1A2 | ||
| -164 A>C (*1F); rs762551 | Sense: 5′-CCCAGAAGTGGAAACTGAGA-3′ | RFLP with ApaI |
| Anti-sense: 5′-GGGTTGAGATGGAGACATTC-3′ |
Direct sequencing was performed using the Big Dye terminator cycle sequencing kit and an automated DNA sequencer (ABI 3100, Applied Biosystems).
Pharmacodynamic evaluation. Aggregation studies were performed within 2 hours after blood collection, at 37°C, by using a photometric method on a 4-channel aggregometer (Regulest, Amneville, France). A 280-μL aliquot of platelet-rich plasma was incubated for 3 minutes at 37°C and was then stirred at 1100 rpm for 2 minutes before adding 20 μL saline or ADP (Sigma Aldrich). A final concentration of 10 μM ADP was chosen because it induces complete platelet aggregation in a manner independent of thromboxane A2 production.17 Platelet aggregation to 5 μM ADP was also measured. Platelets aggregate when the agonist is added, thereby leading to an increase in light transmission, which is recorded for 5 minutes. Aggregation was expressed as the maximal percent change in light transmission from baseline, using platelet-poor plasma as reference (arbitrarily 100%).
We also assessed the level of phosphorylated VASP (vasodilator-stimulated phosphoprotein), a good index of P2Y12 activity.18,19 P2Y12 is a Gi-coupled receptor whose activation reduces the platelet cyclic adenosine monophosphate (cAMP) level by inhibiting adenyl cyclase. The decrease in cAMP production leads to a reduction in the activation of specific protein kinases, which can no longer phosphorylate VASP. The level of phosphorylated VASP, which can be measured by flow cytometry, is thus a good index of inhibition of the ADP-P2Y12 interaction by clopidogrel. VASP was measured in whole blood on days 1 (baseline) and 7 of clopidogrel intake, using a flow cytometric assay (Platelet VASP; Diagnostica Stago, Biocytex, Asnières, France) adapted to a Cy-Flow apparatus (Partec, Münster, Germany). Results were expressed as a platelet reactivity index (PRI, %) calculated from the mean fluorescence intensity (MFI) of samples incubated with PGE1 alone or with both PGE1 and ADP simultaneously, using the following formula: (MFI PGE1 – MFI PGE1+ADP/MFI PGE1) × 100.
Platelet function was measured blindly to genotyping data.
Statistical analysis
The effect of the studied genotypes on clopidogrel response (assessed by 10 μM ADP–induced platelet aggregation) across time was first evaluated by using Friedman test. If the global test was significant, the effect of the identified genotypes on pharmacodynamic parameters (ADP-induced platelet aggregation and the VASP platelet reactivity index) was assessed only at baseline and on day 7 by using the Wilcoxon test to limit the number of statistical tests. Frequencies of categorical variables were compared among groups with Fisher exact test.
All analyses were done with SAS statistical software version 9.1 (SAS, Cary, NC). A P value less than .05 was considered significant.
Results
Genotyping results
Table 2 summarizes the characteristics of the 4 selected functional SNPs and the prevalence of each genotype in the 28 subjects. All allelic frequencies were in keeping with those previously reported in white populations (dbSNP). The distribution of the CYP genetic variants did not deviate from Hardy-Weinberg equilibrium.
Position, effects, and frequencies of screened SNPs in the 28 subjects
Allele . | Nucleotide change* . | Effect of the mutant allele . | Enzyme activity in vivo . | Genotype frequency† . | Allele frequency‡ . |
|---|---|---|---|---|---|
| CYP2C19*2 | 681G>A (rs4244285) | 681A: cryptic splicing site | None | GG (20); GA (8); AA (0) | G (85.7); A (14.3) |
| CYP3A5*3 | 6986G>A (rs776746) | 6986G: alternative splicing | None | GG (27); GA (1); AA (0) | G (98.2); A (1.8) |
| CYP2B6*5 | 1459C>T (rs3211371) | Amino acid substitution; Arg487Cys | Decreased | CC (23); CT (5); TT (0) | C (91.1); T (8.9) |
| CYP1A2*1F | -164A>C (rs762551) | Promoter regulation? | Decreased | AA (13); AC (13); CC (2) | A (69.7); C (30.3) |
Allele . | Nucleotide change* . | Effect of the mutant allele . | Enzyme activity in vivo . | Genotype frequency† . | Allele frequency‡ . |
|---|---|---|---|---|---|
| CYP2C19*2 | 681G>A (rs4244285) | 681A: cryptic splicing site | None | GG (20); GA (8); AA (0) | G (85.7); A (14.3) |
| CYP3A5*3 | 6986G>A (rs776746) | 6986G: alternative splicing | None | GG (27); GA (1); AA (0) | G (98.2); A (1.8) |
| CYP2B6*5 | 1459C>T (rs3211371) | Amino acid substitution; Arg487Cys | Decreased | CC (23); CT (5); TT (0) | C (91.1); T (8.9) |
| CYP1A2*1F | -164A>C (rs762551) | Promoter regulation? | Decreased | AA (13); AC (13); CC (2) | A (69.7); C (30.3) |
Numbers in parentheses are dbSNP accession nos.
Number of patients is given parenthetically.
Numbers in parentheses are percentages.
Clopidogrel responsiveness according to CYP polymorphisms
None of the selected SNPs in CYP2B6, CYP3A5, and CYP1A2 influenced the clopidogrel response (data not shown). In contrast, the response to clopidogrel was strongly influenced by the CYP2C19 genotypic status (P < .003 by Friedman test). Twenty of the 28 subjects were CYP2C19 wild-type homozygotes (*1/*1), and the other 8 subjects were CYP2C19 (*1/*2) heterozygotes. At baseline, platelet aggregation in the presence of 10 μM ADP did not differ significantly according to the CYP2C19 genotype (76.2% ± 6.8% in *1/*1 subjects and 77.2% ± 4.3% in *1/*2 subjects; P = .63; Figure 1A). During treatment with clopidogrel 75 mg once daily, platelet aggregation in response to 10 μM ADP gradually decreased in *1/*1 subjects, reaching 48.9% ± 14.9% on day 7 (P < .001 vs baseline), whereas it did not change in *1/*2 subjects (71.8% ± 14.6%, P = .22 vs baseline and P < .003 vs *1/*1 subjects; Figure 1B). Figure 2 shows the influence of the CYP2C19 genotype on changes in 10 μM ADP–induced platelet aggregation at the different time points. Similar results were obtained when platelet aggregation was induced by 5 μM ADP (data not shown).
The association between the allelic variant CYP2C19*2 and lower pharmacodynamic response to clopidogrel was confirmed by VASP phosphorylation studies. At baseline, the platelet reactivity index (PRI) did not differ significantly according to the CYP2C19 genotype (72.6% ± 7.1% in *1/*1 subjects and 78.7% ± 6.9% in *1/*2 subjects). On day 7, the PRI had fallen significantly in both groups, but to a lesser extent in *1/*2 subjects (42.9% ± 16.6% in *1/*1 subjects and 58.2% ± 12.6% in *1/*2 subjects; intergroup comparison: P < .02).
Stratification of the subjects into quartiles of 10 μM ADP–induced platelet aggregation on day 7 relative to baseline was used to identify subjects as “poor responders” to clopidogrel, as follows (mean ± SD, [range]): first quartile: 100% ± 11% (87%-123%), second quartile: 81% ± 3% (76%-86%), third quartile: 63% ± 4% (57%-68%), fourth quartile: 45% ± 10% (24%-53%). Five of the 7 subjects belonging to the first quartile of poor responders carried the CYP2C19*2 allele. Two *1/*2 subjects belonged to the second quartile and 1 to the third quartile.
Finally, the subjects were further genotyped for other functional (*3, *4, *5, *6) CYP2C19 variants that were not initially studied, because of their low expected allelic frequencies (< 3%) in whites. Only the CYP2C19*4 allele was detected, in a CYP2C19 *1/*1 subject and in the heterozygous state. This subject belonged to the third quartile of clopidogrel responsiveness, and reclassification of this subject in the CYP2C19*2 group did not modify the previous results.
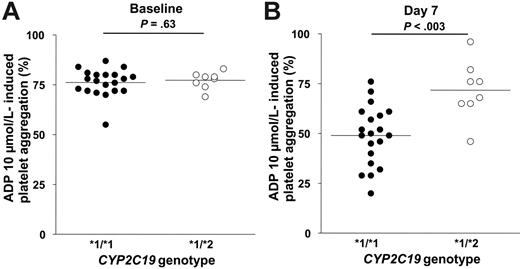
Platelet aggregation (absolute values) in response to 10 μM ADP according to the CYP2C19*2 genotype. Absolute values at baseline (A) and after 6 days (day 7) of clopidogrel 75 mg/d (B). The line indicates the mean value.
Discussion
We report evidence that the *2 allelic variant encoding a deficient drug-metabolizing enzyme CYP2C1920 is associated with impaired responsiveness to a 7-day oral course of 75 mg/d clopidogrel in young healthy white men, as assessed ex vivo in terms of platelet aggregation in the presence of ADP. This influence was confirmed by measuring VASP phosphorylation before and after clopidogrel administration: values fell less markedly in subjects carrying one *2 allele, reflecting weaker inhibition of P2Y12 ADP receptors.18,19,21 Of interest, our results are in line with preliminary data showing that the CYP2C19*2 polymorphism reduces ADP-induced platelet aggregation 4 and 24 hours after a single oral dose of 300 mg clopidogrel, this effect being due to a lower exposure to the active metabolite of clopidogrel.22 We have now shown that the difference in ADP-induced platelet aggregation between genotypes persists after 7-day administration of the standard marketed dose of 75 mg clopidogrel, indicating that the part of low pharmacodynamic response to clopidogrel due to the CYP2C19*2 polymorphism cannot be reversed by repeated dosing.
The active metabolite of clopidogrel, which irreversibly blocks platelet ADP P2Y12 receptors, arises from complex biochemical reactions23 involving several CYP isoforms.2,24,25 Variability in the catalytic activity of these isoforms may therefore affect the pharmacodynamic action of clopidogrel. Lau et al26 have reported that CYP3A4 metabolic activity is associated with between-subject variability in clopidogrel responsiveness. Both our results and those of Brandt et al22 now underline the role of CYP2C19 in the clopidogrel metabolic activation process and raise the possibility that this effect may also be shared by other thienopyridines, such as prasugrel.27 The relative contribution of these 2 CYP isoforms on clopidogrel response should deserve further investigations.
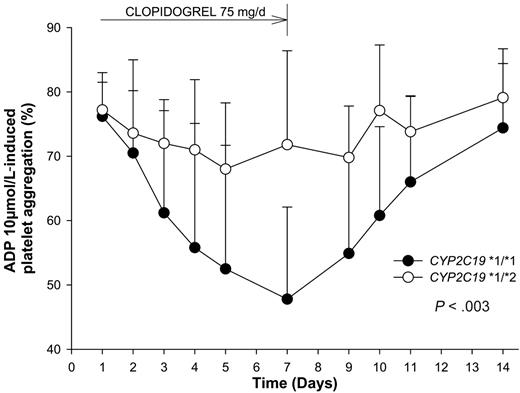
Time course of ex vivo platelet aggregation (mean ± SD) in response to 10 μM ADP, during and after the 7-day course of clopidogrel 75 mg/d, according to the CYP2C19*2 genotype (P < .003 by Friedman test).
We did not determine the CYP2C19 activity phenotype, as several studies have established that CYP2C19 genotyping identifies more than 90% of poor metabolizers, indicating good genotype-phenotype agreement.15,16 CYP2C19 catalytic efficiency is mainly influenced by the 681G>A polymorphism in exon 5 (CYP2C19*2, allelic frequency 15% in whites), which creates an aberrant splice site and results in a truncated and nonfunctional enzyme.20 Our finding that clopidogrel responsiveness is reduced in subjects carrying a single *2 allele is in keeping with previous reports of a “gene-dose effect” of the CYP2C19*2 allele on the pharmacokinetics/pharmacodynamics of proton pump inhibitors, which are also CYP2C19 substrates.15,28 Our results suggest that CYP2C19 *1/*2 subjects are not totally refractory to clopidogrel but that they respond less well than CYP2C19 *1/*1 subjects. Indeed, we found that VASP phosphorylation in response to clopidogrel was reduced but not abolished in CYP2C19 *1/*2 subjects, compared with CYP2C19 *1/*1 subjects. This suggests that clopidogrel is still partly converted into the active metabolite in *1/*2 subjects as suggested also by Brandt et al.22 It remains to be determined whether this is also the case in CYP2C19 *2/*2 homozygotes, or whether clopidogrel responsiveness is totally abolished in these subjects. Of interest, 2 of the 7 subjects who had the weakest responses to clopidogrel did not carry functional variants associated with CYP2C19 deficiency, implying that genetic or nongenetic factors other than loss-of-function CYP2C19 polymorphisms may also influence clopidogrel responsiveness. We did not observe any effect of other tested polymorphisms on ADP-induced platelet response to clopidogrel. However we observed low allelic frequencies for 2 of them, and our study was sufficiently powered to detect factors that only strongly affect clopidogrel response (as observed with CYP2C19*2 variant). Thus we cannot exclude a minor influence of these other polymorphisms on clopidogrel response.
This proof-of-concept pharmacogenetic study took place in strictly controlled conditions and involved only a small number of healthy volunteers, thereby creating optimal conditions to investigate the influence of genetic factors on clopidogrel responsiveness, notably because of normal baseline platelet function and a lack of potentially interfering treatments. Although the sample size of this study was small, it provided a 95% power to detect the observed difference of 22% in ADP-induced aggregation (absolute values) between genotype groups with an alpha risk of 5% and a standard deviation of 14.9%.
In conclusion, this study shows that the CYP2C19*2 mutant allele is a major genetic determinant of the pharmacodynamic response to standard-dose clopidogrel (75 mg/d) in healthy young white men. Further studies are required to investigate the influence of CYP2C19 functional polymorphisms on the response to clopidogrel in the clinical setting, particularly following a loading dose29 or with doses higher than 75 mg/d, and especially on the risk of recurrent thrombotic events during clopidogrel therapy.
Prepublished online as Blood First Edition Paper, June 13, 2006; DOI 10.1182/blood-2006-04-013052.
Supported by a grant from Programme Hospitalier de Recherche Clinique (Ministère chargé de la Santé, PHRC RBM 02-58, sponsor: INSERM).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank the nursing staff of the Clinical Investigation Center who participated in the protocol.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal