Abstract
During hematopoietic differentiation of human embryonic stem cells (hESCs), early hematopoietic progenitors arise along with endothelial cells within the CD34+ population. Although hESC-derived hematopoietic progenitors have been previously identified by functional assays, their phenotype has not been defined. Here, using hESC differentiation in coculture with OP9 stromal cells, we demonstrate that early progenitors committed to hematopoietic development could be identified by surface expression of leukosialin (CD43). CD43 was detected on all types of emerging clonogenic progenitors before expression of CD45, persisted on differentiating hematopoietic cells, and reliably separated the hematopoietic CD34+ population from CD34+CD43–CD31+KDR+ endothelial and CD34+CD43–CD31–KDR– mesenchymal cells. Furthermore, we demonstrated that the first-appearing CD34+CD43+CD235a+CD41a+/–CD45– cells represent precommitted erythro-megakaryocytic progenitors. Multipotent lymphohematopoietic progenitors were generated later as CD34+CD43+CD41a–CD235a–CD45– cells. These cells were negative for lineage-specific markers (Lin–), expressed KDR, VE-cadherin, and CD105 endothelial proteins, and expressed GATA-2, GATA-3, RUNX1, C-MYB transcription factors that typify initial stages of definitive hematopoiesis originating from endothelial-like precursors. Acquisition of CD45 expression by CD34+CD43+CD45–Lin– cells was associated with progressive myeloid commitment and a decrease of B-lymphoid potential. CD34+CD43+CD45+Lin– cells were largely devoid of VE-cadherin and KDR expression and had a distinct FLT3highGATA3lowRUNX1lowPU1highMPOhighIL7RAhigh gene expression profile.
Introduction
During human and mouse embryogenesis, primary hematopoietic cells are generated in the yolk sac and para-aortic splanchnopleura/aortagenital ridges-mesonephros (AGM) region; however, only cells generated in theAGM region are believed to contribute to hematopoietic stem cells.1 Both extraembryonic and intraembryonic hematopoietic cells develop in close association with endothelial cells from common hematoendothelial precursors, which were identified within early embryonic and embryonic stem cell (ESC)–derived cell populations expressing endothelial markers (VEGF-R2 [KDR], VE-cadherin, CD31, Tie2).2-8 These precursors are of particular interest for studies on the divergence of endothelial and hematopoietic cell lineages and establishment of hematopoietic stem cells; however, their identification requires a reliable separation of the earliest lineage-committed progeny, because hematopoietic and endothelial derivatives may share a common phenotype at early stages of development and still may not express typical lineage-specific markers. For example, in the mouse embryo, CD45, the most specific marker of hematopoietic lineage, is not expressed on the earliest hematopoietic progenitors arising in the yolk sac and AGM region; however, these progenitors can be identified by expression of CD41 molecule, which is specific for megakaryocytic lineage in adults.3,9-12
Hematopoietic differentiation of human ESCs (hESCs) reproduces many aspects of embryonic hematopoiesis and provides an in vitro model to elucidate mechanisms of early hematopoietic commitment,13,14 practically inaccessible in the human embryo.15 Several hESC hematopoietic differentiation systems based on either coculture with stromal cells16-18 or formation of embryoid bodies19-21 have been established. Recently, we described hESC differentiation in coculture with OP9 stromal cells that resulted in a highly efficient generation of hematopoietic progenitors after 4 to 5 days of coculture.18 We and others have demonstrated that hematopoietic clonogenic progenitors arise within the CD34+ cell population before CD45+ cells emerge, suggesting that the first hematopoietic progenitors in humans, as in mice, cannot be identified by CD45 expression.16,18,20,21
In the present study, we evaluated expression of specific hematopoietic markers (CD41a, CD43, CD45, CD235a) during hESC differentiation in OP9 coculture. We demonstrate that hESC-derived hematopoietic progenitors could be identified by surface expression of leukosialin (CD43). Selection of CD43+ cells reliably separated CD34+ hematopoietic cells from CD34+CD43–KDR+CD31+ endothelial cells. Also, CD43 was expressed on erythroid progenitors lacking CD34 expression, thus allowing complete isolation of CD34+ and CD34– hematopoietic progenitors from hESC/OP9 cocultures. Whereas CD41a was detected on CD43+ cells before CD45, isolation of CD41a+ cells demonstrated that these cells are already committed to erythro-megakaryocytopoiesis. In addition, we found that the first multipotent lymphohematopoietic progenitors have CD34+CD43+CD45–Lin– phenotype, coexpress endothelial proteins KDR, VE-cadherin, and CD105, and have FLT3lowGATA3highRUNX1high gene expression profile. Acquisition of CD45 expression by CD34+CD43+CD45–Lin– cells was associated with advanced differentiation toward the myeloid cell lineage.
Materials and methods
Cell lines
H1 and H9 hESC lines22 were obtained from WiCell Research Institute (Madison, WI), and maintained in an undifferentiated state by weekly passages on mouse embryonic fibroblasts as described.23 Mouse OP9 stromal cell line was kindly provided by Dr Toru Nakano (Osaka University, Osaka, Japan). OP9 was maintained on gelatin-coated plastic in αMEM (Gibco-Invitrogen, Carlsbad, CA) supplemented with 20% defined FBS (HyClone Laboratories, Logan, UT). Mouse MS-5 stromal cell line was obtained from DSMZ (Braunschweig, Germany) and maintained in αMEM supplemented with 5% heat-inactivated FBS (Gibco-Invitrogen).
hESC differentiation in OP9 coculture
Hematoendothelial hESC differentiation in OP9 coculture was performed as previously described.18 Briefly, overgrown OP9 cultures were prepared by feeding and prolonged culture of confluent OP9 cells for 4 days. hESCs prepared in suspension of small cell aggregates were added to OP9 cells in αMEM supplemented with 10% FBS (HyClone Laboratories) and 100 μM monothioglycerol (MTG; Sigma, St Louis, MO). hESC/OP9 cocultures were incubated up to 9 days in standard conditions (37°C, 5% CO2, > 95% humidity). Half the medium was replaced with fresh medium on days 4, 6, and 8. For cell harvesting, hESC/OP9 monolayers were dispersed by successive enzymatic treatment with collagenase IV (Gibco-Invitrogen; 1 mg/mL) for 20 minutes at 37°C, and 0.05% trypsin-0.5 mM EDTA (Gibco-Invitrogen) for 15 minutes at 37°C. Cells were resuspended by pipetting, washed twice in PBS-5%FBS, filtered through a 70-μm cell strainer (BD Falcon, Bedford, MA), and used for further analysis and cell sorting.
Cell sorting
Isolation of CD34+ cells and subsequent separation of CD34+CD43+ and CD34+KDR+/– subpopulations was performed by multiparameter magnet-activated cell sorting (MACS) using microbeads, MidiMACS magnet, and LS+ columns from Miltenyi Biotech (Bergisch Gladbach, Germany). A single-cell suspension from hESC/OP9 cocultures was incubated with basic (nonconjugated) microbeads, washed with PBS-5%FBS, and passed through a MidiMACS/LS+ unit to remove cells that bind the beads/column nonspecifically. CD34+ cells were isolated using CD34 Multisort microbeads and further processed to remove the antibody-tagged magnetic label as recommended by the manufacturer. Isolated CD34+ cells were stained with CD43-FITC and KDR-PE monoclonal antibodies (mAbs). The CD34+CD43+ subpopulation was isolated by anti-FITC microbeads, and the CD43– fraction was separated into CD34+CD43–KDR+ and CD34+CD43–KDR– subpopulations by anti-PE microbeads. In some experiments, CD43+ cells were directly isolated using CD43 microbeads. As determined by fluorescence-activated cell sorting (FACS) analysis, purity of MACS-isolated CD34+ subpopulations was more than 95%.
For sorting of CD43+ subsets, total CD43+ cells were isolated from hESC/OP9 cocultures by positive MACS selection using CD43-FITC mAb/anti-FITC microbeads labeling. MACS-enriched CD43+ cells were stained with CD41a-PE, CD235a-PE, and CD45-APC mAbs, and CD43+ subsets (CD43+CD41a/CD235a+CD45–, CD43+CD41a/CD235a–CD45–, CD43+CD41a/CD235a–CD45+) were separated on a FACSVantage cell sorter (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA) using cell gating by scatter parameters (live cells) and positive FITC staining (total CD43+ cells). Purity of sorted fractions as verified by FACS analysis was more than 98%.
FACS analysis
For cell surface staining, cells were prepared in PBS-2%FBS containing 0.05% sodium azide, 1 mM EDTA, and 1% mouse serum (Sigma), and labeled with multicolor mAb combinations. For intracellular staining, cells were fixed and permeabilized using Fix&Perm reagents (Caltag-Invitrogen, Carlsbad, CA). Samples were analyzed using FACSCalibur flow cytometer (BDIS) and FlowJo software (Tree Star, Ashland, OR) as previously described.18 All mAbs used in this study (Table S1, available at the Blood website; see the Supplemental Materials link at the top of the online article) were verified for nonreactivity with OP9 cells.
Endothelial cell culture and assays
To reveal endothelial cells in CD34+ subpopulations, cells were plated onto gelatin-coated 6-well plates at 2 × 105 cells/well in 3 mL complete medium containing endothelial serum-free medium (ESFM; Gibco-Invitrogen) supplemented with 5% FBS (Gibco-Invitrogen), 20 ng/mL basic fibroblast growth factor (bFGF; Invitrogen, Carlsbad, CA) and 1:100 dilution of endothelial cell growth factor (acidic FGF + heparin; Sigma). Endothelial cells were further expanded on fibronectin-coated dishes in complete medium without FBS. In some experiments, endothelial cultures were treated overnight with 25 ng/mL tumor necrosis factor (TNF; PeproTech, Rocky Hill, NJ) and examined for ICAM-1/VCAM-1 expression by flow cytometry. For immunofluorescent staining, endothelial cultures were prepared on fibronectin-coated chamber glass slides (BD Falcon), fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with Image-iT-FX signal enhancer (Molecular Probes-Invitrogen, Eugene, OR). Slides were stained with indicated antibodies and examined on a DMIRB fluorescent microscope (Leica Microsystems, Bannockburn, IL) equipped with MagnaFire camera/software (Optronics, Goleta, CA).
For acetylated low-density lipoprotein (Ac-LDL) uptake assay, endothelial cultures were incubated with 10 μg/mL DiI-Ac-LDL conjugate (Biomedical Technologies, Stoughton, MA) for 4 hours at 37°C, washed 3 times with PBS (with Ca/Mg), and inspected under fluorescent microscope or dispersed and analyzed by FACS to determine percentage of Ac-LDL–incorporating cells. Parallel cultures incubated at 4°C were used as a control.
For vascular tube formation, growth factor–reduced Matrigel (BD Falcon) was added into a 24-well plate (0.5 mL/well) and allowed to solidify for 1 hour at 37°C. Cells prepared in ESFM with 40 ng/mL VEGF165 (PeproTech) were plated onto a gel matrix (5 × 104 cells/well in 0.5 mL medium) and incubated 12 hours at 37°C.
Hematopoietic colony-forming assays and lymphomyeloid cultures
Hematopoietic clonogenic assays were performed using MethoCult GF+ complete methylcellulose medium with FBS and cytokines (SCF, G-CSF, GM-CSF, IL-3, IL-6, EPO) and MegaCult serum-free collagen assay with cytokines (TPO, IL-3, IL-6; StemCell Technologies, Vancouver, BC, Canada). MegaCult medium formulated for detection of megakaryocytic (Mk) colony-forming cells (CFCs) was additionally supplemented with 75 ng/mL SCF (PeproTech) and 5 U/mL EPO (StemCell Technologies) to enable simultaneous detection of erythroid CFCs. Cell-plating densities for CFC assays were optimized according to a day of hESC differentiation or cell subsets tested (Table S2). Colonies were scored after 14 to 21 days of incubation according to morphologic criteria as erythroid (E), granulocyte/macrophage (GM), macrophage (M), and mixed (Mix) colonies containing erythroid and nonerythroid cells. Multilineage Mix colonies were identified as large, often irregular, multicentric colonies containing myeloid (granulocytes and macrophages) and erythro-megakaryocytic component (GEMM). Identification of Mk colonies in collagen gels was performed by CD41a immunostaining according to the protocol supplied with MegaCult kit.
For lymphoid and myeloid differentiation, CD43+ subsets were seeded on irradiated (50 Gy) MS-5 stromal monolayers at a density of 103 cells/well of 6-well plates in 4 mL complete αMEM containing 10% FBS (HyClone Laboratories), 100 μM MTG, and either myeloid (SCF, 50 ng/mL; G-CSF, 20 ng/mL; IL-3, 10 ng/mL) or lymphoid (SCF, 50 ng/mL; Flt-3L, 50 ng/mL; IL-7, 20 ng/mL; IL-3, 5 ng/mL) cytokine combinations (all cytokines were from PeproTech). Every 4 days, the plates were gently agitated and half the medium containing nonadherent cells was replaced with fresh complete medium without IL-3. At indicated time points, nonadherent cells were collected and adherent cells were dispersed as described for hESC/OP9 cocultures. Nonadherent and adherent cells were pooled and assayed for myeloid CFCs by MethoCult GF+ assay, total CD43+ cells by FACS, and lymphoid transcripts by reverse transcription–polymerase chain reaction (RT-PCR).
Gene expression analysis by RT-PCR
Total RNA from hESCs, stromal cells, and cocultures was isolated with RNAwiz (Ambion, Austin, TX). Total RNA from sorted cell fractions was isolated using Perfect RNA Eukaryotic mini isolation kit (Eppendorf, Hamburg, Germany). All RNA samples were treated with DNAse using DNAfree reagents (Ambion). cDNA was prepared from 1 μg total RNA by oligo-dT15–primed RT with StrataScript RT kit (Stratagene, La Jolla, CA). Quantitative PCR (qPCR) was performed using Brilliant SYBR Green QPCR master mix (Stratagene). Real-time PCR detection was performed on ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) and the mean minimal cycle threshold values (Ct) from duplicate reactions were derived. All qPCR products were analyzed on 2% agarose gels to confirm the specificity of detection. Quantification of target genes was performed in comparison to the reference GAPDH gene as described24 and expressed as a ratio (R; fold differences to GAPDH). GAPDH was used as a reference gene because its expression remains constant during ESC differentiation.25 Relative expression in the group of n samples to be compared was calculated as Rn/ΣR(n) (contribution of each R value to the pooled R values across the group). All primers are shown in Table S3.
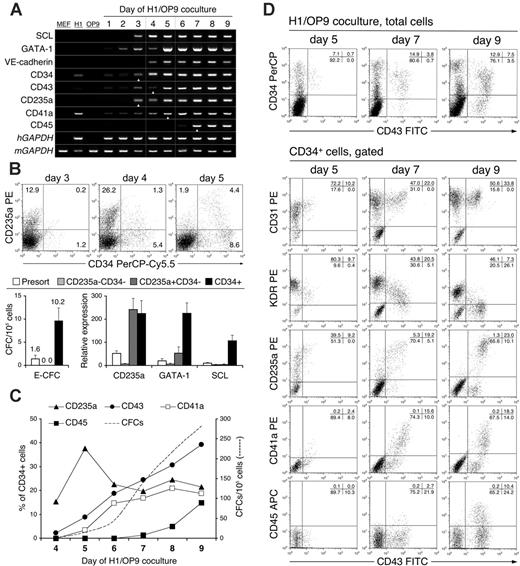
Kinetic analysis of hematopoietic development in H1/OP9 coculture. (A) Gene expression analysis of hematopoiesis-inductive transcription factors (SCL, GATA1) and hematoendothelial markers by RT-PCR. Triangular white pointers indicate the first day when a surface expression of respective hematopoietic markers was detected by flow cytometry. Dotted vertical lines show the timeframe of CFC emergence. Human- and mouse-specific GAPDH primers were used for positive control of human (hGAPDH) and MEF/OP9 (mGAPDH) RNA, respectively. (B) Early CD235a+CD34– cells in H1/OP9 coculture. Representative FACS analysis shows a burstlike CD235a expression and gradual emergence of CD34+CD235a+/– cells during 3 to 5 days of H1/OP9 coculture. Values within dot plots indicate percentage of cells in respective quadrants; 20 000 FACS events are displayed. Bar graphs show CFC potential and relative expression of CD235a, GATA-1, and SCL mRNA in FACS-sorted CD235a–CD34–, CD235a+CD34–, and CD34+ cells on day 4 of H1/OP9 coculture. CFCs and mRNA levels were determined by MethoCult GF+ assay and qRT-PCR, respectively. The relative expression of each GAPDH-normalized target gene was calculated in comparison with undifferentiated H1 cells using the 2ΔΔCt method. Results are the mean ± SD from 3 independent experiments. (C) Expression of hematopoietic markers during H1/OP9 coculture was analyzed by FACS within gated CD34+ cells and represented as a percentage of CD34+ cells (left y-axis). Dashed trend line shows total CFC counts (right y-axis). Results are the means from 3 independent experiments. (D) Representative FACS analysis of CD43+ cells in H1/OP9 coculture. Values within dot plots indicate percentage of cells in respective quadrants; 20 000 (total H1/OP9 cells), 5000 (gated CD34+ cells, day 5), and 10 000 (gated CD34+ cells, days 7, 9) FACS events are displayed.
Kinetic analysis of hematopoietic development in H1/OP9 coculture. (A) Gene expression analysis of hematopoiesis-inductive transcription factors (SCL, GATA1) and hematoendothelial markers by RT-PCR. Triangular white pointers indicate the first day when a surface expression of respective hematopoietic markers was detected by flow cytometry. Dotted vertical lines show the timeframe of CFC emergence. Human- and mouse-specific GAPDH primers were used for positive control of human (hGAPDH) and MEF/OP9 (mGAPDH) RNA, respectively. (B) Early CD235a+CD34– cells in H1/OP9 coculture. Representative FACS analysis shows a burstlike CD235a expression and gradual emergence of CD34+CD235a+/– cells during 3 to 5 days of H1/OP9 coculture. Values within dot plots indicate percentage of cells in respective quadrants; 20 000 FACS events are displayed. Bar graphs show CFC potential and relative expression of CD235a, GATA-1, and SCL mRNA in FACS-sorted CD235a–CD34–, CD235a+CD34–, and CD34+ cells on day 4 of H1/OP9 coculture. CFCs and mRNA levels were determined by MethoCult GF+ assay and qRT-PCR, respectively. The relative expression of each GAPDH-normalized target gene was calculated in comparison with undifferentiated H1 cells using the 2ΔΔCt method. Results are the mean ± SD from 3 independent experiments. (C) Expression of hematopoietic markers during H1/OP9 coculture was analyzed by FACS within gated CD34+ cells and represented as a percentage of CD34+ cells (left y-axis). Dashed trend line shows total CFC counts (right y-axis). Results are the means from 3 independent experiments. (D) Representative FACS analysis of CD43+ cells in H1/OP9 coculture. Values within dot plots indicate percentage of cells in respective quadrants; 20 000 (total H1/OP9 cells), 5000 (gated CD34+ cells, day 5), and 10 000 (gated CD34+ cells, days 7, 9) FACS events are displayed.
Results
CD43 is the earliest and predominant pan-hematopoietic marker expressed by differentiating hESCs in OP9 coculture
We have previously demonstrated that early hematopoietic progenitors generated in hESC/OP9 coculture arise within the CD34+ cell population; however, CD34+ cells are heterogeneous and include at least endothelial cells.18 To define the earliest and specific marker for hematopoietic progenitors, we analyzed expression of CD45, CD43, CD41a (GPIIb), and CD235a (glycophorin A) during hESC/OP9 coculture. In humans, both CD45 and CD43 are considered as pan-hematopoietic markers,26,27 though CD45 is not expressed on platelets and cells of the erythroid lineage,27 whereas CD43 is expressed on platelets and erythroid progenitors, but it is not detected on mature erythrocytes and B-cell subsets.26,28 CD41a and CD235a are known markers of megakaryocytic and erythroid cell lineages, respectively.29,30
Using PCR and flow cytometry, we found that undifferentiated hESCs did not express CD235a and CD45 mRNA (Figure 1A) or protein (not shown). Whereas CD34, CD43, and CD41a mRNAs were detected in undifferentiated hESCs (Figure 1A), no surface or intracellular expression of these proteins was detected by flow cytometry (not shown).
The first hematopoietic marker detected in hESC/OP9 coculture was CD235a. CD235a+ cells emerged abruptly in relatively high numbers on day 3 of differentiation (Figure 1B) coincidently with up-regulation of CD34 and CD43 mRNA, and induction of SCL and up-regulation of GATA1 transcription factors (Figure 1A), which are known primary inducers of hematopoietic commitment.13,31,32 However, almost all day-3 CD235a+ cells were CD34–, and only a few CD34+CD235a+/– cells could be detected by flow cytometry (Figure 1B). Both CD235a+CD34– and CD34+ cells continued to increase on day 4, whereas a significant drop in CD235a+CD34– and a rise in CD34+CD235a+ cells were observed on day 5. Comparative analysis of isolated CD235a+CD34– and CD34+ cells revealed that SCL and GATA1 were predominantly expressed in CD34+ cells and only CD34+ cells contained the first detectable on day-4 CFC-Es (Figure 1B). Moreover, CD235a+CD34– cells demonstrated morphologic heterogeneity and were lacking typical erythroblastoid cells (Figure S1). Thus, these results confirm our previous observation that first hESC-derived hematopoietic progenitors are confined to CD34+ cells. It is possible that early expression of CD235a on CD34– cells is related to promiscuous gene expression during hESC differentiation.
The first CD43+ cells were detected on days 4 to 5 of differentiation as a subpopulation of CD34+ cells expressing the highest density of CD235a (Figure 1C-D). Their emergence strictly correlated with the appearance of the first CFC-Es. The first CD41a+ and CD45+ cells were detected as subpopulations of CD43+ cells on day 5 and day 7, respectively. On day 9, almost all CD235a+, CD41a+, and CD45+ cells were found within a more numerous CD43+ population (Figure 1C-D). Notably, early CD43+ cells expressed endothelial markers KDR (VEGF-R2) and CD31 (PECAM-1). KDR was down-regulated on CD43+ cells along with advanced differentiation, whereas CD31 was stably expressed (Figure 1D).
To reveal hematopoietic markers expressed by clonogenic progenitors, we evaluated the CFC potential of cells from hESC/OP9 coculture after depletion of CD34+, CD43+, CD235a+, CD41a+, and CD45+ cells. As shown in Table 1, only depletion of CD43+ cells resulted in removal of all CFC types throughout hESC/OP9 coculture. CD34+ depletion led to removal of the CFC-Mix/GM/Ms, whereas CFC-Es were progressively detected in CD34– fractions after 6 days of hESC/OP9 coculture. These results indicated that erythroid progenitors down-regulated CD34 expression with advanced maturation, but retained CD43 expression. Separation of CD41a+, CD235a+, and CD45+ cells resulted in partial, mostly erythroid (CD41a, CD235a) or myeloid (CD45) CFC depletion.
CFC depletion by antibodies against hematopoietic markers
. | CFC depletion, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Depleted cell subset, day of differentiation . | CFC-E . | CFC-Mix . | CFC-GM . | CFC-M . | |||
| CD34+ | |||||||
| 5 | 100 | NA | NA | NA | |||
| 6 | 96.8 ± 2.6 | 100 | 100 | 94.2 ± 11.6 | |||
| 7 | 62.3 ± 16.2 | 98.0 ± 3.5 | 97.7 ± 4.1 | 96.1 ± 3.6 | |||
| 8 | 37.8 ± 17.3 | 99.6 ± 0.8 | 99.1 ± 3.5 | 99.4 ± 2.3 | |||
| CD43+ | |||||||
| 5 | 98.3 ± 3.3 | NA | NA | NA | |||
| 6 | 100 | 100 | 99.9 ± 1.9 | 90.5 ± 13.2 | |||
| 7 | 100 | 99.3 ± 1.2 | 96.3 ± 5.0 | 96.4 ± 2.6 | |||
| 8 | 98.8 ± 1.4 | 100 | 97.3 ± 2.4 | 99.4 ± 1.0 | |||
| CD235a+ | |||||||
| 5 | 100 | NA | NA | NA | |||
| 6 | 100 | 69.7 ± 26.3 | 35.8 ± 22.6 | 38.1 ± 28.1 | |||
| 7 | 100 | 60.1 ± 7.6 | 19.0 ± 9.2 | 11.6 ± 8.3 | |||
| 8 | 99.0 ± 1.5 | 19.4 ± 7.7 | 5.2 ± 2.5 | 11.8 ± 5.4 | |||
| CD41a+ | |||||||
| 5 | 55.8 ± 18.0 | NA | NA | NA | |||
| 6 | 87.4 ± 9.6 | 20.8 ± 16.1 | 37.7 ± 23.7 | 35.5 ± 27.3 | |||
| 7 | 93.2 ± 9.3 | 31.6 ± 10.7 | 9.0 ± 5.6 | 12.7 ± 9.6 | |||
| 8 | 87.6 ± 13.3 | 4.1 ± 3.5 | 7.8 ± 6.6 | 11.3 ± 8.2 | |||
| CD45+ | |||||||
| 5 | <1 | NA | NA | NA | |||
| 6 | <1 | <1 | <1 | <1 | |||
| 7 | 4.7 ± 4.2 | 50.3 ± 6.5 | 46.2 ± 16.9 | 49.9 ± 17.5 | |||
| 8 | <1 | 87.4 ± 11.7 | 56.4 ± 13.1 | 75.0 ± 9.3 | |||
. | CFC depletion, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Depleted cell subset, day of differentiation . | CFC-E . | CFC-Mix . | CFC-GM . | CFC-M . | |||
| CD34+ | |||||||
| 5 | 100 | NA | NA | NA | |||
| 6 | 96.8 ± 2.6 | 100 | 100 | 94.2 ± 11.6 | |||
| 7 | 62.3 ± 16.2 | 98.0 ± 3.5 | 97.7 ± 4.1 | 96.1 ± 3.6 | |||
| 8 | 37.8 ± 17.3 | 99.6 ± 0.8 | 99.1 ± 3.5 | 99.4 ± 2.3 | |||
| CD43+ | |||||||
| 5 | 98.3 ± 3.3 | NA | NA | NA | |||
| 6 | 100 | 100 | 99.9 ± 1.9 | 90.5 ± 13.2 | |||
| 7 | 100 | 99.3 ± 1.2 | 96.3 ± 5.0 | 96.4 ± 2.6 | |||
| 8 | 98.8 ± 1.4 | 100 | 97.3 ± 2.4 | 99.4 ± 1.0 | |||
| CD235a+ | |||||||
| 5 | 100 | NA | NA | NA | |||
| 6 | 100 | 69.7 ± 26.3 | 35.8 ± 22.6 | 38.1 ± 28.1 | |||
| 7 | 100 | 60.1 ± 7.6 | 19.0 ± 9.2 | 11.6 ± 8.3 | |||
| 8 | 99.0 ± 1.5 | 19.4 ± 7.7 | 5.2 ± 2.5 | 11.8 ± 5.4 | |||
| CD41a+ | |||||||
| 5 | 55.8 ± 18.0 | NA | NA | NA | |||
| 6 | 87.4 ± 9.6 | 20.8 ± 16.1 | 37.7 ± 23.7 | 35.5 ± 27.3 | |||
| 7 | 93.2 ± 9.3 | 31.6 ± 10.7 | 9.0 ± 5.6 | 12.7 ± 9.6 | |||
| 8 | 87.6 ± 13.3 | 4.1 ± 3.5 | 7.8 ± 6.6 | 11.3 ± 8.2 | |||
| CD45+ | |||||||
| 5 | <1 | NA | NA | NA | |||
| 6 | <1 | <1 | <1 | <1 | |||
| 7 | 4.7 ± 4.2 | 50.3 ± 6.5 | 46.2 ± 16.9 | 49.9 ± 17.5 | |||
| 8 | <1 | 87.4 ± 11.7 | 56.4 ± 13.1 | 75.0 ± 9.3 | |||
Indicated cell subsets were depleted from hESC/OP9 cocultures by negative MACS technique using FITC/PE-conjugated specific mAbs and anti-FITC/PE secondary microbeads. Isotype-matched control mAbs were used for nonspecific depletion control. Depletion of cell subsets was in the range of 85% to 97% as determined by flow cytometry. MACS-processed cell samples were tested for erythroid (E), mixed (Mix), and myeloid (GM, M) CFCs by MethoCult GF+ assay. CFC depletion (%) was calculated by the following formula: 1 – (CFC counts in specific mAb-depleted sample – CFC counts in isotype control mAb-treated sample) × 100. Results are the mean ± SD of 4 independent experiments with H1 (n = 2) and H9 (n = 2) cells. By negative selection analysis, CFC depletion values (%) reflect the proportion of CFCs expressing a depletory marker. CD43 was the only cell marker expressed by all CFC types throughout the hESC/OP9 coculture.
NA indicates not applicable (no CFC detection in the isotype control mAb-treated samples).
These data demonstrate that CD43 is a pan-hematopoietic marker of the earliest clonogenic progenitors and differentiating hematopoietic cells and can be used for selection of the entire hematopoietic population generated in hESC/OP9 coculture.
CD43 discriminates hESC-derived hematopoietic from endothelial cells
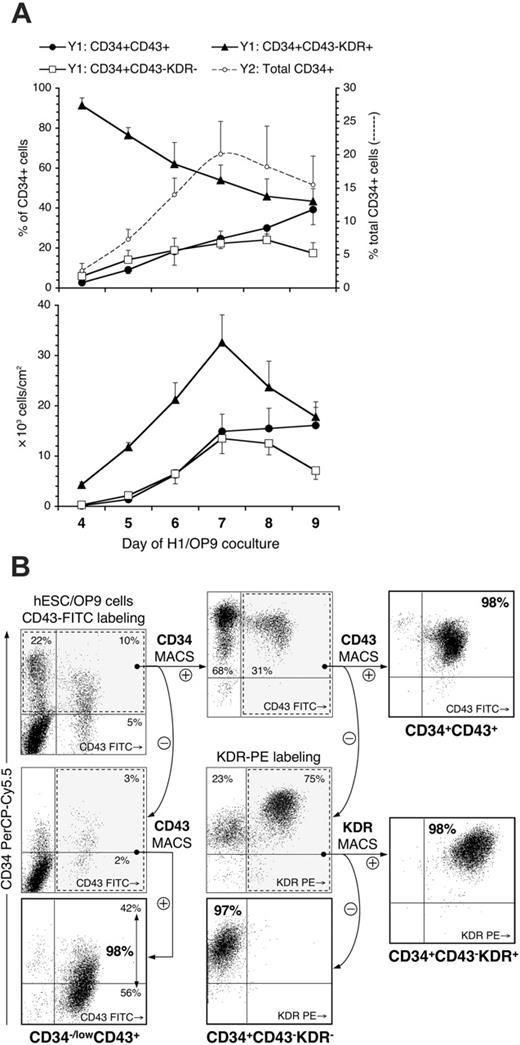
Because CD43+ cells retained KDR expression up to day 7 of differentiation (Figure 1D), it was unclear whether CD43 discriminates hematopoietic from endothelial cells. FACS analysis revealed that based on the expression of KDR and CD43, CD34+ cells could be subdivided into 3 major subsets: (1) CD34+CD43+, (2) CD34+CD43–KDR+, and (3) CD34+CD43–KDR– (Figure 1D). As shown in Figure 2A, the majority of early CD34+ cells (day 4) were KDR+ and CD43–. With advanced differentiation, CD34+CD43+ and CD34+CD43–KDR– subsets gradually increased, whereas a proportion of CD34+CD43–KDR+ cells decreased. CD34+ subsets were isolated on days 6 and 9 of differentiation as depicted in Figure 2B and assayed for CFC potential or cultured with endothelial growth factors. As expected, positive selection of CD43+ cells resulted in recovery of all CFC types from the CD34+ population (Figure 3A). The number of CFCs within CD43– fractions remained negligible. In the same time, only CD34+CD43–KDR+ cells readily formed the monolayer of VE-cadherin+ cells when cultured in endothelial conditions (Figure 3C). CD34+CD43+ cells remained in suspension and did not give rise to adherent cells up to 10 days of endothelial culture. CD34+CD43–KDR– cells produced neither VE-cadherin+ nor proliferating cells in endothelial conditions. Morphologically, the majority of CD34+CD43+ cells demonstrated a high nuclear-to-cytoplasm ratio, characteristic for hematopoietic blasts; however, more mature erythroid cells could be also found (Figure 3D). CD34+CD43–KDR+ cells had typical endothelial morphology. Analysis of gene expression by qRT-PCR revealed that CD34+CD43+ cells expressed other hematopoietic markers (CD235a, CD41a, CD45) and transcription factors essential for hematopoietic commitment (SCL, LMO2, RUNX1, C-MYB, GATA2) and specification of erythro-megakaryocytic (SCL, LMO2, GATA1, NF-E2) and lymphomyeloid (C-MYB, PU1, IKAROS) lineages. In contrast, predominant expression of VE-cadherin, Flt-1, KDR, and TIE2 in CD34+CD43–KDR+ cells reflected their endothelial potential (Figure 3B).
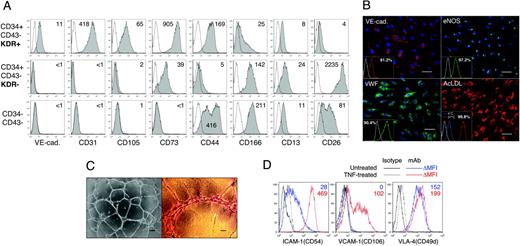
To confirm that CD34+CD43–KDR+ cells represent endothelial cells, we evaluated their phenotype and functional potential. As shown in Figure 4A, isolated CD34+CD43–KDR+ cells were positive for endothelial markers VE-cadherin and CD31. They also possessed a distinctive expression of CD90 and CXCR4 (not shown) and α4-integrin (CD49d, VLA-4; Figure 4D). In culture, these cells expressed markers of mature endothelial cells, eNOS and VWF, and were capable of efficient Ac-LDL incorporation (Figure 4B). When plated on Matrigel matrix, CD34+CD43–KDR+ cells formed typical vascular tubes (Figure 4C). Another feature of endothelial cells is the ability to up-regulate expression of VCAM-1 and ICAM-1 adhesion molecules in response to proinflammatory factors.33 As shown in Figure 4D, CD34+CD43–KDR+ cells up-regulated ICAM-1 expression and became VCAM-1+ after treatment with TNF. This effect was specific and was not observed for VLA-4. Collectively, these data provide evidence that CD34+CD43–KDR+ cells isolated after 6 days of hESC/OP9 coculture represent an endothelial cell population.
Developmental kinetics and sorting strategy of CD34+ subsets. (A) Kinetic analysis of CD34+ subsets in H1/OP9 coculture. Indicated CD34+ subsets were analyzed by FACS within gated CD34+ population and are represented on the top graph as percentage of CD34+ cells (left y1-axis). Total CD34+ cells (%) are depicted by dashed trend line (right y2-axis). The bottom graph shows absolute numbers of respective CD34+ subsets. Results are the mean ± SD from 4 independent experiments. (B) Flow diagram of multiparameter MACS separation of CD34+ subsets showing a FACS analysis of target cell populations throughout the sorting procedure (see “Materials and methods” for details). Positive and negative MACS fractions are indicated by arrows marked with + and – circles, respectively. Values within dot plots indicate percentage of cells in respective quadrants. The representative experiment is shown.
Developmental kinetics and sorting strategy of CD34+ subsets. (A) Kinetic analysis of CD34+ subsets in H1/OP9 coculture. Indicated CD34+ subsets were analyzed by FACS within gated CD34+ population and are represented on the top graph as percentage of CD34+ cells (left y1-axis). Total CD34+ cells (%) are depicted by dashed trend line (right y2-axis). The bottom graph shows absolute numbers of respective CD34+ subsets. Results are the mean ± SD from 4 independent experiments. (B) Flow diagram of multiparameter MACS separation of CD34+ subsets showing a FACS analysis of target cell populations throughout the sorting procedure (see “Materials and methods” for details). Positive and negative MACS fractions are indicated by arrows marked with + and – circles, respectively. Values within dot plots indicate percentage of cells in respective quadrants. The representative experiment is shown.
To further evaluate whether CD34+CD43+ and CD34+CD43–KDR+ cells are committed to hematopoietic and endothelial development, respectively, we cultured these cells with fresh OP9 cells in the presence of hematopoietic and endothelial growth factors (SCF, TPO, IL-6, bFGF). No hematopoietic cell production was detected from CD34+CD43–KDR+ cells and no endothelial differentiation was observed from CD34+CD43+ cells isolated as early as day 6 of differentiation (not shown). However, CD34+CD43–KDR+ cells at earlier stages of differentiation (day 5) gave rise to hematoendothelial colonies on coculture with OP9 cells or in feeder-free conditions (Figure 3E). These colonies arose from primary endothelial clusters, which were formed during the first 4 days of the culture and subsequently gave rise to peripheral expansion of hematopoietic cells. In parallel cultures of CD34+CD43+ cells (day 5), such colonies were not observed, indicating that hematoendothelial precursors have primarily endothelial-like characteristics and reside in the CD34+CD43–KDR+ population up to 5 days of differentiation.
The developmental potential of CD34+CD43–KDR– cells remains unclear. These cells showed lack of detectable hematoendothelial potential either in direct assays or on OP9 coculture. Cytospin preparations revealed that CD34+CD43–KDR– cells have a relatively high nuclear-to-cytoplasm ratio, a kidney-shaped or irregular nucleus, and dark blue cytoplasm (Figure 3D). Phenotypically, CD34+CD43–KDR– cells were similar to the majority of CD34–CD43– cells in hESC/OP9 coculture (Figure 4A). However, they could be distinguished by expression of CD73, low expression of CD44, higher expression of aminopeptidase N (CD13), and a unique bright expression of dipeptidyl-peptidase IV (CD26). These features are consistent with cells of mesenchymal origin and indicate that CD34+CD43–KDR– cells may develop in hESC/OP9 coculture through a distinct nonhematoendothelial differentiation pathway.
In addition to CD34+CD43+ cells, we identified a small population of CD34–/lowCD43+ cells (Figure 2B) that expressed CD235a (not shown) and formed only CFC-Es in MethoCult assay (Figure 3A). These features are in agreement with CFC depletion experiments described under “CD43 is the earliest and predominant pan-hematopoietic marker expressed by differentiating hESCs in OP9 culture” and indicate that CD34–/lowCD43+ cells represent erythroid progenitors at an advanced stage of maturation.
Altogether, these results demonstrate that CD43 expression discriminates hematopoietic from endothelial cells and signifies commitment to hematopoietic lineage.
Identification of functionally distinct CD43+ hematopoietic progenitors
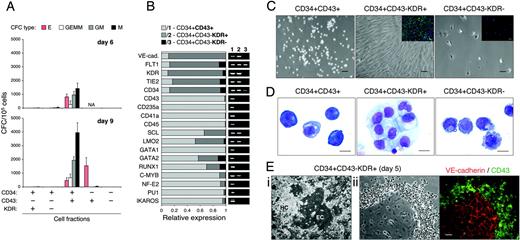
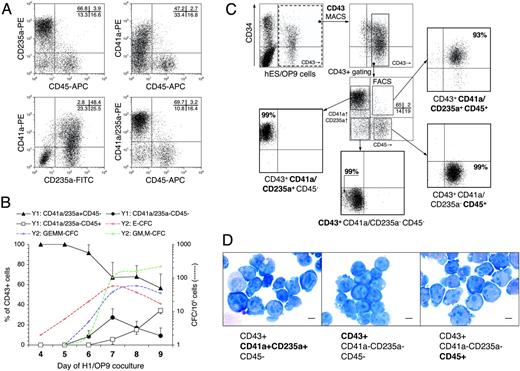
As demonstrated, CD43+ hematopoietic progenitors generated in hESC/OP9 coculture included CD41a+, CD235a+, and CD45+ cells (Figure 1D). Phenotypic analysis of isolated CD43+ cells revealed that a major subset of CD43+ cells coexpressed CD235a and CD41a, whereas the CD43+CD41a–CD235a– subpopulation contained CD45+ and CD45– cells (Figure 5A). Therefore, PE-labeled CD235a and CD41a mAbs were combined and used with APC-labeled CD45 mAb to define 3 main CD43+ subsets: (1) CD43+CD41a/CD235+CD45–, (2) CD43+CD41a/CD235a–CD45–, and (3) CD43+CD41a/CD235a–CD45+ (Figure 5A). Kinetic analysis of CD43+ subsets during hESC/OP9 coculture demonstrated that first CFC-Es appeared coincidently with first CD43+CD41a/CD235a+CD45– cells (days 4-5), first CFC-GEMMs and CFC-GM/Ms emerged along with CD43+CD41a/CD235a–CD45– cells (day 6), and a surge in CFC-GM/Ms on day 7 was associated with the appearance of CD43+CD41a/CD235a–CD45+ cells (Figure 5B). Because no lymphomyeloid or other tested markers (CD11b, CD14, CD2, CD3, CD7, CD19, CD38, CD45RA, HLA-DR) were detected either on CD34+ or CD43+ cells during hESC/OP9 coculture (not shown), we considered CD43+CD41a/CD235a– cells as lineage negative (Lin–) and designated the defined CD43+ subsets as (1) CD43+CD41a/CD235a+, (2) CD43+CD45–Lin–, and (3) CD43+CD45+Lin–, respectively.
Hematopoietic and endothelial potential of CD34+ subsets. (A) CFC potential of MACS-sorted CD34+ subsets with indicated phenotype (+/– chart) was tested using MethoCult GF+ assay. Results are the mean ± SD from 9 independent experiments with H1 (n = 6) and H9 (n = 3) cells. NA indicates not applicable (subset was not detected/sorted). (B) qRT-PCR analysis of CD34+ subsets on day 6 of H1/OP9 coculture. The stacked bar graph shows expression levels of indicated transcripts represented by relative units (see “Materials and methods” for details). qPCR results are the means from 3 independent experiments. Representative agarose gel electrophoresis of qPCR products is shown. (C) Endothelial culture of CD34+ subsets isolated on day 6 of H1/OP9 coculture. Photographs show 5-day culture of indicated CD34+ subsets in endothelial conditions (scale bar represents 50 μm). Insets show VE-cadherin expression (scale bar represents 50 μm). VE-cadherin was stained by anti–VE-cadherin antibody (goat IgG; R&D Systems, Minneapolis, MN) followed by anti–goat IgG–Alexa Fluor-488 conjugate (green fluorescence; Molecular Probes). Cell nuclei were visualized by DAPI staining (blue fluorescence). Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 20×/0.3 objective lens, and were acquired through MagnaFire camera and software (Optronics). Fluorescent images were composed using Adobe Photoshop software (Adobe Systems, San Jose, CA). The representative experiment is shown. Identical results were obtained with CD34+ subsets isolated on day 9 (H1, H9) and day 6 (H9). (D) Wright-stained cytospins of CD34+ subsets isolated on day 9 of H1/OP9 coculture (scale bar represents 10 μm). Images were captured with a Microphot-SA microscope (Nikon, Melville, NY) equipped with a 100×/1.40 oil-immersion objective lens, and were acquired through a DFC320 camera and Firecam 1.3 software (Leica Microsystems). (E) CD34+CD43– KDR+ cells isolated on day 5 of H1/OP9 coculture were cultured 6 days with fresh OP9 cells (i) or without feeder cells (ii) in StemSpan serum-free medium (StemCell Technologies) supplemented with 2% FBS (HyClone Laboratories), Ex-Cyte (1/500; Serological, Norcross, GA), 10 ng/mL bFGF, 50 ng/mL SCF, 10 ng/mL TPO, 20 ng/mL IL-6. Central endothelial clusters (ECs) surrounded by proliferating hematopoietic clusters (HCs) were observed in coculture with OP9 cells (i; scale bar represents 100 μm). Similar hematoendothelial colonies were formed in feeder-free cultures (ii; scale bar represents 50 μm); bright-field (left panel) and fluorescent (right panel) photographs show the same colony stained with anti-CD43 mAb (BD Pharmingen, San Diego, CA) and anti–VE-cadherin antibody (goat IgG; R&D Systems) followed by anti–mouse IgG–Alexa Fluor-488 (green fluorescence) and anti–goat IgG–Alexa Fluor-555 conjugates (red florescence; Molecular Probes). Images captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 10×/0.25 (i) or a 20×/0.3 (ii) objective lens, and were acquired with a MagnaFire camera and software (Optronics). Note a clear separation of the hematopoietic and endothelial cells by CD43 staining: all rounded hematopoietic cells are CD43+, but adherent VE-cadherin+ endothelial cells are CD43–.
Hematopoietic and endothelial potential of CD34+ subsets. (A) CFC potential of MACS-sorted CD34+ subsets with indicated phenotype (+/– chart) was tested using MethoCult GF+ assay. Results are the mean ± SD from 9 independent experiments with H1 (n = 6) and H9 (n = 3) cells. NA indicates not applicable (subset was not detected/sorted). (B) qRT-PCR analysis of CD34+ subsets on day 6 of H1/OP9 coculture. The stacked bar graph shows expression levels of indicated transcripts represented by relative units (see “Materials and methods” for details). qPCR results are the means from 3 independent experiments. Representative agarose gel electrophoresis of qPCR products is shown. (C) Endothelial culture of CD34+ subsets isolated on day 6 of H1/OP9 coculture. Photographs show 5-day culture of indicated CD34+ subsets in endothelial conditions (scale bar represents 50 μm). Insets show VE-cadherin expression (scale bar represents 50 μm). VE-cadherin was stained by anti–VE-cadherin antibody (goat IgG; R&D Systems, Minneapolis, MN) followed by anti–goat IgG–Alexa Fluor-488 conjugate (green fluorescence; Molecular Probes). Cell nuclei were visualized by DAPI staining (blue fluorescence). Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 20×/0.3 objective lens, and were acquired through MagnaFire camera and software (Optronics). Fluorescent images were composed using Adobe Photoshop software (Adobe Systems, San Jose, CA). The representative experiment is shown. Identical results were obtained with CD34+ subsets isolated on day 9 (H1, H9) and day 6 (H9). (D) Wright-stained cytospins of CD34+ subsets isolated on day 9 of H1/OP9 coculture (scale bar represents 10 μm). Images were captured with a Microphot-SA microscope (Nikon, Melville, NY) equipped with a 100×/1.40 oil-immersion objective lens, and were acquired through a DFC320 camera and Firecam 1.3 software (Leica Microsystems). (E) CD34+CD43– KDR+ cells isolated on day 5 of H1/OP9 coculture were cultured 6 days with fresh OP9 cells (i) or without feeder cells (ii) in StemSpan serum-free medium (StemCell Technologies) supplemented with 2% FBS (HyClone Laboratories), Ex-Cyte (1/500; Serological, Norcross, GA), 10 ng/mL bFGF, 50 ng/mL SCF, 10 ng/mL TPO, 20 ng/mL IL-6. Central endothelial clusters (ECs) surrounded by proliferating hematopoietic clusters (HCs) were observed in coculture with OP9 cells (i; scale bar represents 100 μm). Similar hematoendothelial colonies were formed in feeder-free cultures (ii; scale bar represents 50 μm); bright-field (left panel) and fluorescent (right panel) photographs show the same colony stained with anti-CD43 mAb (BD Pharmingen, San Diego, CA) and anti–VE-cadherin antibody (goat IgG; R&D Systems) followed by anti–mouse IgG–Alexa Fluor-488 (green fluorescence) and anti–goat IgG–Alexa Fluor-555 conjugates (red florescence; Molecular Probes). Images captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 10×/0.25 (i) or a 20×/0.3 (ii) objective lens, and were acquired with a MagnaFire camera and software (Optronics). Note a clear separation of the hematopoietic and endothelial cells by CD43 staining: all rounded hematopoietic cells are CD43+, but adherent VE-cadherin+ endothelial cells are CD43–.
FACS-isolated CD43+ subsets (Figure 5C) were examined for morphology, CFC potential, and gene expression profile. All subsets displayed morphology similar to hematopoietic blasts: large nucleus with multiple, often elongated, prominent nucleoli and scant amount of basophilic cytoplasm (Figure 5D). However, some distinctive features could be noticed. CD43+CD45–Lin– cells were smaller and had the highest nuclear-to-cytoplasm ratio. CD43+CD45+Lin– cells were larger and had more plentiful cytoplasm, often with a single large light-pink granule. CD43+CD41a/CD235a+ cells were variable in size and contained occasional erythroblastoid cells (Figure 5D).
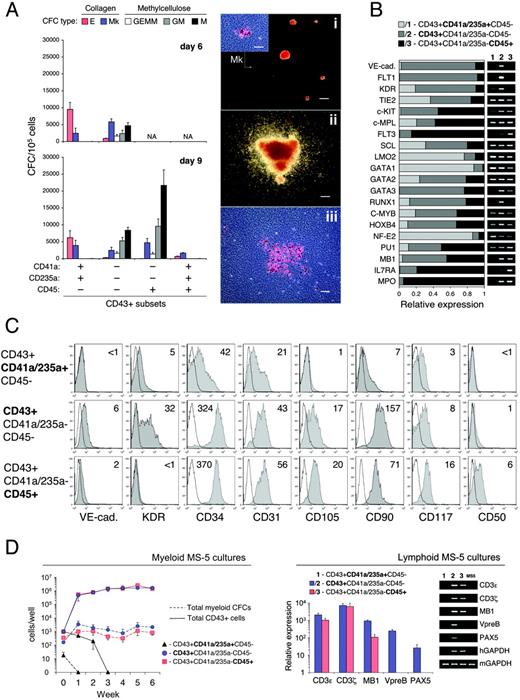
CFC potential of CD43+ cell subsets was tested using FBS-containing methylcellulose medium and serum-free collagen assay adapted for the simultaneous detection of CFC-Es and CFC-Mks. As shown in Figure 6A, CD43+CD41a/CD235a+ cells yielded CFC-Es and CFC-Mks, confirming their restricted erythro-megakaryocytic potential. Accordingly, in this population, qRT-PCR analysis revealed the highest expression levels of GATA1 and NF-E2 transcription factors (Figure 6B), which play a critical role in the erythro-megakaryocytic lineage commitment and differentiation.34,35 In addition, a minor CD45+ subpopulation expressing CD41a/CD235+ (Figure 5C) also produced only CFC-Es and CFC-Mks (Figure 6A).
In contrast to CD43+CD41a/CD235+ cells, multilineage CFC potential was strictly associated with CD43+CD45–/+Lin– subsets. Notably, all CFC-GEMMs and CFC-GM/Ms first detectable on day 6 of hESC/OP9 coculture were recovered by selection of the rare CD43+CD45–Lin– cells (Figure 6A). Gene expression analysis revealed the highest levels of SCF-R (c-KIT), transcription factors associated with definitive hematopoietic potential (RUNX1, C-MYB, HOXB4), and markers of lymphomyeloid commitment (PU1, MB1, IL7RA) in CD43+CD45–/+Lin– subsets (Figure 6B). Moreover, FLT3 and GATA3 were restricted to these populations. By phenotype, CD43+CD45–/+Lin– subsets could be distinguished from CD43+CD41a/CD235+ cells by endoglin (CD105) expression, and higher expression of CD90 and CD34 (Figure 6C). Remarkably, CD43+CD45–Lin– cells contained KDR+ subpopulation and expressed VE-cadherin and FLT1 endothelial markers, which were all down-regulated in CD43+CD45+Lin– cells (Figure 6B-C). CD43+CD41a/CD235+ cells retained a low level of KDR expression, but VE-cadherin, FLT1, and CD105 were almost undetectable in this population (Figure 6B-C). CD50 (ICAM-3), suggested as an early marker of hematopoietic progenitors in fetal bone marrow CD34+CD38– population,36 was weakly expressed only on CD45+ cells.
Endothelial phenotype and function of CD34+CD43–KDR+ cells isolated after 6 days of H1/OP9 coculture. (A) FACS analysis of KDR+ and KDR– fractions of CD34+CD43– cells isolated on day 9 of H1/OP9 coculture. Phenotype of CD34+CD43– KDR– cells was compared with phenotype of CD34–CD43– cells obtained after depletion of CD34+ and CD43+ cells. Plots show isotype control (open) and specific mAb (tinted) histograms. Values within plots indicate specific mean fluorescence intensity (ΔMFI) calculated by formula: linear-scaled MFI of specific mAb-stained cells – linear-scaled MFI of isotype control mAb-treated cells. The representative experiment is shown. Similar results were obtained in 5 independent experiments with H1- and H9-derived CD34+CD43–KDR+/– cells isolated on day 6 (n = 2) and day 9 (n = 3) of differentiation. (B) CD34+CD43–KDR+ cells were cultured 7 days in endothelial expansion conditions and examined for markers of mature endothelial cells. Immunofluorescent staining was performed with primary antibodies against VE-cadherin (goat IgG; R&D Systems), von Willebrand factor (VWF; rabbit IgG; Sigma) and endothelial NO synthetase (eNOS; mouse IgG1; BD Pharmingen) followed by respective secondary antibody against goat IgG-Alexa Fluor-555 (red fluorescence), rabbit IgG-Alexa Fluor-488 (green fluorescence), and mouse IgG-Alexa Fluor-488 (Molecular Probes). Negative controls were done using appropriate primary IgG controls (Sigma). Cell nuclei were visualized by DAPI staining (blue fluorescence). Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 20×/0.3 objective lens, and were acquired with a MagnaFire camera and software (Optronics). Fluorescent images were composed using Adobe Photoshop software. Ac-LDL uptake was assessed by incubation with DiI-Ac-LDL conjugate as described in “Materials and methods.” Scale bar represents 50 μm. Insets show FACS analysis of respective surface (VE-cadherin) and intracellular (eNOS, VWF) markers in parallel cultures, or instant FACS profiles of cells incubated with DiI-Ac-LDL at 37°C (Ac-LDL uptake) versus 4°C (control Ac-LDL binding). (C) Vascular tubes formation by CD34+CD43– KDR+ cells (scale bar represents 200 μm, left panel; and 50 μm, right panel). Images were captured with an inverted DMIRB micrscope (Leica Microsystems) equipped with a 5×/0.12 (left) or 20×/0.3 (right) objective lens, and were acquired through a MagnaFire camera and software (Optronics). (D) TNF-induced up-regulation of ICAM-1 and induction of VCAM-1 expression in CD34+CD43–KDR+ endothelial cultures. Numbers within plots indicate ΔMFI values for untreated (blue) and TNF-treated (red) cells. VLA-4 staining was used as a control. A representative example of 3 independent experiments is shown.
Endothelial phenotype and function of CD34+CD43–KDR+ cells isolated after 6 days of H1/OP9 coculture. (A) FACS analysis of KDR+ and KDR– fractions of CD34+CD43– cells isolated on day 9 of H1/OP9 coculture. Phenotype of CD34+CD43– KDR– cells was compared with phenotype of CD34–CD43– cells obtained after depletion of CD34+ and CD43+ cells. Plots show isotype control (open) and specific mAb (tinted) histograms. Values within plots indicate specific mean fluorescence intensity (ΔMFI) calculated by formula: linear-scaled MFI of specific mAb-stained cells – linear-scaled MFI of isotype control mAb-treated cells. The representative experiment is shown. Similar results were obtained in 5 independent experiments with H1- and H9-derived CD34+CD43–KDR+/– cells isolated on day 6 (n = 2) and day 9 (n = 3) of differentiation. (B) CD34+CD43–KDR+ cells were cultured 7 days in endothelial expansion conditions and examined for markers of mature endothelial cells. Immunofluorescent staining was performed with primary antibodies against VE-cadherin (goat IgG; R&D Systems), von Willebrand factor (VWF; rabbit IgG; Sigma) and endothelial NO synthetase (eNOS; mouse IgG1; BD Pharmingen) followed by respective secondary antibody against goat IgG-Alexa Fluor-555 (red fluorescence), rabbit IgG-Alexa Fluor-488 (green fluorescence), and mouse IgG-Alexa Fluor-488 (Molecular Probes). Negative controls were done using appropriate primary IgG controls (Sigma). Cell nuclei were visualized by DAPI staining (blue fluorescence). Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 20×/0.3 objective lens, and were acquired with a MagnaFire camera and software (Optronics). Fluorescent images were composed using Adobe Photoshop software. Ac-LDL uptake was assessed by incubation with DiI-Ac-LDL conjugate as described in “Materials and methods.” Scale bar represents 50 μm. Insets show FACS analysis of respective surface (VE-cadherin) and intracellular (eNOS, VWF) markers in parallel cultures, or instant FACS profiles of cells incubated with DiI-Ac-LDL at 37°C (Ac-LDL uptake) versus 4°C (control Ac-LDL binding). (C) Vascular tubes formation by CD34+CD43– KDR+ cells (scale bar represents 200 μm, left panel; and 50 μm, right panel). Images were captured with an inverted DMIRB micrscope (Leica Microsystems) equipped with a 5×/0.12 (left) or 20×/0.3 (right) objective lens, and were acquired through a MagnaFire camera and software (Optronics). (D) TNF-induced up-regulation of ICAM-1 and induction of VCAM-1 expression in CD34+CD43–KDR+ endothelial cultures. Numbers within plots indicate ΔMFI values for untreated (blue) and TNF-treated (red) cells. VLA-4 staining was used as a control. A representative example of 3 independent experiments is shown.
The sustained proliferation and establishment of long-term lymphomyeloid cultures are distinctive features of definitive hematopoietic progenitors.1 To address the lymphoid and long-term myeloid potential, we cultured isolated CD43+ subsets on MS-5 stromal cells in conditions that support differentiation of hESC-derived CD34+ cells into B and natural killer (NK) cells, granulocytes, and macrophages.18 As shown in Figure 6D, in MS-5 cocultures supplemented with myeloid cytokine combination (SCF, G-CSF, IL-3), CD43+CD45–/+Lin–, but not CD43+CD41a/CD235a+ cells displayed vigorous proliferation associated with retention of myeloid CFCs up to 6 weeks of culture. In MS-5 cocultures with SCF, Flt3-L, and IL-7, commitment to lymphoid cell lineages determined by expression of NK cell (CD3E, CD3Z) and B-cell (MB1, VPREB, PAX5) specific transcripts was also restricted to CD43+CD45–/+Lin– subsets.
Although both CD43+CD45–Lin– and CD43+CD45+Lin– cells contained multilineage progenitors, CD45+ cells displayed several features indicating their advanced lineage-restricted specification, predominantly toward myeloid pathway: (1) CD45+ cells were highly enriched in myeloid CFCs (Figure 6A) and expressed early marker of myeloid commitment, myeloperoxidase (MPO; Figure 6B); (2) CD90, GATA-3 and RUNX1 were down-regulated in CD45+ cells, whereas FLT3 was strongly up-regulated (Figure 6B-C); (3) critical markers of B-lymphoid commitment PAX5 and VPREB, which were reproducibly detected in MS-5 cocultures with CD43+CD45–Lin– cells, were undetectable in parallel cultures with CD45+ cells, whereas NK-specific transcripts (CD3E, CD3Z) were highly expressed in both cultures (Figure 6D).
Thus, we identified 2 major populations of hematopoietic progenitors generated during one-step hESC/OP9 coculture: (1) CD43+CD235a+CD41a+CD45– precommitted erythro-megakaryocytic, and (2) CD43+CD45–/+Lin– multilineage progenitors. Based on CD45 expression, multilineage progenitors could be subdivided on the (1) early CD43+CD45–Lin– progenitors with lymphomyeloid potential, and (2) late CD43+CD45+Lin– progenitors undergoing progressive myeloid commitment.
Discussion
We report a novel observation that CD43 defines early hematopoietic progenitors and discriminates hematopoietic from endothelial cells at hESC differentiation in vitro. CD43 (also known as leukosialin, sialophorin) is one of the most prevalent leukocyte transmembrane sialoglycoproteins37 expressed exclusively on cells of hematopoietic lineage, including hematopoietic stem cells,38 but excluding mature erythrocytes and B-cell subsets.26,28 CD43 has a highly conserved across species cytoplasmic domain and mucinlike extracellular domain, which is extensively O-glycosylated.39,40 The cytoplasmic domain of CD43 interacts with cytoskeletal proteins and transmits signals that regulate a variety of intracellular signal transduction pathways involved in cell activation, proliferation, and survival.41 Notably, cross-linking of CD43 induces apoptosis of bone marrow clonogenic progenitors, but not hematopoietic stem cells.42 The high level of glycosylation and net negative charge explain antiadhesive properties of the CD43 molecule, which have been demonstrated in a number of studies.43,44 CD43 also transmits signals enabling other ligand-receptor interactions to promote cell adhesion,45-47 and may function as a ligand for ICAM-1 and E-selectin molecules expressed on endothelial cells.48-50 It has been proposed that CD43 can act as a gateway to facilitate certain cell contacts.41 Early expression of CD43 on hESC-derived hematopoietic progenitors reported in the present study may also indicate a possible role of CD43 in hematopoietic development, including acquisition of antiadhesive properties by emerging hematopoietic cells.
CD43+ cell subsets: definition, kinetic profile, sorting and morphology. (A) The phenotype of CD43+ cells isolated on day 8 of H1/OP9 coculture. CD41a and CD235a were coexpressed (bottom left dot plot), and both in opposite manner to CD45 (top dot plots). Combination of CD41a-PE, CD235a-PE, and CD45-APC mAbs (bottom right dot plot) defines 3 major CD43+ subsets: (1) CD43+CD41a/CD235+CD45–, (2) CD43+CD41a/CD235a–CD45–, and (3) CD43+CD41a/CD235a– CD45+. Values within plots indicate percentage of cells in respective quadrants. Representative analysis is shown. (B) Kinetic analysis of indicated CD43+ subsets in H1/OP9 coculture represented as percentage of total CD43+ cells (left y1-axis). Dashed trend lines show parallel kinetics of indicated CFC types (right y2-axis). Results are the mean ± SD from 3 independent experiments. (C) Sorting strategy used for isolation of CD43+ subsets. A representative example of 7 independent experiments is shown (H1, n = 5; H9, n = 2). (D) Wright-stained cytospins of FACS-sorted CD43+ subsets (scale bar represents 5 μm). Images were captured with a Microphot-SA microscope (Nikon, Melville, NY) equipped with a 100×/1.40 oil-immersion objective lens, and were acquired through a DFC320 camera and Firecam 1.3 software (Leica Microsystems).
CD43+ cell subsets: definition, kinetic profile, sorting and morphology. (A) The phenotype of CD43+ cells isolated on day 8 of H1/OP9 coculture. CD41a and CD235a were coexpressed (bottom left dot plot), and both in opposite manner to CD45 (top dot plots). Combination of CD41a-PE, CD235a-PE, and CD45-APC mAbs (bottom right dot plot) defines 3 major CD43+ subsets: (1) CD43+CD41a/CD235+CD45–, (2) CD43+CD41a/CD235a–CD45–, and (3) CD43+CD41a/CD235a– CD45+. Values within plots indicate percentage of cells in respective quadrants. Representative analysis is shown. (B) Kinetic analysis of indicated CD43+ subsets in H1/OP9 coculture represented as percentage of total CD43+ cells (left y1-axis). Dashed trend lines show parallel kinetics of indicated CFC types (right y2-axis). Results are the mean ± SD from 3 independent experiments. (C) Sorting strategy used for isolation of CD43+ subsets. A representative example of 7 independent experiments is shown (H1, n = 5; H9, n = 2). (D) Wright-stained cytospins of FACS-sorted CD43+ subsets (scale bar represents 5 μm). Images were captured with a Microphot-SA microscope (Nikon, Melville, NY) equipped with a 100×/1.40 oil-immersion objective lens, and were acquired through a DFC320 camera and Firecam 1.3 software (Leica Microsystems).
In mice, the earliest-appearing multipotent hematopoietic progenitors in yolk sac and AGM or derived from ESCs in vitro are CD45–3,9,10 and can be identified by expression of CD41.9-12,51 Additionally, MAC-1 (CD11b) and α4-integrin (CD49d) were proposed as markers of early hematopoietic precursors.52-54 We demonstrated that the earliest hESC-derived multipotent hematopoietic progenitors expressed CD43 but not CD41a and CD11b. Although human hematopoietic progenitors were CD49d+ (not shown), this molecule was also expressed on hESC-derived endothelial cells (Figure 4D); thus, it was not useful for discrimination between these 2 lineages. We have not found reports related to expression of CD43 on hematopoietic cells during early embryonic or ESC-derived hematopoiesis in mice. However, striking similarities in CD43 expression on human and mouse bone marrow hematopoietic stem cells have been reported,38 and therefore it is reasonable to expect that expression of CD43 during hematopoietic development in mice will follow the pattern that we described here for hESCs.
Emergence of clonogenic hematopoietic progenitors before CD45 expression has also been observed following hESC differentiation on S17 stromal cells16 and embryoid body differentiation.20,21 It is likely that similar to the OP9 system, early CD45– hematopoietic progenitors described in other differentiation systems might be identified by expression of CD43. The question remains whether CD43 expression on hematopoietic progenitors before CD45 seen during hESC differentiation in vitro recapitulate events occurring in vivo. CD43+CD45–Lin– hematopoietic blast cells morphologically similar to hESC-derived CD43+CD45–Lin– hematopoietic progenitors have been described in human embryonic and fetal liver,55 suggesting that these cells are an integral part of hematopoietic development in vivo.
Phenotypic and functional analysis of CD43+ subsets. (A) CFC potential of FACS-sorted CD43+ subsets on day 6 and day 9 of hESC/OP9 cocultures. CFC-E/Mks and CFC-GEMM/GM/Ms were determined by serum-free MegaCult collagen assay and FBS-containing MethoCult GF+ methylcellulose assay, respectively. Results are the mean ± SD from 9 independent experiments (H1, n = 6; H9, n = 3). NA indicates not applicable (subset was not detected/sorted). Photographs show typical E and small Mk colonies detected in CD43+CD41a/CD235a+CD45–/+ subsets (i, scale bar represents 200 μm; inset shows Mk colony stained with anti-CD41a mAb, scale bar represents 50 μm) and multilineage GEMM (ii, scale bar represents 200 μm) and large Mk colonies (iii, scale bar represents 50 μm) detected in CD43+CD41a/CD235a–CD45–/+ subsets. Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 5×/0.12 (i-iii) or a 20×/0.3 (inset) objective lens, and were acquired with a MagnaFire camera and software (Optronics). (B) qRT-PCR analysis of FACS-sorted CD43+ subsets on day 8 of H1/OP9 coculture. The stacked bar graph shows expression levels of indicated transcripts represented by relative units (see “Materials and methods” for details). Results are the means of 2 independent experiments. Representative agarose gel electrophoresis of qPCR products is shown. (C) FACS analysis of CD43+ subsets. CD43+ cells were isolated on day 8 of H1/OP9 coculture by direct CD43 MACS microbeads. Color-matching combinations of CD43, CD41a, CD235a, CD45, and indicated mAbs were used for CD43+ subset gating and analysis. Plots show isotype control (open) and specific mAb (tinted) histograms. Values within plots indicate ΔMFI values. Representative analysis of 3 independent experiments is shown. (D) Lymphoid and myeloid differentiation of FACS-sorted CD43+ subsets in coculture with MS-5 stromal cells. CD43+ subsets were isolated on day 8 of hESC/OP9 cocultures and cultured with MS-5 cells in presence of cytokines supporting either lymphoid or myeloid differentiation (see “Materials and methods” for details). Lymphoid MS-5 cultures were examined for expression of NK cell (CD3E, CD3Z) and B-cell (MB1, VPREB, PAX5) specific transcripts by qRT-PCR on the fourth week of culture. The relative expression of each GAPDH-normalized target gene was calculated in comparison with isolated CD43+ cells before coculture (MB1, very low levels of CD3E/Z, but no detectable VPREB and PAX5 were found in CD43+ cells before coculture). Results are the mean ± SD from 3 independent experiments with H1 (n = 2) and H9 (n = 1) cells. A representative agarose gel of qPCR products is shown. Myeloid MS-5 cocultures were examined for total CD43+ cells and myeloid CFCs (GM/M) during 6 weeks of culture. Results are the means ± SD from 4 independent experiments (H1, n = 2; H9, n = 2).
Phenotypic and functional analysis of CD43+ subsets. (A) CFC potential of FACS-sorted CD43+ subsets on day 6 and day 9 of hESC/OP9 cocultures. CFC-E/Mks and CFC-GEMM/GM/Ms were determined by serum-free MegaCult collagen assay and FBS-containing MethoCult GF+ methylcellulose assay, respectively. Results are the mean ± SD from 9 independent experiments (H1, n = 6; H9, n = 3). NA indicates not applicable (subset was not detected/sorted). Photographs show typical E and small Mk colonies detected in CD43+CD41a/CD235a+CD45–/+ subsets (i, scale bar represents 200 μm; inset shows Mk colony stained with anti-CD41a mAb, scale bar represents 50 μm) and multilineage GEMM (ii, scale bar represents 200 μm) and large Mk colonies (iii, scale bar represents 50 μm) detected in CD43+CD41a/CD235a–CD45–/+ subsets. Images were captured with an inverted DMIRB microscope (Leica Microsystems) equipped with a 5×/0.12 (i-iii) or a 20×/0.3 (inset) objective lens, and were acquired with a MagnaFire camera and software (Optronics). (B) qRT-PCR analysis of FACS-sorted CD43+ subsets on day 8 of H1/OP9 coculture. The stacked bar graph shows expression levels of indicated transcripts represented by relative units (see “Materials and methods” for details). Results are the means of 2 independent experiments. Representative agarose gel electrophoresis of qPCR products is shown. (C) FACS analysis of CD43+ subsets. CD43+ cells were isolated on day 8 of H1/OP9 coculture by direct CD43 MACS microbeads. Color-matching combinations of CD43, CD41a, CD235a, CD45, and indicated mAbs were used for CD43+ subset gating and analysis. Plots show isotype control (open) and specific mAb (tinted) histograms. Values within plots indicate ΔMFI values. Representative analysis of 3 independent experiments is shown. (D) Lymphoid and myeloid differentiation of FACS-sorted CD43+ subsets in coculture with MS-5 stromal cells. CD43+ subsets were isolated on day 8 of hESC/OP9 cocultures and cultured with MS-5 cells in presence of cytokines supporting either lymphoid or myeloid differentiation (see “Materials and methods” for details). Lymphoid MS-5 cultures were examined for expression of NK cell (CD3E, CD3Z) and B-cell (MB1, VPREB, PAX5) specific transcripts by qRT-PCR on the fourth week of culture. The relative expression of each GAPDH-normalized target gene was calculated in comparison with isolated CD43+ cells before coculture (MB1, very low levels of CD3E/Z, but no detectable VPREB and PAX5 were found in CD43+ cells before coculture). Results are the mean ± SD from 3 independent experiments with H1 (n = 2) and H9 (n = 1) cells. A representative agarose gel of qPCR products is shown. Myeloid MS-5 cocultures were examined for total CD43+ cells and myeloid CFCs (GM/M) during 6 weeks of culture. Results are the means ± SD from 4 independent experiments (H1, n = 2; H9, n = 2).
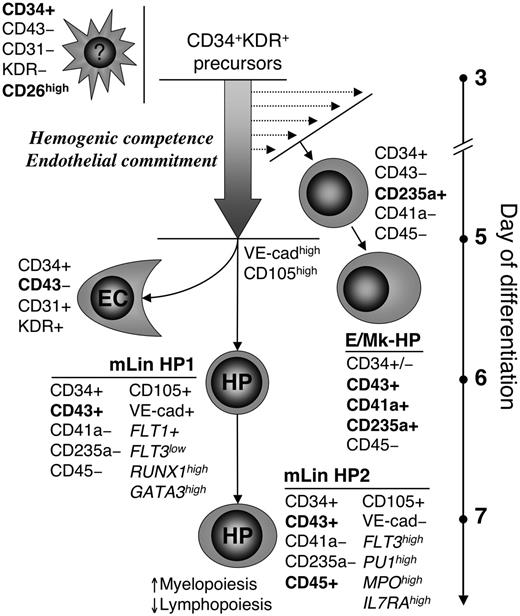
Studies in mouse ESC/OP9 cocultures have demonstrated a step-wise divergence of lineage-restricted primitive hematopoietic progenitors from VEGF-R2+VE-cadherin– mesodermal precursors and multilineage definitive progenitors from later VEGF-R2+VE-cadherin+ endothelial-like precursors.7,31,56,57 Using CD43 as the earliest pan-hematopoietic marker, we found a similar 2-step divergence of hematopoietic elements in hESC/OP9 coculture. In fact, CD43+CD41a/CD235a+ erythro-megakaryocytic progenitors develop first on day 4 to 5 of differentiation followed by CD43+CD45–Lin– multilineage progenitors on day 6 (Figures 1D and 5B). CD43+CD45–Lin– multilineage progenitors retain expression of VE-cadherin, KDR, and CD105 endothelium-associated molecules, highly suggesting that they are derived from endothelial-like precursors. In contrast, CD43+CD41a/CD235a+ cells retain expression of only KDR, pointing to earlier precursors for this lineage. It can be suggested that erythro-megakaryocytic progenitors diverge from the earliest CD34+KDR+ precursors at their pre-endothelial commitment stage, most likely through CD34+CD235a+ intermediates. During subsequent endothelial commitment, CD34+KDR+ precursors up-regulate VE-cadherin and CD105 expression and acquire the capacity to generate multilineage hematopoietic progenitors and endothelial cells (Figure 7). Although our study identifies CD43 as a marker of committed hematopoietic progenitors, phenotypic features that discriminate hemogenic from committed endothelial cells are currently unclear, and whether VE-cadherin, CD105, or other endothelial markers can separate hemogenic endothelium from earlier CD34+KDR+ hemogenic precursors remains to be determined. Prospective identification of these precursors will facilitate studies of primitive versus definitive hematopoiesis in hESC/OP9 coculture. Preliminary analysis of single erythroid colonies generated from H1-derived CD43+CD41a/CD235a+ cells shows that all colonies express embryonic hemoglobin (ζ/ϵ-chains); however, about 40% colonies on day 6 express adult hemoglobin (β-chain), and this proportion increases up to 90% on day 9. Thus, primitive erythroid progenitors (ζ/ϵ+β–) may predominate on early days (4-6) of differentiation, whereas definitive ones (β+)58 progressively contribute to CD43+CD41a/CD235a+ population on later days. Because the first-appearing CD43+ cells already express CD235a and seem to originate from CD34+CD235a+CD43– cells (Figure 1D), this transient population that peaked on day 5 (Figure 1C) may represent immediate precursors of primitive hematopoiesis in hESC/OP9 coculture.
In the present study, we defined CD43+CD45– Lin– cells as the earliest multilineage definitive progenitors developed from hESCs in vitro. These cells have lymphomyeloid potential and molecular phenotype (SCL+GATA2+GATA3highRUNX1highC-MYB+HOXB4+) consistent with emerging hematopoietic precursors identified in human and mouse AGM region.10,59 Acquisition of CD45 expression by these cells was accompanied with dramatic increase of myeloid clonogenic progenitors, loss of B-lymphoid potential, down-regulation of GATA3 and RUNX1, and up-regulation of MPO, PU1, and FLT3, that altogether signify a progressive myeloid commitment in CD45+ cells (findings summarized in Figure 7). It is possible that myeloid propensity of CD45+ cells was only influenced by differentiation conditions in OP9 culture. However, recent observations in mouse embryo at day 10.5 postcoitus demonstrated that multipotent precursors from the AGM region that generate lymphoid cells and possess long-term repopulating potential differ from their fetal liver and adult counterpart by lack or low level of CD45 expression, whereas CD45+ cells represent already committed myeloid cells.10 To our knowledge, presence of CD45– hematopoietic precursors in human yolk sac or AGM has not been described yet. Because ESC differentiation in vitro recapitulates many aspects of embryonic development,9,60,61 we presume that the sequence of hESC hematopoietic development described here reflects, at least in some degree, events occurring in vivo in human yolk sac and AGM region.
A model of hematoendothelial differentiation in hESC/OP9 coculture. Hematopoietic and endothelial cells develop from early precursors identified by a CD34+KDR+(CD43–) phenotype. These precursors appear at day 3 of differentiation and retain hematoendothelial potential up to day 5, but after 6 days, CD34+CD43– KDR+ cells constitute a population of committed endothelial cells (ECs). CD43 is identified as a specific marker of early hematopoietic progenitors. Two types of CD43+ hematopoietic progenitors are identified in hESC/OP9 coculture: (1) CD43+CD41a/CD235a+ erythro-megakaryocytic progenitors (E/Mk-HP) first detectable on day 4 of differentiation, and (2) CD43+CD41a/CD235a– multilineage (mLin) progenitors (HP1) appeared 2 days later. Emergence of E/Mk-HP before mLin HP1 and residual expression of VE-cadherin, FLT1, and CD105 endothelial markers by HP1 cells may reflect a step-wise endothelial commitment of CD34+KDR+ hematoendothelial precursors (block arrow): CD34+KDR+ precursors at initial pre-endothelial commitment stage are only competent to generate E/Mk-HP through CD235+ intermediates, whereas multipotent HP1 are derived from CD34+KDR+ precursors with an endothelial phenotype (VE-cadherinhighCD105high). HP1 have lymphomyeloid potential and a gene expression profile found in the most immature hematopoietic progenitors. HP1 transition to CD45+ stage (HP2) is associated with progressive myeloid commitment and a decrease of lymphoid potential. CD34+CD43–KDR–CD26high cells arise along with the first CD34+KDR+ cells and may comprise more than 20% of total CD34+ cells in hESC/OP9 cocultures. These cells are devoid of detectable hematoendothelial potential.
A model of hematoendothelial differentiation in hESC/OP9 coculture. Hematopoietic and endothelial cells develop from early precursors identified by a CD34+KDR+(CD43–) phenotype. These precursors appear at day 3 of differentiation and retain hematoendothelial potential up to day 5, but after 6 days, CD34+CD43– KDR+ cells constitute a population of committed endothelial cells (ECs). CD43 is identified as a specific marker of early hematopoietic progenitors. Two types of CD43+ hematopoietic progenitors are identified in hESC/OP9 coculture: (1) CD43+CD41a/CD235a+ erythro-megakaryocytic progenitors (E/Mk-HP) first detectable on day 4 of differentiation, and (2) CD43+CD41a/CD235a– multilineage (mLin) progenitors (HP1) appeared 2 days later. Emergence of E/Mk-HP before mLin HP1 and residual expression of VE-cadherin, FLT1, and CD105 endothelial markers by HP1 cells may reflect a step-wise endothelial commitment of CD34+KDR+ hematoendothelial precursors (block arrow): CD34+KDR+ precursors at initial pre-endothelial commitment stage are only competent to generate E/Mk-HP through CD235+ intermediates, whereas multipotent HP1 are derived from CD34+KDR+ precursors with an endothelial phenotype (VE-cadherinhighCD105high). HP1 have lymphomyeloid potential and a gene expression profile found in the most immature hematopoietic progenitors. HP1 transition to CD45+ stage (HP2) is associated with progressive myeloid commitment and a decrease of lymphoid potential. CD34+CD43–KDR–CD26high cells arise along with the first CD34+KDR+ cells and may comprise more than 20% of total CD34+ cells in hESC/OP9 cocultures. These cells are devoid of detectable hematoendothelial potential.
In summary, we identified CD43 as the earliest pan-hematopoietic marker during hESC differentiation in vitro and described functionally distinct populations of hematopoietic progenitors generated from hESCs. These findings provide a means for direct detection and prospective analysis of hESC-derived hematopoietic progenitors and are important for further studies of hematoendothelial divergence in hESCs.
Prepublished online as Blood First Edition Paper, June 6, 2006; DOI 10.1182/blood-2006-02-003327.
Supported by Defense Advanced Research Projects Agency (DARPA) grant 7003-UWM and National Institutes of Health (NIH) grant P51 RR000167 to the National Primate Research Center, University of Wisconsin–Madison. I.I.S. is supported by NIH grant HD44067.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Toru Nakano for providing OP9 cells, Joel Puchalski and Kathy Schell for FACS sorting, Maryna Gumenyuk for cytologic analysis, and Deborah Faupel and Amanda Paus for editorial assistance. All DARPA-funded research used NIH-approved stem cell lines.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal