Abstract
Hereditary combined vitamin K–dependent (VKD) coagulation factor deficiency is an autosomal recessive bleeding disorder associated with defects in either the γ-carboxylase, which carboxylates VKD proteins to render them active, or the vitamin K epoxide reductase (VKORC1), which supplies the reduced vitamin K cofactor required for carboxylation. Such deficiencies are rare, and we report the fourth case resulting from mutations in the carboxylase gene, identified in a Tunisian girl who exhibited impaired function in hemostatic VKD factors that was not restored by vitamin K administration. Sequence analysis of the proposita did not identify any mutations in the VKORC1 gene but, remarkably, revealed 3 heterozygous mutations in the carboxylase gene that caused the substitutions Asp31Asn, Trp157Arg, and Thr591Lys. None of these mutations have previously been reported. Family analysis showed that Asp31Asn and Thr591Lys were coallelic and maternally transmitted while Trp157Arg was transmitted by the father, and a genomic screen of 100 healthy individuals ruled out frequent polymorphisms. Mutational analysis indicated wild-type activity for the Asp31Asn carboxylase. In contrast, the respective Trp157Arg and Thr591Lys activities were 8% and 0% that of wild-type carboxylase, and their compound heterozygosity can therefore account for functional VKD factor deficiency. The implications for carboxylase mechanism are discussed.
Introduction
Hereditary combined vitamin K–dependent (VKD) factor deficiency is a bleeding disorder characterized by the reduced activities of the procoagulant factors II, VII, IX, and X and anticoagulant proteins C, S, and Z.1-8 The inheritance of the disease is autosomal recessive and is due to mutations in the genes for either the γ-carboxylase9-12 or the vitamin K epoxide reductase (VKORC1).13 The carboxylase converts clusters of Glus to γ-carboxylated Glus (Glas) in the Gla domains of VKD proteins, which renders them active by generating a calcium-binding module that binds either to anionic phospholipids that become exposed on cell surfaces or to hydroxyapatite in the extracellular matrix.14,15 The carboxylase uses reduced vitamin K (KH2) as a cofactor to drive Glu carboxylation, and the KH2 becomes oxygenated to a vitamin K epoxide (KO) product that must be recycled for continuous carboxylation. Recycling is accomplished by VKORC1, which is the target of anticoagulant therapy with coumarin derivatives like warfarin that block KH2 regeneration and consequently inhibit VKD protein carboxylation. Both VKORC1 and the carboxylase are integral membrane enzymes that reside in the endoplasmic reticulum (ER), where the VKD hemostatic factors are modified during their secretion from the cell. The concerted action of these 2 enzymes can therefore explain why congenital defects in either the carboxylase or VKORC1 lead to combined functional deficiency of the VKD factors.
The interactions between VKD proteins and the carboxylase are complex and not well understood.16,17 All VKD proteins have a domain, usually a propeptide, that confers high-affinity binding to the carboxylase and consequent conversion of the multiple Glus to Glas by a processive mechanism.18,19 Allosteric changes caused by VKD substrate binding regulate carboxylation: Binding of the propeptide and Glu residues to the carboxylase is reciprocally modulated,20,21 and binding of both domains increases carboxylase affinity for KH2.22 Consequently, VKD factor association with carboxylase results in more efficient vitamin K utilization, which is significant because the availability of KH2 appears to regulate carboxylation.23-26 In the absence of VKD substrate binding, KH2 epoxidation to KO does not occur,27 and this regulation prevents the unfettered formation of highly reactive and undesirable vitamin K intermediates. At present, the residues that make up the carboxylase active site to facilitate the reaction are largely unknown, due in part to the lack of a crystal structure or homology with other proteins that might indicate functional residues. Structure-function relationships for VKORC1 are even less well defined, because the gene for this enzyme has only recently been identified.13,28
Mutations associated with hereditary combined VKD coagulation factor deficiency are rare. The carboxylase is encoded by a single gene,29 and only 3 naturally occurring missense mutations have previously been identified: Leu394Arg,10 which is impaired in both Glu binding and propeptide binding,30 Trp501Ser,9,11 which shows decreased propeptide binding,31 and Arg485Pro,12 whose functional analysis has not yet been reported. In all 3 cases, vitamin K supplementation resulted in at least partial restoration of VKD factor function, consistent with the effect of VKD protein binding on carboxylase affinity for vitamin K. Two carboxylase gene mutations in noncoding regions are also known that include a splice site mutation in intron 212 and a short deletion affecting a putative cis-acting element in the promoter of the gene.32 In the case of VKORC1, only the missense mutation Arg98Trp has been identified,13 which appears to cause a large decrease in VKORC1 activity.
We report a case of combined deficiency in VKD hemostatic factors that is associated with compound heterozygosity in the carboxylase gene, which was identified in a Tunisian family. No mutations were found in the VKORC1 gene. Sequence analysis revealed 3 new heterozygous missense mutations, Asp31Asn, Trp157Arg, and Thr591Lys, and functional analysis indicated that the VKD factor deficiency phenotype is due to compound heterozygosity in Trp157Arg and Thr591Lys. The relevance of these residues to carboxylase function is discussed.
Patient, materials, and methods
Coagulation assay
Blood samples collected by venipuncture into 0.4% sodium citrate, at a ratio of 9:1, were centrifuged at 3000g for 20 minutes at 4°C before freezing at –20°C. The activities of factors II, V, VII, IX, and X were measured in one-stage clotting assays using Diagnostica Stago (Parsippany, NJ) reagents. Antigen levels of factors II and IX were determined by an enzyme-linked immunosorbent assay (ELISA). Protein C and S activities were assayed by the chronometric method from Diagnostica Stago. The prothrombin time (PT) and kaolin-activated partial thromboplastin time (APTT) were performed using the Neoplastin CI5 or C.K. Prest kits (both from Diagnostica Stago), respectively, on an STA instrument.
Case report
The proposita, the second female offspring of asymptomatic parents of Tunisian origin, was admitted at 2 years of age with acute infection, ecchymosis, severe epistaxis, and gingivorrhagia that required blood transfusion. The patient presented with developmental retardation and stunted growth (–3 or –2 standard deviations for weight and size, respectively) and exhibited facial dysmorphia that was likely due to skeletal anomalies. Malabsorption was ruled out by a normal duodenal biopsy. Both the PT and APTT were prolonged, and coagulation testing revealed defective activities of the VKD coagulation factors but normal activity of factor V (Table 1). Intramuscular injections of high doses of vitamin K (ie, 10 mg/d phylloquinone for 2 weeks) did not modify either the PT or APTT of the proposita (Table 1). The coagulation values of her parents and a sister were within the normal range; her brother appeared affected, consistent with a severe bleeding episode from his lip that had required infusion of PPSB (a plasma fraction enriched in the VKD factors II, VII, IX, and X) when he was 1 year old. The brother was subsequently diagnosed for combined deficiency in VKD hemostatic factors (Table 1). At age 11, the proposita developed heart failure due to incomplete closure of the ventricular septum, and incomplete septal closure was also observed in the affected brother (at 1½ years) during a medical examination. An older sibling died at 4 days old without any diagnosis.
Coagulation factor values in the proposita and family members
Factors . | Patient . | Brother . | Mother . | Father . | Sister . |
|---|---|---|---|---|---|
| II:C, % | 9 | 14 | 119 | 97 | 86 |
| V:C, % | 87 | 47 | 74 | 85 | 97 |
| VII:C, % | 6 | 7 | 100 | 68 | 84 |
| IX:C, % | 7 | — | 92 | 85 | — |
| X:C, % | 5 | 7 | 82 | 109 | 116 |
| II Ag, % | 60 | — | 110 | 80 | 80 |
| IX Ag, % | 73 | 70 | — | — | — |
| PC, % | < 1 | < 1 | 89 | 112 | 88 |
| PS, % | < 1 | < 1 | 44 | 40 | 87 |
| PT, s | 49.5/11.5 | > 60/11.5 | 12.7/11.5 | 13.7/11.5 | 13.7/11.5 |
| PT, % | 10 | < 10 | 80 | 70 | 70 |
| PT, s* | 48/11.5 | — | — | — | — |
| PT, %* | 9 | — | — | — | — |
| APTT, s | 60/30 | 56/30 | 36/30 | 38/30 | 33/30 |
| APTT, s* | 60/30 | — | — | — | — |
| AT, % | 102 | 102 | 86 | 84 | 103 |
Factors . | Patient . | Brother . | Mother . | Father . | Sister . |
|---|---|---|---|---|---|
| II:C, % | 9 | 14 | 119 | 97 | 86 |
| V:C, % | 87 | 47 | 74 | 85 | 97 |
| VII:C, % | 6 | 7 | 100 | 68 | 84 |
| IX:C, % | 7 | — | 92 | 85 | — |
| X:C, % | 5 | 7 | 82 | 109 | 116 |
| II Ag, % | 60 | — | 110 | 80 | 80 |
| IX Ag, % | 73 | 70 | — | — | — |
| PC, % | < 1 | < 1 | 89 | 112 | 88 |
| PS, % | < 1 | < 1 | 44 | 40 | 87 |
| PT, s | 49.5/11.5 | > 60/11.5 | 12.7/11.5 | 13.7/11.5 | 13.7/11.5 |
| PT, % | 10 | < 10 | 80 | 70 | 70 |
| PT, s* | 48/11.5 | — | — | — | — |
| PT, %* | 9 | — | — | — | — |
| APTT, s | 60/30 | 56/30 | 36/30 | 38/30 | 33/30 |
| APTT, s* | 60/30 | — | — | — | — |
| AT, % | 102 | 102 | 86 | 84 | 103 |
Coagulation (C) and antigen (Ag) levels are indicated as the percent of normal values.
PC indicates protein C; PS, protein S; AT, antithrombin activity; and —, not done.
The PT and APTT times were also measured after intramuscular phylloquinone administration (2 weeks at 10 mg/d).
Sequence analysis of the carboxylase and VKORC1 genes
The genomic DNA of the proposita and 4 members of her family was isolated from frozen peripheral blood, as described.33 Blood collection was achieved with informed written consent from all donors in accordance with the Helsinki protocol. The study protocol was approved by the Ethics Committee of Hôpital Habib Thamenr, Tunis, Tunisia. The 13-kb gene encoding the carboxylase and 5-kb gene encoding VKORC1 were entirely sequenced using the polymerase chain reaction (PCR) and subsequent direct sequencing of the PCR products. The primers and PCR conditions for the carboxylase and VKORC1 genes are summarized, respectively, in Tables 2 and S1 (Table S1 is available on the Blood website; see the Supplemental Table link at the top of the online article).
Primers and PCR conditions used to sequence the carboxylase gene
Gene region . | PCR conditions . | Primer names . | Primer sequences . |
|---|---|---|---|
| Exon 1 | 94°C/1 min, TD 70/65°C/30 s, 2°C↘, 72°C/1 min (2% DMSO) | E-1-U | GCGTCCTCAACTCGGCGTCACTC |
| E-1-L | CTCCACCTCAAATCAAAGAAATC | ||
| Exon 2 | 94°C/1 min, TD 61/55°C/30 s, 2°C↘, 72°C/1 min | E-2-U | GAGCTGTTGGTGCAGTGATTTCT |
| E-2-L | AGAGATTGTCATTCTCCACTCT | ||
| Exon3/4/5 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-3-U | CCAATGACCAACTCCCCTAT |
| E-5-L | TCCTCCCTCTGTCCTAAAAT | ||
| Exon 6 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-6-U | TGTAACTCAGGAGCATGGATTC |
| E-6-L | CATTACTGAGAGAGATGAGTCACCT | ||
| Exon 7 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-7-U | GCTGTGAATGTGCTTTGATGTG |
| E-7-L | AAGCCCCAGTCCTCTTATC | ||
| Exon 8 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-8-U | AGGCCCAGCCAAACTCCT |
| E-8-L | CTCACACTGACCCCATCC | ||
| Exon 9/10 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-9-U | GCTGATTCCCCTCTGTGCTG |
| E-10-L | AACCAGCTATGCCCACAAC | ||
| Exon 11 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-11-U | GGTGGCTGTGATGTCCTTAGAA |
| E-11-L | CCCCATGGCAGAGTGAAC | ||
| Exon 12 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-12-U | GCCATGGGGTGGGATGATGAAC |
| E-12-L | CAGGCAACTGACAAGGGA | ||
| Exon 13 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-13-U | AGAAAGAAGCCAAGAGTCAT |
| E-13-L | GGCTAGAACATCATTCATAACC | ||
| Exon 14 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-14-U | CTAGCTGGCAGAAGAGGAGTT |
| E-14-L | AGAATGGCAGGAAAAGATACC | ||
| Exon 15 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-15-U | GGCTGTTCCTACCCTATCC |
| E-15-L | ACCTCCCCTTCTGCTCACC | ||
| P1 | 94°C/30 s, TD 65/55°C/30 s, 1°C↘, 72°C/30 s (1.5 mM MgCl2) | 1U | GGGCTCAAGCGAACCTC |
| 379L | CTCCCCGACCCCATTAGT | ||
| P2 | 94°C/15 s, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | 284U | GCGTCCTCAACTCGGCGTCACTC |
| 1124L | GGGCTCCACCTCAAATCAAAGAAATC | ||
| In1 | 94°C/15 s, 55°C/30 s, 72°C/1 min | 969U | ACTGTAGTCTGAGGGGTTCTGG |
| 2261L | TAGTCACATTTTGGGCTGGTTA | ||
| 6 kb | 94°C/15 s, TD 70/62°C/30 s, 68°C/4 min* (3 mM MgCl2) | 724U | GCAGAGCAATGGCGGTGTC |
| 6065L | AGGGAGGCAGCGGAGAGTGGT | ||
| In5-6 | 94°C/30 s, 55°C/30 s, 72°C/30 s (1.5 mM MgCl2) | 5812U | AGATGTGCCCAGGATAGAT |
| 6511L | GGCAAGAATAAAATAAGGAG | ||
| 4 kb | 94°C/15 s, TD 66/58°C/30 s, 2°C↘, 68°C/3 min* (3 mM MgCl2) | 6261U | TTCTTCCTTGGTGCCTGATACTGTC |
| 10364L | AGCCTCTCCTCACTTTCCTCCATAC | ||
| 3.5 kb | 94°C/15 s, 60°C/30 s, 68°C/2.5 min (1 mM MgCl2) | 10470U | GGGATGATGGTGGTAAAGGT |
| 13595L | GGAGGAGTGGGGGAGAGTAT | ||
| 3′ NC | 94°C/30 s, 55°C/30 s, 72°C/30 s (1.5 mM MgCl2) | 13321U | GAAGGGGAGGTAAAGTAAGAAT |
| 13676L | AATCCTGGAGTAGACACAATCA |
Gene region . | PCR conditions . | Primer names . | Primer sequences . |
|---|---|---|---|
| Exon 1 | 94°C/1 min, TD 70/65°C/30 s, 2°C↘, 72°C/1 min (2% DMSO) | E-1-U | GCGTCCTCAACTCGGCGTCACTC |
| E-1-L | CTCCACCTCAAATCAAAGAAATC | ||
| Exon 2 | 94°C/1 min, TD 61/55°C/30 s, 2°C↘, 72°C/1 min | E-2-U | GAGCTGTTGGTGCAGTGATTTCT |
| E-2-L | AGAGATTGTCATTCTCCACTCT | ||
| Exon3/4/5 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-3-U | CCAATGACCAACTCCCCTAT |
| E-5-L | TCCTCCCTCTGTCCTAAAAT | ||
| Exon 6 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-6-U | TGTAACTCAGGAGCATGGATTC |
| E-6-L | CATTACTGAGAGAGATGAGTCACCT | ||
| Exon 7 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-7-U | GCTGTGAATGTGCTTTGATGTG |
| E-7-L | AAGCCCCAGTCCTCTTATC | ||
| Exon 8 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-8-U | AGGCCCAGCCAAACTCCT |
| E-8-L | CTCACACTGACCCCATCC | ||
| Exon 9/10 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-9-U | GCTGATTCCCCTCTGTGCTG |
| E-10-L | AACCAGCTATGCCCACAAC | ||
| Exon 11 | 94°C/1 min, 55°C/30 s, 72°C/1 min | E-11-U | GGTGGCTGTGATGTCCTTAGAA |
| E-11-L | CCCCATGGCAGAGTGAAC | ||
| Exon 12 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-12-U | GCCATGGGGTGGGATGATGAAC |
| E-12-L | CAGGCAACTGACAAGGGA | ||
| Exon 13 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-13-U | AGAAAGAAGCCAAGAGTCAT |
| E-13-L | GGCTAGAACATCATTCATAACC | ||
| Exon 14 | 94°C/1 min, TD 52/48°C/30 s, 2°C↘, 72°C/1 min | E-14-U | CTAGCTGGCAGAAGAGGAGTT |
| E-14-L | AGAATGGCAGGAAAAGATACC | ||
| Exon 15 | 94°C/1 min, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | E-15-U | GGCTGTTCCTACCCTATCC |
| E-15-L | ACCTCCCCTTCTGCTCACC | ||
| P1 | 94°C/30 s, TD 65/55°C/30 s, 1°C↘, 72°C/30 s (1.5 mM MgCl2) | 1U | GGGCTCAAGCGAACCTC |
| 379L | CTCCCCGACCCCATTAGT | ||
| P2 | 94°C/15 s, TD 65/55°C/30 s, 2°C↘, 72°C/1 min | 284U | GCGTCCTCAACTCGGCGTCACTC |
| 1124L | GGGCTCCACCTCAAATCAAAGAAATC | ||
| In1 | 94°C/15 s, 55°C/30 s, 72°C/1 min | 969U | ACTGTAGTCTGAGGGGTTCTGG |
| 2261L | TAGTCACATTTTGGGCTGGTTA | ||
| 6 kb | 94°C/15 s, TD 70/62°C/30 s, 68°C/4 min* (3 mM MgCl2) | 724U | GCAGAGCAATGGCGGTGTC |
| 6065L | AGGGAGGCAGCGGAGAGTGGT | ||
| In5-6 | 94°C/30 s, 55°C/30 s, 72°C/30 s (1.5 mM MgCl2) | 5812U | AGATGTGCCCAGGATAGAT |
| 6511L | GGCAAGAATAAAATAAGGAG | ||
| 4 kb | 94°C/15 s, TD 66/58°C/30 s, 2°C↘, 68°C/3 min* (3 mM MgCl2) | 6261U | TTCTTCCTTGGTGCCTGATACTGTC |
| 10364L | AGCCTCTCCTCACTTTCCTCCATAC | ||
| 3.5 kb | 94°C/15 s, 60°C/30 s, 68°C/2.5 min (1 mM MgCl2) | 10470U | GGGATGATGGTGGTAAAGGT |
| 13595L | GGAGGAGTGGGGGAGAGTAT | ||
| 3′ NC | 94°C/30 s, 55°C/30 s, 72°C/30 s (1.5 mM MgCl2) | 13321U | GAAGGGGAGGTAAAGTAAGAAT |
| 13676L | AATCCTGGAGTAGACACAATCA |
The gene regions correspond to the PCR fragments shown in Figure 1A. The positions within the gene are indicated by the oligonucleotide primer number (U, upper; L, lower). PCR was performed for 30 cycles using the “touchdown” (TD) method: After a 94°C denaturation step, a hybridization step was performed (30 seconds, initial and final temperatures are indicated) with 2°C temperature decrements (↘) every 3 cycles. The last 15 cycles were performed at the final temperatures indicated. The products were then extended at 72°C for 1 minute, except for the 3.5 kb, 4 kb, and 6 kb fragments, which were extended at 68°C for 2.5, 3, and 5 minutes, respectively.
NC indicates noncoding sequence.
The extensions during the final 15 cycles of TD for the 4 kb and 6 kb fragments were 68°C for 5 and 4 minutes, respectively.
mRNA analysis
Reverse transcriptase–PCR was used to analyze the size and amount of carboxylase mRNA in the proposita. Total RNA was isolated from platelets and used as the template (20 ng) for first strand–specific cDNA synthesis in a 2.5-μL reaction mixture that contained PCR buffer (10 mM Tris [pH 8.3], 50 mM KCl), 5 mM MgCl2, 2.5 μM oligo(dT)16, 1 mM of each dNTP, 10 units of RNAse inhibitor, and 25 units of MuLV reverse transcriptase (Applied Biosystems, Weiterstadt, Germany). The reaction mixture was successively incubated for 10 minutes at room temperature, 30 minutes at 42°C, 10 minutes at 95°C, and 10 minutes at 4°C. The reaction product then served as the template for PCR in a 12.5 μL reaction mixture containing PCR buffer, 2 mM MgCl2, 0.15 μM of the forward and reverse primers (Proligo, St Louis, MO), and 1.25 units of AmpliTaq gold DNA polymerase (Applied Biosystems). PCR was conducted for 41 cycles (1 minute at 94°C, 1 minute at 66°C, and 1 minute at 72°C) followed by an extension step for 10 minutes at 72°C. Amplification products were then electrophoresed on 1.5% (wt/vol) agarose gels stained by ethidium bromide and were also directly sequenced.
Screen for polymorphisms
The genomic DNA of 100 unrelated controls (50 each from the Tunisian or French population) was isolated from frozen blood (Nucleospin Blood Quick Pure; Macherey-Nagel, Easton, PA) and screened for 3 carboxylase mutations (Asp31Asn, Trp157Arg, and Thr591Lys) identified in the proposita. Differential hybridization PCR was used to search for the 1235G>A mutation in exon 2 and the 11134C>A mutation in exon 13 that corresponded to the amino acid mutations Asp31Asn and Thr591Lys, respectively. Primer 5′ ACTCTCAACCAAATTGCTCCCA was used in combination with primers 5′ TCAGGGCCCAGGCAGG or 5′ TCAGGGCCCAGGCAGA, which are specific for 1235G or A in exon 2. Amplification generates a 179 bp fragment with the following conditions: 95°C for 1 minute, 62°C to 52°C for 30 seconds, and 72°C for 1 minute. The second set of PCR reactions used primer 5′ GGGGATGATGGTGGTAAAGGTG with either 5′ AAGAAGGGCTAGGTGATGTCGT or 5′ AAGAAGGGCTAGGTGATGTCTT, which are specific for 11134C or A in exon 13, to amplify a 686 bp fragment using the following conditions: 95°C for 1 minute, 72°C to 66°C for 30 seconds, and 72°C for 1 minute. The resulting fragments were analyzed by 1.5% (wt/vol) agarose gel electrophoresis. The third mutation, 1811T>C in exon 4, which corresponds to the amino acid mutation Trp157Arg, was screened in healthy individuals by PCR followed by sequencing. The primers 5′ TGTGCCGCTTCCCCTTGCTG and 5′ GCCATTTTGCTTTGTGTCAT were used to generate a 753 bp PCR fragment (94°C for 1 minute, 65°C to 55°C for 30 seconds, and 72°C for 1 minute), which was then directly sequenced.
Construction and analysis of mutant carboxylase enzymes
Generation of carboxylase cDNAs with the individual mutations Asp31Asn, Trp157Arg, or Thr591Lys was accomplished using the Quick Change XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions and using the following oligonucleotide primers: Asp31Asn: (upper) GATCTCAGGGCCCAGGCAGAACAGCCGAATAGGGAAACTCTTG and (lower) CAAGAGTTTCCCTATTCGGCTGTTCTGCCTGGGCCCTGAGATC; Trp157Arg: (upper) CTCCTGGACAAGACATCACGGAACAACCACTCCTAT and (lower) ATAGGAGTGGTTGTTCCGTGATGTCTTGTCCAGGAG; Thr591Lys: (upper) GTACCATAAGGTGTATAAGACATCACCTAGCCCTTCTTGC and (lower) GCAAGAAGGGCTAGGTGATGTCTTATACACCTTATGGTAC.
Briefly, a full-length carboxylase cDNA34 was hybridized to the indicated primers and subjected to Pfu polymerase (Stratagene) extension. After 20 cycles of amplification, parental wild-type strands were digested with the methyl-sensitive restriction enzyme DpnI, and amplified mutant strands were self-ligated and used to transform DH5α Escherichia coli. A BamHI fragment containing the full-length cDNA was then isolated and subcloned into the baculovirus expression vector pBacPak9 (Invitrogen), and the carboxylase cDNA was entirely sequenced on both strands. The plasmid was cotransfected into SF21 insect cells with Bsu36I-digested viral DNA (BacPAK6; Clontech, Palo Alto, CA), and virus was isolated from individual plaques and amplified. Virally infected SF21 cells were screened for carboxylase by preparing lysates, as before,35 followed by Western analysis using an antibody against the C terminus of the carboxylase.34 Two independent viral isolates of each mutant were subsequently tested for carboxylase activity by measuring [14C]-CO2 incorporation into the peptide Boc-Glu-Glu-Leu-Ome 35 and for epoxidase activity by measuring conversion of KH2 to KO.36
Results
Combined deficiency in VKD coagulation factors in the proposita is associated with compound heterozygosity of mutations Asp31Asn, Trp157Arg, and Thr591Lys in the carboxylase gene
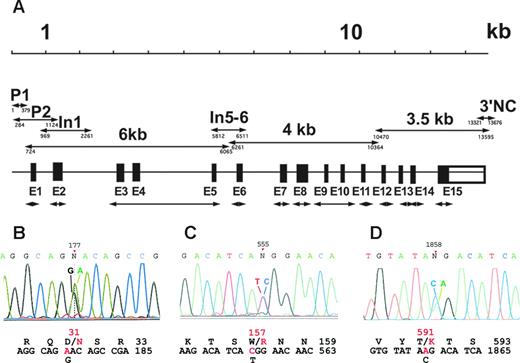
Combined deficiency in VKD factors in a 2-year-old girl was diagnosed based on frequent bleeding, prolonged PT and APTT, and severe defects in the activities of both procoagulant and anticoagulant VKD factors (Table 1). Factors II, VII, IX, and X and proteins C and S showed activities ranging from less than 1% to 9%, while the activity of factor V, which is not a VKD protein, was normal. The combined deficiency prompted us to analyze the genes for both VKORC1 and the carboxylase. Sequence analysis of the entire 5 kb VKORC1 gene did not detect any mutations in the proposita (Table S1). In contrast, the 13 kb carboxylase gene sequence (Figure 1A) revealed 3 heterozygous base substitutions: a 1235G>A transition (corresponding to 177G>A in the mRNA) in exon 2, a 1811T>C transition (555T>C in the mRNA) in exon 4, and a 11134C>A transversion (1858C>A in the mRNA) in exon 13 (Figure 1B-D). These same heterozygous substitutions were found in carboxylase mRNA isolated from platelets and analyzed by reverse transcriptase–PCR, indicating comparative expression levels of both alleles. The substitutions predicted the 3 amino acid mutations Asp31Asn, Trp157Arg, and Thr591Lys (Figure 1B-D), none of which have been reported to date.
PCR strategy and identification of carboxylase coding substitutions. (A) The entire carboxylase gene was sequenced using PCR fragments generated with the primers that are indicated in Table 2. The double-headed arrows indicate exonic (E1-E15) and other fragments (whose nucleotide positions at the 5′ and 3′ ends are shown below the lines) whose names are the same as those used in Table 2. (B-D) Electropherograms corresponding to the 3 new mutations identified in this work are shown: (B) Asp31Asn in exon 2, (C) Trp157Arg in exon 4, and (D) Thr591Lys in exon 13. The positions of the mutations are indicated on top of the electropherograms using the mRNA numbering for the carboxylase (GenBank accession M81592). Wild-type and mutant nucleotides are indicated on top of the corresponding peaks, and wild-type (black) and mutant (red) nucleotide and amino acid sequences are shown below the electropherogram.
PCR strategy and identification of carboxylase coding substitutions. (A) The entire carboxylase gene was sequenced using PCR fragments generated with the primers that are indicated in Table 2. The double-headed arrows indicate exonic (E1-E15) and other fragments (whose nucleotide positions at the 5′ and 3′ ends are shown below the lines) whose names are the same as those used in Table 2. (B-D) Electropherograms corresponding to the 3 new mutations identified in this work are shown: (B) Asp31Asn in exon 2, (C) Trp157Arg in exon 4, and (D) Thr591Lys in exon 13. The positions of the mutations are indicated on top of the electropherograms using the mRNA numbering for the carboxylase (GenBank accession M81592). Wild-type and mutant nucleotides are indicated on top of the corresponding peaks, and wild-type (black) and mutant (red) nucleotide and amino acid sequences are shown below the electropherogram.
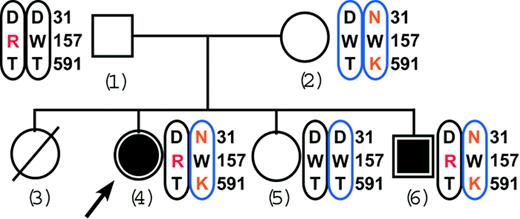
The Tunisian family pedigree shows cosegregation of the 3 carboxylase mutations with VKD coagulation factor deficiency. Affected and unaffected subjects are indicated by solid or open symbols, respectively. The arrow points to the proposita. The female indicated by the hatched circle was dead in early infancy for unknown reasons. Mutated residues are in red, and the chromosomal colors indicate maternal (blue) or paternal (black) inheritance.
The Tunisian family pedigree shows cosegregation of the 3 carboxylase mutations with VKD coagulation factor deficiency. Affected and unaffected subjects are indicated by solid or open symbols, respectively. The arrow points to the proposita. The female indicated by the hatched circle was dead in early infancy for unknown reasons. Mutated residues are in red, and the chromosomal colors indicate maternal (blue) or paternal (black) inheritance.
These 3 mutations were not found in the genomes of 100 healthy French or Tunisian individuals, thus ruling out frequent polymorphisms. Two known polymorphisms in the proposita's carboxylase gene were identified that have previously been reported37 : 9167T>C in exon 9, which is a silent polymorphism, and 8762G>A in exon 8, which is present in roughly half of the population and results in a Gln325Arg substitution that does not affect activity (K.W.H., R.L.S., and K.L.B., unpublished results, June 2005). In addition, a novel polymorphism at position 5338 in intron 4-5 was identified that corresponds to a homozygous G>A transition. To determine whether this substitution affected either the size or amount of carboxylase mRNA, reverse transcriptase–PCR products of platelet RNA from the proposita were compared with that of healthy subjects. This analysis did not reveal any differences, indicating that this intron mutation did not contribute to the defect in VKD factor function. The combined results, then, strongly indicate that the VKD factor deficiency in the proposita is highly likely to be associated with coding mutations in the carboxylase gene.
The combined deficiency in the function of VKD factors cosegregates with mutations Asp31Asn, Trp157Arg, and Thr591Lys
The analysis of family members (Figure 2) showed that the mother, who was asymptomatic, was a compound heterozygote for the mutations Asp31Asn and Thr591Lys like her affected daughter, the proposita. However, the mother was homozygous for the wild-type Trp157 residue, suggesting that Asp31Asn and Thr591Lys were carried by the same allele. This interpretation was confirmed by the analysis of the father, who was unaffected as well and homozygous for the wild-type residues Asp31 and Thr591 but heterozygous for the mutant residue Trp157Arg. We thus conclude that the proposita is a compound heterozygote, with Asp31Asn and Thr591Lys being coallelic and of maternal origin and with Trp157Arg being derived from the paternal allele. Analysis of the proposita's siblings supported this conclusion: The unaffected sister tested normal for both alleles while the clinically affected brother was identical to the proposita, carrying all 3 mutations. Thus, the segregation pattern of the Asp31Asn, Thr591Lys, and Trp157Arg mutations confirmed the autosomal recessive inheritance of combined deficiency of the VKD coagulation factors in this family (Figure 2).
Functional analysis of the carboxylase mutants
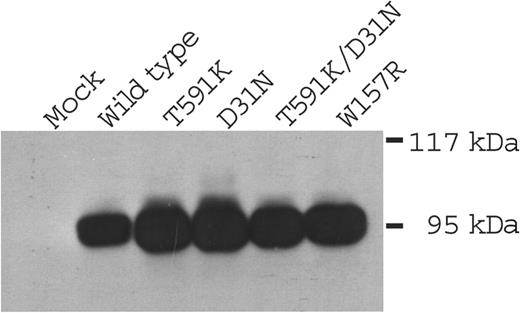
Wild-type and mutant carboxylases were expressed in insect cells, which do not contain endogenous activity but synthesize active enzyme that is exogenously introduced.38 Similar levels of carboxylase protein were observed following infection of insect cells with baculoviruses containing either wild-type or mutated carboxylases (Figure 3), indicating that the mutations did not impair expression. When tested for activity in a carboxylase assay that measures [14C]-CO2 incorporation into a Glu-containing peptide substrate, the Asp31Asn mutant showed wild-type activity (Table 3). In contrast, the Trp157Arg mutant exhibited activity that was only 8% to 10% that of wild-type, and the Thr591Lys mutant, as well as a double mutant containing both the Thr591Lys and Asp31Asn substitutions, did not show detectable activity. The carboxylase drives Glu carboxylation through epoxidation of KH2 to KO. In the wild-type enzyme these 2 activities are coupled (ie, showing an approximate 1:1 stoichiometry).39 To determine if any of the mutants showed differences in epoxidation versus carboxylation, epoxidase activity was measured. The ratio of epoxidase to carboxylase activity in Asp31Asn and Trp157Arg was similar to that of wild-type enzyme, indicating that the coupling mechanism was unaffected. The actual epoxidase value for Trp157Arg was substantially less than that of wild-type enzyme, and neither Thr591Lys nor Thr591Lys/Asp31Asn showed detectable activity (Table 3). The Trp157Arg and Thr591Lys mutations, then, can account for the combined deficiency in the function of VKD factors.
Trp157Arg and Thr591Lys show impaired carboxylase and epoxidase activities
Sample . | Epoxidase activity, pmol/h × 10-2, % . | . | Carboxylase activity, pmol/h × 10-2 . | Epoxidation-carboxylation ratio . |
|---|---|---|---|---|
| Wild type | 27.2 | 97 | 25.0 | 1.1 |
| Wild type | 28.1 | 100 | 24.7 | 1.1 |
| Asp31Asn | 29.1 | 104 | 26.2 | 1.1 |
| Asp31Asn | 29.1 | 104 | 27.0 | 1.1 |
| Trp157Arg | 2.2 | 8 | 1.8 | 1.2 |
| Trp157Arg | 2.7 | 10 | 1.8 | 1.5 |
| Thr591Lys | ND | 0 | 0.2 | — |
| Thr591Lys | ND | 0 | 0.2 | — |
| Asp31Asn/Thr591Lys | ND | 0 | 0.2 | — |
| Asp31Asn/Thr591Lys | ND | 0 | 0.2 | — |
| Mock | ND | 0 | 0.2 | — |
| Mock | ND | 0 | 0.2 | — |
Sample . | Epoxidase activity, pmol/h × 10-2, % . | . | Carboxylase activity, pmol/h × 10-2 . | Epoxidation-carboxylation ratio . |
|---|---|---|---|---|
| Wild type | 27.2 | 97 | 25.0 | 1.1 |
| Wild type | 28.1 | 100 | 24.7 | 1.1 |
| Asp31Asn | 29.1 | 104 | 26.2 | 1.1 |
| Asp31Asn | 29.1 | 104 | 27.0 | 1.1 |
| Trp157Arg | 2.2 | 8 | 1.8 | 1.2 |
| Trp157Arg | 2.7 | 10 | 1.8 | 1.5 |
| Thr591Lys | ND | 0 | 0.2 | — |
| Thr591Lys | ND | 0 | 0.2 | — |
| Asp31Asn/Thr591Lys | ND | 0 | 0.2 | — |
| Asp31Asn/Thr591Lys | ND | 0 | 0.2 | — |
| Mock | ND | 0 | 0.2 | — |
| Mock | ND | 0 | 0.2 | — |
Lysates containing equivalent amounts of wild-type or mutant carboxylase protein (as determined by a quantitative Western analysis) were assayed for conversion of KH2 to KO (epoxidase activity) or for incorporation of [14C]-CO2 into a Glu substrate (carboxylase activity). Epoxidase activity was quantitated by reference to a KO standard, and carboxylase activity was determined by using a specific activity of 50 cpm/pmol for [14C]-CO2.
ND indicates not detected; —, not applicable.
Discussion
We report the identification of 3 novel missense mutations in the carboxylase gene associated with combined deficiency of hemostatic VKD factors. The carboxylase mutations could not be accounted for by frequent polymorphisms, and no mutations in the gene for VKORC1 were detected. The proposita is unusual with respect to the previously identified patients with carboxylase missense mutations9-12 because of the compound heterozygosity in the mutations. The mutations Trp157Arg and Thr591Lys, which were derived from the father and mother, respectively (Figure 2), resulted in impaired carboxylation in vitro (Table 3) that can account for the combined VKD factor deficiency. The carboxylase also modifies VKD proteins with nonhemostatic functions that include bone development, apoptosis, and signal transduction.15 While the activities of these nonhemostatic VKD proteins are not typically screened when combined VKD factor deficiency is suspected, they are also very likely affected by carboxylase defects. The proposita and her affected brother both appeared to have skeletal abnormalities, which could be due at least in part to functional deficiencies in VKD proteins like osteocalcin and matrix Gla protein.40 Incomplete closure of the ventricular septum was also observed in both siblings, which appears to be due to poor VKD protein carboxylation, because anatomic abnormalities associated with other syndromes that affect ventricular septal closure41 were not observed.
Expression of carboxylase mutants in insect cells. Lysates from insect cells mock-infected or infected with baculoviruses containing either wild-type or mutant carboxylases were analyzed with Western analysis using anti–C-terminal carboxylase antibody.
Expression of carboxylase mutants in insect cells. Lysates from insect cells mock-infected or infected with baculoviruses containing either wild-type or mutant carboxylases were analyzed with Western analysis using anti–C-terminal carboxylase antibody.
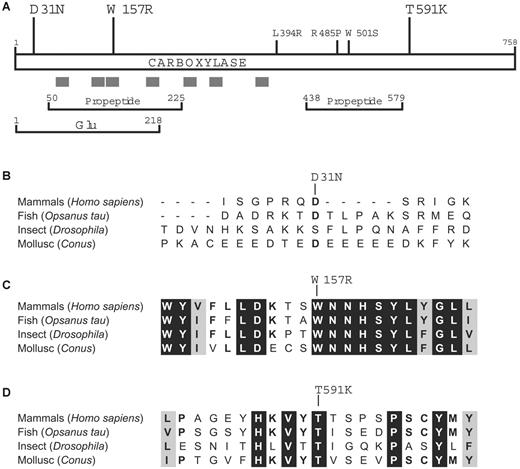
The mutations in the proposita are informative in understanding how carboxylase residues contribute to function, which currently is not well understood. As shown in Figure 4A, all 3 of the newly identified mutations are located away from the 3 previously known missense mutations that cause combined VKD factor deficiency, and 2 residues lie within regions of the carboxylase that have been functionally implicated by cross-linking studies using Glu or propeptide sequences. A third functionally important region, the vitamin K binding site, has not yet been identified but must be closely juxtaposed to the Glu binding site in the 3-dimensional structure to allow the chemical reactivity between vitamin K and Glu that ultimately leads to Glu carboxylation. The carboxylase active site, then, is unusual in accommodating the binding of both a hydrophilic substrate (Glu) and a hydrophobic cofactor (vitamin K), and the structure of the carboxylase reflects this property: The C-terminal half is hydrophilic and located in the lumen of the ER46 while most of the N-terminal half is hydrophobic. Membrane association of N-terminal sequences has been confirmed by the ability of short carboxylase-derived peptides to confer membrane insertion in vitro.47 The carboxylase active site, then, may be similar to that of other enzymes (reviewed by Bracey et al48 ) that contain membrane-bound residues that interact with lumenal (or cytoplasmic) residues to form the active site.
Trp157 and Thr591 are evolutionarily conserved residues. (A) Mutations implicated in combined VKD factor deficiency that were previously identified (small font) or that are identified in this work (large font) are shown along with regions of the carboxylase that are hydrophobic (gray bars) or that cross-link in vitro to propeptide or Glu-containing peptide.42-44 (B-D) The alignment of carboxylases from evolutionarily distant organisms45 is shown for sequences surrounding the residues (single-letter codes) whose mutations were identified in the proposita: Asp31Asn (B), Trp157Arg (C), and Thr591Lys (D). Residues that are identical or similar in all proteins are highlighted in black or gray, respectively.
Trp157 and Thr591 are evolutionarily conserved residues. (A) Mutations implicated in combined VKD factor deficiency that were previously identified (small font) or that are identified in this work (large font) are shown along with regions of the carboxylase that are hydrophobic (gray bars) or that cross-link in vitro to propeptide or Glu-containing peptide.42-44 (B-D) The alignment of carboxylases from evolutionarily distant organisms45 is shown for sequences surrounding the residues (single-letter codes) whose mutations were identified in the proposita: Asp31Asn (B), Trp157Arg (C), and Thr591Lys (D). Residues that are identical or similar in all proteins are highlighted in black or gray, respectively.
Trp157Arg, which had only 8% to 10% activity compared with wild-type enzyme (Table 3), resides within a hydrophobic region suggested by evolutionary comparison to be important to function. Thus, carboxylases that depend upon vitamin K have been identified in distantly related organisms that include Conus, Drosophila, and fish,49-53 and the comparison of these orthologs to the mammalian carboxylases has been valuable in evaluating functional residues (Figure 4B-D). The sequences around Trp157Arg are highly conserved, suggesting functional relevance, and the hydrophobicity and number of aromatic amino acids in this region raise the question of whether residue Trp157 is part of the vitamin K binding site. The proposita did not respond to vitamin K supplementation (Table 1), which contrasts with the previously identified patients with carboxylase mutations.9-12 Those patients had mutations (Leu394Arg and Trp501Ser) that were subsequently shown to decrease VKD substrate binding.30,31 Because VKD substrate binding regulates carboxylase affinity for vitamin K,22 this decrease impaired vitamin K binding (ie, causing an increase in Km for vitamin K), which can explain why these patients responded to vitamin K supplementation. The lack of response of the proposita, then, suggests that the Trp157Arg mutation impairs vitamin K catalysis rather than binding. In this regard, the change in charge caused by the substitution of an Arg for Trp is notable: Previous studies54 indicate that the carboxylase active site provides an unusual environment for the deprotonation of the catalytic base that reacts with vitamin K to initiate carboxylation, and this environment would be disrupted by a charged residue. The fact that the proposita did not respond to vitamin K supplementation also suggests that the Trp157Arg carboxylase is normally saturated with vitamin K, due either to the mutation causing a decrease in Km (which has been observed with other carboxylase mutants55 ) or to vitamin K normally being present at saturating levels. The later case would be surprising, because the availability of reduced vitamin K has been shown to regulate carboxylation, at least in cell lines.23-26
The Thr591Lys mutation, which resulted in loss of detectable carboxylase and epoxidase activities (Table 3), lies within the ER lumenal hydrophilic region of the carboxylase (Figure 4A). Sequences surrounding Thr591 do not show extensive evolutionary conservation; however, this residue is invariant in all metazoan carboxylases that have been identified51,52,56 (Figure 4D). This region is completely absent in an ortholog of the VKD carboxylase that is present in the bacterial pathogen Leptospira, which is thought to have been acquired by horizontal transfer of sequences from the mammalian host into the bacterium.45 Adaptation has resulted in a Leptospira enzyme with altered properties from mammalian carboxylase (ie, with no detectable carboxylase activity and unregulated epoxidase activity that occurs in the absence of VKD substrate).45 Epoxidase activity in the Leptospira enzyme but not in Thr591Lys suggests that the function of the Thr591 residue either is in the coupling mechanism or is specific to the carboxylase part of the reaction, with both epoxidation and carboxylation being impaired because of the mechanism regulating their coupling.27 The Thr591Lys mutation is nonconservative and introduces a charged residue, which may account for the dramatic loss of activity that was observed, and studies that test whether more conservative substitutions are as deleterious will be of interest.
The Asp31Asn mutation lies within a region that shows almost no evolutionary conservation (Figure 4B), and the Asp31 residue itself is not well conserved even among mammals (eg, bovine carboxylase contains a Gly instead of an Asp57 ). Asp31Asn exhibited wild-type activity (Table 3), but this mutation was not observed in a screen of 100 healthy individuals, and so either this substitution is a rare polymorphism or Asp31 is important to some function not assessed in the activity assays that were used. The first approximately 60 N-terminal carboxylase residues are thought to be cytoplasmically exposed,47 which likely separates Asp31 from the active site. Topologic separation therefore suggests that the N-terminus is important for some function other than activity, which may or may not involve Asp31. Thus, whether the Asp31Asn mutation also contributes to the combined deficiency in VKD factors remains to be established. Future studies that address the functional consequence of all 3 mutations will clearly be of interest in defining how molecular alterations in the carboxylase cause this disease.
Prepublished online as Blood First Edition Paper, May 18, 2006; DOI 10.1182/blood-2005-12-010660.
Supported by grants from the Tunisian Ministry of Superior Education, the Groupe d'Etude de l'Hémostase et de la Thrombose and Société Française d'Hématologie (D.D.), and the National Institutes of Health (HLB 55666) (K.L.B.).
D.D., K.W.H., R.L.S., and S.L.R. performed the research; A. Mrad, Y.G., and R.K. provided biologic assessment of the patient; A. Maherzi collected clinical data; and J.-P.R. and K.L.B. designed the research and cowrote the manuscript.
K.L.B. and J.-P.R. contributed equally to this study.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Raymonde Bredoux for helpful advice on the PCR experiments, Odile Issertial for expert technical assistance in polymorphism analysis, and Narges El Golli for help in the mutagenesis experiments. We also thank Drs Kurt W. Runge and Mark A. Rishavy for helpful comments during the preparation of this manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal