FcγRIIA expressed on neutrophils and monocytes has a fundamental role in combating bacterial infections. In the present study, the requirement of cytosolic phospholipase A2 (cPLA2) for induction of FcγRIIA expression was studied in a model of cPLA2-deficient PLB-985 cells (PLB-D cells). FcγRIIA was acquired only during differentiation of PLB but not of PLB-D cells induced by either 1,25-dihydroxyvitamin D3, retinoic acid, or interferon γ. Addition of prostaglandin E2 (PGE2) to PLB-D cells undergoing differentiation restored the expression of FcγRIIA protein, whereas addition of indomethacin to PLB cells during differentiation inhibited both the production of PGE2 and the expression of FcγRIIA. Inhibition of PKA during PLB differentiation prevented FcγRIIA expression, whereas dibutyryl cAMP (dbcAMP) induced its expression in both PLB and PLB-D cells. CREB phosphorylation and CREB-CRE interaction were detected only in differentiated PLB cells and not PLB-D cells and were inhibited by indomethacin. A reporter gene containing a FcγRIIA gene promoter fragment with the CRE element was sufficient for CREB activation. Our results are the first to show that CREB activation is involved in up-regulation of FcγRIIA expression in myeloid lineages. PGE2 formed via cPLA2 activates CREB through PKA and this process is dependent on development of PGE2 receptor 4.
Introduction
Fcγ receptors (FcγRs) are membrane glycoproteins that bind the Fc domain of IgG, thereby mediating several immunologic processes including humoral and cellular components of the immune system such as phagocytosis, antibody-dependent cellular cytotoxicity, and cytokine production.1-4 Three different classes of FcγRs have been identified: FcγRI, FcγRII, and FcγRIII. FcγRIIA (CD32), a 47-kDa integral glycoprotein, is restricted to cells of the myeloid lineages and megakaryocytes and mediates several functions. Monocytes express, in addition to FcγRIIA, FcγRI (CD64), a 70-kDa glycoprotein.5 FcγRIIIB (CD16), the non-transmembrane FcγR expressed in neutrophils, is a heavily glycosylated protein with an apparent molecular mass of 50 to 80 kDa linked by a glycosyl-phosphatidylinositol anchor to the outer plasma membrane.6 FcγRIIA has particular clinical importance because it plays a role in combating infectious diseases such as Neisseria meningitis, Haemophilus influenzae type b, Staphylococcus aureus, and Malaria.7-9 Its role is even more significant in complement deficiency where phagocytosis depends mainly on FcγRs.10 Understanding the regulation of FcγRIIA gene expression in myeloid cells may provide a means for controlling innate immunity.
The phospholipase A2 (PLA2) superfamily consists of a broad range of enzymes that are defined by their ability to specifically catalyze the hydrolysis of the center (sn-2) ester bond of glycerophospholipids.11-14 The hydrolysis products of the PLA2 reaction are free fatty acid and lysophospholipid. Cytosolic PLA2 (cPLA2) has received much attention as a key regulator of stimulus-initiated eicosanoid and PAF biosynthesis because it selectively releases arachidonic acid (AA).15,16 AA is the major precursor of biologically active eicosanoids, such as prostaglandins and leukotrienes, products catalyzed by cyclo-oxygenase and lypo-oxygenase.17 Among these active eicosanoids, prostaglandin E2 (PGE2) plays an important role in the signal transduction cascade during induction of gene transcription.18-21
We previously established a model of cPLA2-deficient differentiated PLB-985 cells (PLB-D cells) and demonstrated that cPLA2-generated AA is essential for the activation of NADPH oxidase,22 oxidase-associated H+ channel,23 and oxidase-associated diaphorase.24 Using this model we showed that cPLA2 is the sole type of PLA2 responsible for PGE2 production in these cells.25 The model of differentiated PLB-D cells provides a unique tool to determine the involvement of cPLA2 in the induction of FcγRIIA during differentiation of the myeloid PLB cells.
Materials and methods
Transfection and selection of PLB-D clones
Creation of PLB-D clones, using plasmid DNA antisense cPLA2 (1-530)-pcDNA3 or vector alone, was done as described in detail in our previous study.22 The clones were screened by Western blot analysis using rabbit antibodies raised against a GST fusion with cPLA2, as described in our previous study4 to select those that were cPLA2 protein deficient.
Cell culture and induction of differentiation
PLB-985 cells, selected PLB-D clones, or HL-60 cells were grown in a stationary suspension culture in RPMI 1640 medium as described in our previous studies.22,26 Optimal concentrations of 5 × 10-8 M 1,25(OH)2D3 (vitamin D), 0.3 mM dibutyryl cAMP (dbcAMP), 10-6 M retinoic acid (RA), and 500 U/mL interferon γ (IFN-γ) were added to PLB cells or PLB-D cells (2 × 105 cells/mL) at their logarithmic growth phase to induce differentiation.22,23
Immunofluorescence analysis of complement and FcγRs
The surface expressions of complement receptors (CD11b and CD35) and FcγRs (CD16, CD32, and CD64) were determined by mixing 1 mL undifferentiated or differentiated PLB or PLB-D cells at 1 × 106 cells/mL and 10 μg of the specific antibodies at 4°C for 30 minutes. For FcγRIIA analysis we used the IV.3 Fab (Medarex, Annandale, NJ) directed against FcRγIIA followed by cross-linking with goat antimouse F(ab′)2 fragments as done in our earlier study27 and FITC-conjugated donkey F(ab′)2 anti-goat IgG antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). In addition, we used the FITC-conjugated monoclonal anti FcγRIIA (IV.3, Medarex), which gave identical results. All the results presented in the manuscript were performed with the FITC-conjugated monoclonal anti FcγRIIA. The negative isotype-matched control was the FITC-conjugated mouse IgG2b monoclonal immunoglobulin isotype control (BD Biosciences PharMingen, San Diego, CA).
Mouse anti-human CD64 and mouse anti-human CD16 were purchased from Serotec (Oxford, United Kingdom); mouse anti-human CD11b receptor (C3b) was from Dako (Glostrup, Denmark); and mouse anti-human CD35 receptor (E-11) was from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies against CD16 do not distinguish between FcγRIIIA and FcγRIIIB. 1,25(OH)2D3 induces differentiation toward the monocyte phenotype, which expresses FcγRIIIA. For negative isotype-matched control receptors the mouse IgG1 negative control from Serotec was used. FITC-conjugated F(ab)2 goat anti-mouse IgG antibodies from Jackson ImmunoResearch Laboratories were used as second antibody. The cells were analyzed by flow microfluorometry on a fluorescence-activated cell sorter (FACS; Becton Dickinson, Mountain View, CA). For each sample 10 000 light scatter-gated viable cells were analyzed. The median (median of fluorescence intensity) was calculated by subtracting the nonspecific fluorescence.
Reverse transcription and polymerase chain reaction
Total cellular RNA was extracted and cDNA was prepared as exactly as described in detail in our previous study.28 cDNA was amplified via polymerase chain reaction (PCR) using Thermus aquaticus DNA polymerase under conditions found to amplify cDNA molecules in a linear fashion. FcγRIIA was detected by amplification of a 762-bp specific to FcγRIIA but not to FcγRIIB/IIC29 using an upstream primer: 5′-ATGTCTCAGAATGTATGTCCCAGA-3′ and a downstream primer: 5′-CTCAAATTGGGCAGCCTTCAC-3′. To confirm the results, a fraction of 557 bp was amplified using primers specific to FcγRIIA, which do not match with FcγRIIB or with FcγRIIC as determined by blast search: the upstream primer: 5′-GCTTCTGCAGACAGTCAAGC-3′ and a downstream primer: 5′-GAAGAGCT GCCCATGCTG-3′.
For detection of the PGE2 receptors (EP receptors) we used published primers.17 PCR amplification was performed in a microprocessor-controlled incubation system (Crocodile II; Appligene, Plessanton, CA). The reaction was carried out with 1 μM 5′ and 3′ primers in 50 μL reaction mixture using a step program for FcγRIIA: 94°C, 1 minute; 55°C, 30 seconds; 72°C, 2 minutes (25 cycles) and for EPs: 94°C, 1 minute; 63°C, 1 minute 10 seconds; 72°C, 1 minute 40 seconds (EP4, 31 cycles; EP1, EP2, and EP3, 40 cycles). A 10-μL sample of the completed reaction mixture was run on a 2% agarose gel stained with ethidium bromide.
Determination of PGE2
Prostaglandin levels were determined in supernatants of growth medium by radioimmunoassay using commercial kits (NEN Life Science Products, Beverly, MA). The samples were immediately stored at -70°C and analyzed within 1 week from the experiments.
Monocyte separation
Separation of monocytes was performed by Ficoll-Hypaque and Percoll gradients as described in our previous study.28 Monocytes were cultured in RPMI 1640 at concentration of 2 × 106/mL.
Phagocytosis
Phagocytosis was assessed as described previously.28 Cells (5 × 105) were suspended in RPMI 1640 containing 10% heat-inactivated FCS and incubated with 1 mg/mL opsonized zymosan (OZ) or zymosan particles at 37°C for different time durations. Subsequently, the cells were smeared and stained with differential Wright-Giemsa. Phagocytosis was determined under the microscope in at least 100 cells and defined as percent of cells containing more than 2 phagocytized particles. OZ was prepared by incubation of 20 mg/mL zymosan with human pooled serum for 60 minutes at 37°C and then washed 3 times with PBS.
Preparation of nuclear protein extract
Gel mobility shift assay
EMSAs were performed using the designed double-stranded oligonucleotides containing the consensus sequence for CREB (5′-TGAGGACTGACGACAGCTGCAC-3′) end-labeled with (γ-32P) ATP by T4 polynucleotide kinase and used as probes for EMSA. Nuclear extracts (10 μg protein) were preincubated with 0.02 A260 units of poly (d(I-C)) for 20 minutes on ice in nuclear extraction buffer. The extracts were then incubated for an additional 30 minutes at room temperature with 50 to 60 pg (50 000-75 000 cpm) of 32P-labeled, double-stranded oligonucleotide. Specificity of CREB binding was estimated by competition with unlabeled wild-type CREB or a mutant oligomer 5′-TGAGGACTGTTGACAGCTGCAC-3′ (mut CREB) with 50-fold molar excess, added to parallel samples during the preincubation period. The complexes were separated on a 7% nondenaturing polyacrylamide gel in 0.5 × TBE buffer with a constant current of 20 to 25 mA, for 2 to 3 hours at 4°C. The gel was dried and exposed overnight to Kodak X-Omat LS film (Eastman Kodak, Rochester, NY).
Immunoblot analysis
For immunoblot detection of ERK, p38 MAP kinase, JNK, and their phosphorylated proteins, cell lysates were prepared as described in detail in our previous studies.22,27 Protein (50 μg) for ERK and JNK and 10 μg protein for p38 MAP kinase from cell lysates were separated by electrophoresis on 7.5% polyacrylamide SDS gels. The resolved proteins were electrophoretically transferred to nitrocellulose, which was stained with Ponsue red to detect protein banding, and then blocked in 5% milk in TBS (10 mM Tris,135 mM NaCl, pH 7.4). Immunoblot determination was done as described before27 using primary antibodies against ERK, P-ERK, JNK, P-JNK, p38 MAP kinase, and P-p38 MAP kinase proteins (Santa Cruz Biotechnology), CREB, and P-CREB proteins (Cell Signaling Technology, Beverly, MA) for overnight incubation at 4°C and second antibody, peroxidase conjugated goat antirabbit or antimouse (Amersham Biosciences, Buckinghamshire, United Kingdom) for 1 hour at room temperature and developed using the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences). For immunoblot detection of CREB, the nuclei fractions of 2 × 106 cells were immediately solubilized in electrophoresis sample buffer and processed for separation on 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Plasmid constructions
The sequence of the FcγRIIA promoter was retrieved from McKenzie and coworkers.31 The promoter region was screened for cAMP responsive element (CRE) by the TF program at http://www.cbrc.jp/research/db/TFSEARCH.html.32 The plasmid pGL3-ΔFcγRIIA-Luc was constructed by cloning a 550-bp fragment of the human FcγRIIa gene promoter (-560 to -10)31 into the SacI and XhoI restriction sites of the vector pGL3-Luc. The FcγRIIA promoter fragment containing the CRE was amplified by PCR and purified, using primers containing the SacI and XhoI sites: 5′-TATAGAGCTCTGAGACGGAGTCTCGCTCTTTCG-3′ and 5′-TATACTCGA GAGCACTGTGCCAACGTCCAGTG-3′. PLB-985 cells (2.5 × 107) in logarithmic growth were transfected by electroporation at 250 V and 960 mF in a Gene Pulser Unit (Bio-Rad, Melville, NY) in 0.6 mL culture medium with 48 μg pGL3-ΔFcγRIIA-Luc or pGL3-Luc and with 12 μg Renilla luciferase expression vector (P-RL-null vector, Promega, Madison, WI) as an internal standard to adjust for transfection efficiency. The cells were incubated in RPMI containing 20% FCS for 4 hours and than diluted with RPMI 10% FCS to 2 × 105 cells/mL (within the linear range) and treated with the cAMP analog, dbcAMP, for the desired time duration. Cell extracts were prepared for luciferase reporter assay (Dual Luciferase Reporter Assay System, Promega) according to the manufacturer's instructions. Aliquots of 20 μL cell extract were used for each luciferase activity assay using TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA).
Statistical analysis
The mean differences were analyzed by Student t test.
Results
Expression of complement and FcγRs on PLB and PLB-D cells
The surface expression of complement and FcγRs was studied in PLB and PLB-D cells differentiated toward the monocyte lineage by 1,25(OH)2D3 by FACS analysis. The means ± SEM of 3 different experiments are summarized in Table 1. Undifferentiated PLB cells or PLB-D cells expressed appreciable and similar levels of CR1 but only basal levels of CR3. Both cell types expressed high levels of the 2 complement receptors after differentiation, as shown in our previous studies for CR3.22 Basal levels of FcγRI were expressed in undifferentiated PLB or PLB-D cells and this receptor did not develop during differentiation. Basal levels of FcγRIIA were detected in undifferentiated PLB and PLB-D cells. However, induction of differentiation caused a significant increase (P < .001) in the expression of FcγRIIA only in parent PLB cells but not in PLB-D cells. In differentiated PLB-D cells there was even a slight decrease of the FcγRIIA surface expression. The constitutive level of FcγRIII in undifferentiated cells did not change in differentiating PLB and PLB-D cells. These results suggest a role for cPLA2 in up-regulation of FcγRIIA expression during differentiation of PLB cells by 1,25(OH)2D3.
C3bR and FcγR expression in differentiated PLB and PLB-D cells
Type of receptor . | PLB, median . | PLB-D, median . |
|---|---|---|
| FcγRI | ||
| Undifferentiated | 8.7 ± 1.5 | 9.1 ± 2.5 |
| Vitamin D | 8.1 ± 1.8 | 8.5 ± 1.9 |
| FcγRIIA | ||
| Undifferentiated | 11.5 ± 2.5 | 9.4 ± 2.1 |
| Vitamin D | 31.1 ± 4.2 | 8.5 ± 2.1 |
| FcγRIII | ||
| Undifferentiated | 26.4 ± 2.1 | 24.9 ± 5.4 |
| Vitamin D | 27.5 ± 3.8 | 26.3 ± 3.1 |
| CR1 | ||
| Undifferentiated | 23.6 ± 3.9 | 18.9 ± 0.9 |
| Vitamin D | 60.8 ± 6.5 | 52.5 ± 6.9 |
| CR3 | ||
| Undifferentiated | 8.9 ± 4.5 | 9.2 ± 2.8 |
| Vitamin D | 54.4 ± 6.3 | 55.4 ± 7.2 |
Type of receptor . | PLB, median . | PLB-D, median . |
|---|---|---|
| FcγRI | ||
| Undifferentiated | 8.7 ± 1.5 | 9.1 ± 2.5 |
| Vitamin D | 8.1 ± 1.8 | 8.5 ± 1.9 |
| FcγRIIA | ||
| Undifferentiated | 11.5 ± 2.5 | 9.4 ± 2.1 |
| Vitamin D | 31.1 ± 4.2 | 8.5 ± 2.1 |
| FcγRIII | ||
| Undifferentiated | 26.4 ± 2.1 | 24.9 ± 5.4 |
| Vitamin D | 27.5 ± 3.8 | 26.3 ± 3.1 |
| CR1 | ||
| Undifferentiated | 23.6 ± 3.9 | 18.9 ± 0.9 |
| Vitamin D | 60.8 ± 6.5 | 52.5 ± 6.9 |
| CR3 | ||
| Undifferentiated | 8.9 ± 4.5 | 9.2 ± 2.8 |
| Vitamin D | 54.4 ± 6.3 | 55.4 ± 7.2 |
Cells were differentiated for 4 days with 1,25(OH)2D3. Results are mean ± SEM of 3 independent experiments. The mean ± SEM of negative controls for FcγRIIA in undifferentiated PLB and PLB-D cells was 5.2 ± 0.6 and 5.8 ± 0.7, respectively, and in differentiated cells it was 4.6 ± 0.6 and 4.7 ± 0.43, respectively. The levels of FcγRIIA were not significantly different in undifferentiated PLB and PLB-D cells from the negative controls (P = .06 and P = .17, respectively) and in differentiated PLB-D cells (P = .15). The mean ± SEM of negative controls for all other receptors in undifferentiated PLB and PLB-D cells was 4.7 ± 0.3 and 4.7 ± 0.4, respectively, and in differentiated cells it was 4.7 ± 0.6 and 4.7 ± 0.5, respectively. The levels of FcγRI or CR3 were not significantly different in undifferentiated PLB and PLB-D cells from their negative controls (P > .19)
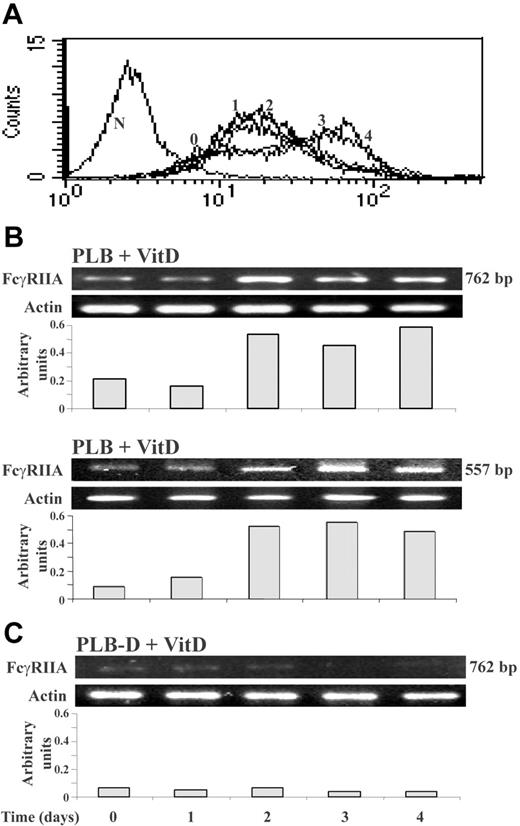
The kinetics of FcγRIIA surface protein and mRNA expression in PLB cells during 4 days of differentiation by 1,25(OH)2D3 was determined. As shown in Figure 1A, a significant increase in FcγRIIA expression was detected from day 3 of differentiation, which did not significantly change by day 4. A significant increase of FcγRIIA mRNA level was detected after 2 days of differentiation (Figure 1B), which preceded FcγRIIA protein expression. FcγRIIA mRNA was undetectable in PLB-D cells during the 4 days of differentiation (Figure 1C). These results indicate that cPLA2 is required to up-regulate FcγRIIA mRNA levels during differentiation of PLB cells by 1,25(OH)2D3.
To study whether the requirement of cPLA2 for FcγRIIA expression is general or specific only to differentiation induced by 1,25(OH)2D3, other agonists for differentiation were used. As determined by FACS analysis FcγRIIA was acquired during 4 days of differentiation by RA or by IFN-γ (median, 28.6 ± 3.1 or 23.9 ± 4.2, respectively), but not in PLB-D cells (median, 9.3 ± 3.8 or 12.5 ± 2.7, respectively). The levels of FcγRI, FcγRIII, CR1, and CR3 were similar in PLB and PLB-D cells after differentiation by RA or IFN-γ (data not shown).
PGE2 is required for the induction of FcγRIIA expression during differentiation of PLB cells by 1,25(OH)2D3
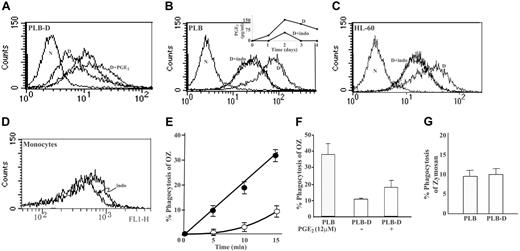
In our previous study25 we have shown that cPLA2 is the sole PLA2 isotype responsible for the production of PGE2 in differentiated PLB cells, because differentiated PLB-D cells did not produce any PGE2. To determine whether PGE2 regulates the expression of FcγRIIA induced by 1,25(OH)2D3, it was added to PLB-D cells during differentiation. As shown in Figure 2A, addition of PGE2 (12 μM) each day during 4 days of differentiation of PLB-D cells with 1,25(OH)2D3 caused a significant (P < .001) restoration of FcγRIIA expression (median, 19.4 ± 2.2 compared with 7.6 ± 2.1 in PLB-D differentiated cells with 1,25(OH)2D3 alone). Addition of PGE2 (12 μM) each day during differentiation of normal PLB cells by 1,25(OH)2D3 did not change the effect of 1,25(OH)2D3 on FcγRIIA expression (data not shown). PGE2 by itself did not induce any expression of FcγRIIA in PLB-D cells (median, 9.3 ± 1.6, data not shown for simplicity). Because PGE2 caused this restoration of FcγRIIA expression during differentiation of PLB-D, it is suggested that PGE2 is required for induction of FcγRIIA. The only partial restoration of FcγRIIA expression by PGE2 added once a day is probably due to their short half-life time. Thus, it could not completely mimic the effect of physiologic exposure to PGE2 during differentiation, which is continuously released and thus continuously exerts its effect resulting in higher expression of the FcγRIIA. To further study the role of PGE2, the effect of indomethacin, a COX inhibitor, was studied during differentiation of PLB cells. Addition of 30 μM indomethacin every day during the 4 days of differentiation with 1,25(OH)2D3 totally prevented the induction of FcγRIIA expression (Figure 2B). It has been reported that extracellular PGE2 signals gene expression by binding to its receptors on the plasma membranes.17 Thus, we studied whether PGE2 is secreted to the growth media during differentiation of PLB cells with 1,25(OH)2D3. As shown in the insert in Figure 2B, PGE2 was detected in the growth media with highest levels at day 2 of differentiation, which coincided with the appearance of FcγRIIA mRNA (Figure 1B). Addition of 30 μM indomethacin every day during the 4 days of differentiation by 1,25(OH)2D3 inhibited the secretion of PGE2,which is in correlation with the inhibition of FcγRIIA expression (Figure 2C). The requirement of PGE2 for up-regulation of FcγRIIA surface expression during differentiation was not restricted to the PLB cell line only, because FcγRIIA acquirement during differentiation of HL-60 cells was also totally inhibited by daily addition of 30 μM indomethacin (Figure 2D). Furthermore, daily addition of indomethacin to cultured peripheral blood monocytes, which express high level of FcγRIIA, caused a marked reduction (between 20% and 40%, depending on blood donor) in the expression of FcγRIIA (Figure 2E).
Kinetics of FcγRIIA surface protein expression and mRNA expression during differentiation of PLB and PLB-D cells by 1,25(OH)2D3. (A) Immunofluorescence analysis of FcγRIIA surface protein expression during the 4 days of differentiation of PLB cells by 1,25(OH)2D3 (histograms labeled from 0-4 days). The left plot (N) represents the negative control. The ordinate and the abscissa represent the cell number and the fluorescence intensity in a logarithmic scale, respectively. The patterns were confirmed by 3 repeated experiments. (B-C) RT-PCR analysis of FcγRIIA mRNA during 4 days of differentiation of PLB or PLB-D cells by 1,25(OH)2D3. The amplified products of FcγRIIA (762 bp) and (557 bp) and the corresponding products of β-actin (269 bp) are shown. The amplification of 557 bp and of 762 bp gave identical results in PLB-D cells. The intensity of each band of FcγRII mRNA was divided by the intensity of each β-actin band after quantification by densitometric scanning. Three other experiments showed similar results.
Kinetics of FcγRIIA surface protein expression and mRNA expression during differentiation of PLB and PLB-D cells by 1,25(OH)2D3. (A) Immunofluorescence analysis of FcγRIIA surface protein expression during the 4 days of differentiation of PLB cells by 1,25(OH)2D3 (histograms labeled from 0-4 days). The left plot (N) represents the negative control. The ordinate and the abscissa represent the cell number and the fluorescence intensity in a logarithmic scale, respectively. The patterns were confirmed by 3 repeated experiments. (B-C) RT-PCR analysis of FcγRIIA mRNA during 4 days of differentiation of PLB or PLB-D cells by 1,25(OH)2D3. The amplified products of FcγRIIA (762 bp) and (557 bp) and the corresponding products of β-actin (269 bp) are shown. The amplification of 557 bp and of 762 bp gave identical results in PLB-D cells. The intensity of each band of FcγRII mRNA was divided by the intensity of each β-actin band after quantification by densitometric scanning. Three other experiments showed similar results.
To determine the effect of the diminished FcγRIIA expression in differentiated PLB-D cells on cell function, phagocytosis of OZ particles mediated by Fcγ and complement receptors was studied. As demonstrated in Figure 2F, phagocytosis of OZ by differentiated PLB-D cells assayed during 15 minutes was significantly lower (P < .001) compared to that by differentiated parent PLB cells. Daily addition of PGE2 (12 μM) to PLB-D cells during differentiation by 1,25(OH)2D3 caused a partial restoration of phagocytosis of OZ (Figure 2F), which coincided with the partial restoration of FcγRIIA surface expression in differentiated PLB-D cells induced by these conditions (Figure 2A). Thus, the diminished expression of FcγRIIA on differentiated PLB-D cells is most likely responsible for the lower initial rate of phagocytosis in these cells. To further support this conclusion, phagocytosis of nonopsonized zymosan particles, which are recognized and bound by mannose receptors, was studied. Phagocytosis of zymosan particles, which is not mediated by Fcγ or complement receptors, was similar in differentiated PLB and PLB-D cells (Figure 2G).
PGE2 effect through PKA activation
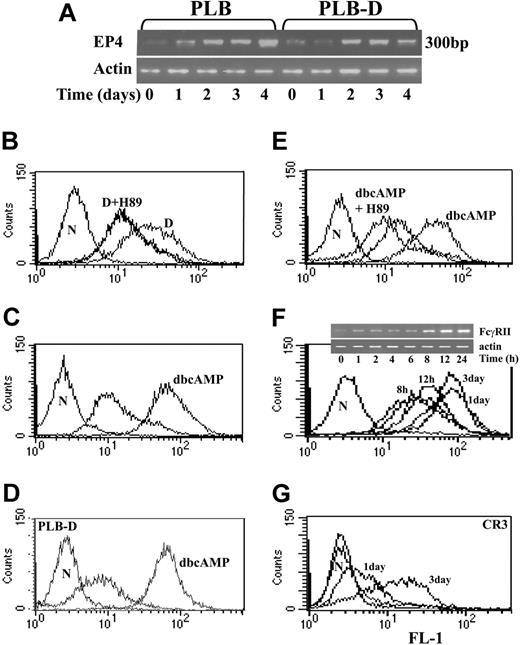
Four different EP receptors have been identified, each of which signals a different pathway in various cell types.17 To explore which pathway is initiated by PGE2 in the induction of FcγRIIA, the expression of these 4 EPs was examined in PLB and PLB-D cells during differentiation with 1,25(OH)2D3 by reverse transcription-PCR (RT-PCR) using specific primers.17 EP4 receptor mRNA expression is very low in undifferentiated cells but was significantly expressed from the second day of differentiation in both PLB and PLB-D cells (Figure 3A). EP1, EP2, or EP3 receptor mRNAs were not detected by RT-PCR in undifferentiated and differentiated PLB and PLB-D cells (data not shown). Because activation of EP4 has been shown to activate PKA,17 the effect of the PKA inhibitor, H-89, was analyzed on FcγRIIA expression during differentiation with 1,25(OH)2D3. Addition of 10 μM H-89 each day to PLB cells during differentiation with 1,25(OH)2D3 totally inhibited the induction of FcγRIIA expression (Figure 3B). Furthermore, the expression of FcγRIIA could be induced in both PLB cells and PLB-D cells by addition of dbcAMP (10 μM), which directly activates PKA (Figure 3C-D) but was totally inhibited in the presence of 10 μM H-89 in PLB cells (Figure 3E) and in PLB-D cells (not shown). FcγRIIA protein surface expression was detected as early as 8 hours after addition of dbcAMP with maximal expression at 24 hours of differentiation, which did not change after 3 days of differentiation (Figure 3F). FcγRIIA mRNA expression was detected as early as 1 hour after addition of dbcAMP (Figure 3F insert). dbcAMP induced FcγRIIA expression significantly faster than 1,25(OH)2D3 (Figure 1A). To study whether the effect of dbcAMP on FcγRIIA expression is attributed to its being a more potent inducer of differentiation22 than 1,25(OH)2D3 or to its direct role in controlling FcγRIIA expression, the kinetics of CR3 expression during differentiation was compared to that of FcγRIIA. CR3 was only slightly expressed at 24 hours of differentiation by dbcAMP and fully expressed after 3 days of differentiation (Figure 3G), in contrast to FcγRIIA, which was fully expressed after 1 day (Figure 3F). These results further support the specific involvement of PKA in signaling the up-regulation of FcγRIIA expression during differentiation.
Involvement of PGE2 on the induction of FcγRIIA surface protein expression during differentiation and on phagocytosis by differentiated cells. (A) The effect of PGE2 on induction of FcγRIIA surface expression: induction of differentiation of PLB-D by 1,25(OH)2D3 alone (D) or with addition of 12 μM PGE2 every day during 4 days of differentiation (D+PGE2). The unlabeled histogram shows the undifferentiated cells. The left plot (N) represents the negative control. The mean medians of 3 experiments are 9.5 ± 1.9, 7.6 ± 2.1, and 19.4 ± 2.2 for undifferentiated, differentiated with vitamin D (Vit D), and with Vit D with PGE2, respectively. (B) The effect of indomethacin on FcγRIIA surface protein induction and on PGE2 secretion during differentiation of PLB cells. Induction of differentiation by 1,25(OH)2D3 alone (D) or with addition of 30 μM indomethacin every day during 4 days of differentiation (D+Indo), which caused total inhibition, overlapping the undifferentiated cells (unlabeled histogram). The left plot (N) represents the negative control. In the insert the levels in the culture medium during differentiation of PLB cells in the absence (D) or presence of 30 μM indomethacin every day during 4 days of differentiation (D+Indo) are shown. (C) The effect of indomethacin on FcγRIIA surface protein induction during differentiation of HL-60 cells. Induction of differentiation by 1,25(OH)2D3 alone (D) or together with the addition of 30 μM indomethacin every day during 4 days of differentiation (D+Indo), which caused total inhibition, overlapping the undifferentiated cells (unlabeled histogram). The negative control (N) is shown in the left plot. (D) The effect of indomethacin on FcγRIIA expression in cultured monocytes. Indomethacin (30 μM) was added every 12 hours during 4 days of culture (Indo). Medians ± SEM of 5 independent experiments: 557 ± 87 and 410 ± 67 without and with indomethacin, respectively. Histograms in all parts of the figure are representative of 3 experiments unless otherwise indicated. (E) The kinetics of phagocytosis of OZ by PLB cells (•) and PLB-D cells (○) differentiated for 4 days by 1,25(OH)2D3. (F) Phagocytosis of OZ, assayed for 15 minutes, by differentiated PLB-D by 1,25(OH)2D3 alone or with daily addition of 12 μM PGE2 during 4 days of differentiation. Phagocytosis by differentiated PLB cells is represented by the dotted column. (G) Phagocytosis of zymosan particles by differentiated PLB cells or PLB-D cells by 1,25(OH)2D3, assayed for 30 minutes. The results are the mean ± SE from 3 experiments in each assay.
Involvement of PGE2 on the induction of FcγRIIA surface protein expression during differentiation and on phagocytosis by differentiated cells. (A) The effect of PGE2 on induction of FcγRIIA surface expression: induction of differentiation of PLB-D by 1,25(OH)2D3 alone (D) or with addition of 12 μM PGE2 every day during 4 days of differentiation (D+PGE2). The unlabeled histogram shows the undifferentiated cells. The left plot (N) represents the negative control. The mean medians of 3 experiments are 9.5 ± 1.9, 7.6 ± 2.1, and 19.4 ± 2.2 for undifferentiated, differentiated with vitamin D (Vit D), and with Vit D with PGE2, respectively. (B) The effect of indomethacin on FcγRIIA surface protein induction and on PGE2 secretion during differentiation of PLB cells. Induction of differentiation by 1,25(OH)2D3 alone (D) or with addition of 30 μM indomethacin every day during 4 days of differentiation (D+Indo), which caused total inhibition, overlapping the undifferentiated cells (unlabeled histogram). The left plot (N) represents the negative control. In the insert the levels in the culture medium during differentiation of PLB cells in the absence (D) or presence of 30 μM indomethacin every day during 4 days of differentiation (D+Indo) are shown. (C) The effect of indomethacin on FcγRIIA surface protein induction during differentiation of HL-60 cells. Induction of differentiation by 1,25(OH)2D3 alone (D) or together with the addition of 30 μM indomethacin every day during 4 days of differentiation (D+Indo), which caused total inhibition, overlapping the undifferentiated cells (unlabeled histogram). The negative control (N) is shown in the left plot. (D) The effect of indomethacin on FcγRIIA expression in cultured monocytes. Indomethacin (30 μM) was added every 12 hours during 4 days of culture (Indo). Medians ± SEM of 5 independent experiments: 557 ± 87 and 410 ± 67 without and with indomethacin, respectively. Histograms in all parts of the figure are representative of 3 experiments unless otherwise indicated. (E) The kinetics of phagocytosis of OZ by PLB cells (•) and PLB-D cells (○) differentiated for 4 days by 1,25(OH)2D3. (F) Phagocytosis of OZ, assayed for 15 minutes, by differentiated PLB-D by 1,25(OH)2D3 alone or with daily addition of 12 μM PGE2 during 4 days of differentiation. Phagocytosis by differentiated PLB cells is represented by the dotted column. (G) Phagocytosis of zymosan particles by differentiated PLB cells or PLB-D cells by 1,25(OH)2D3, assayed for 30 minutes. The results are the mean ± SE from 3 experiments in each assay.
Involvement of CREB in the induction of FcγRIIA transcription
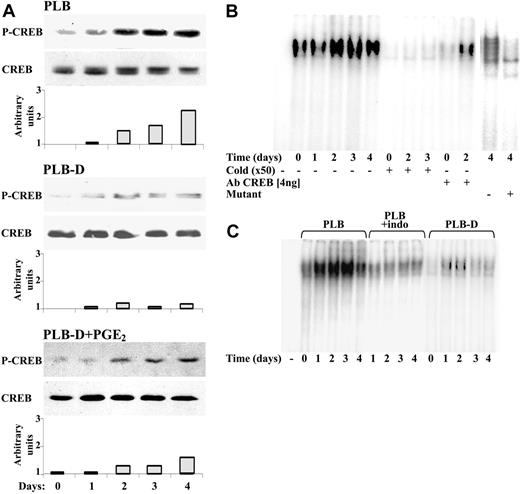
Using the TFSEARCH program, we found a single nuclear cAMP-responsive element binding (CREB) site in the FcγRIIA promoter, extending from -97 to -75.31 This element was not found in either the promoter of FcγRI or FcγRIII. As demonstrated in Figure 4A, a significant increase in CREB phosphorylation on Ser133 was detected from day 2 of differentiation by 1,25(OH)2D3, which coincided with the appearance of FcγRIIA mRNA (Figure 1B). In contrast, no increase in CREB phosphorylation on Ser133 was detected during differentiation of PLB-D cells. However, daily addition of PGE2 during differentiation of PLB-D by 1,25(OH)2D3, which caused a partial but significant restoration of the FcγIIA (Figure 2A), caused also an elevation in CREB phosphorylation that was lower than that detected in PLB cells in accordance with the partial restoration of the receptor (Figure 4A). DNA-binding activity of CREB was analyzed by EMSA, using as a probe the designed double-stranded oligonucleotides containing a CREB consensus sequence from the FcγRIIA promoter end labeled with 32P. Protein-DNA-binding activation was determined by the intensity of the radiolabeled bands on the films. As shown in Figure 4B, significant levels of CREB-DNA complexes could be detected from the second day of differentiation of PLB cells by 1,25(OH)2D3. Fifty-fold of the unlabeled CRE consensus oligonucleotides, but not mutated oligonucleotides, competed for the binding of CREB, demonstrating the specificity of CREB-DNA binding. Specific polyclonal antibodies against CREB inhibited the CREB-DNA complex, indicating that CREB specifically binds to the FcγRIIA promoter. No elevation in activation of CREB-DNA binding was detected during differentiation of PLB-D cells or during differentiation of parent PLB cells in the presence of (30 μM) indomethacin (Figure 4C).
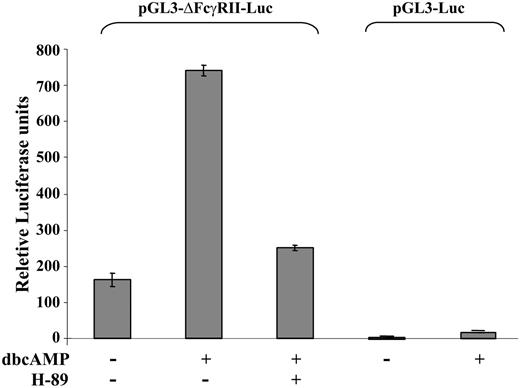
To study whether CREB acts alone or in concert with other known transcription factors, undifferentiated PLB cells were transiently transfected with pGL3-ΔFcγRIIA-Luc plasmid (described in “Materials and methods”). Addition of 10 μM dbcAMP activated the expression of the reporter gene, which was significantly inhibited (P < .001) in the presence of the PKA inhibitor, 10 μM H-89 (Figure 5).
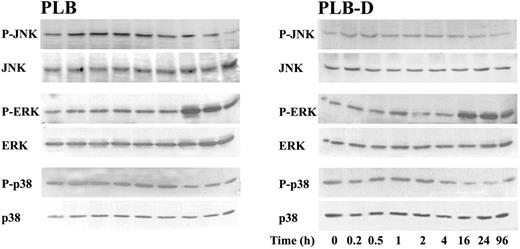
Because CREB can also be phosphorylated by different MAP kinases,33 the activation of the 3 MAP kinases, ERK, p38, and JNK, was studied in PLB and PLB-D cells during differentiation with 1,25(OH)2D3. A similar pattern of activation of the 3 MAP kinases, analyzed by their phosphorylated forms, was observed in PLB and PLB-D cells (Figure 6). Slight phosphorylation of the 3 MAP kinases was detected in undifferentiated cells. In both cell types an immediate and significant increase in JNK phosphorylation was detected, reaching maximal levels at 2 hours and then decreasing. A high phosphorylation of ERK was detected from 16 hours and a constitutive phosphorylation of p38 MAP kinase in undifferentiated cells did not change during differentiation.
Involvement of PKA in induction of FcγRII expression during differentiation. (A) EP4 mRNA: representative RT-PCR analysis of EP4 mRNA during 4 days of differentiation of PLB cells or PLB-D cells by 1,25(OH)2D3. Amplification of β-actin was done as a control. Three other experiments showed similar results. (B) The effect of PKA inhibitor on FcγRIIA expression detected by immunofluorescent analysis induced by 1,25(OH)2D3. Induction of differentiation of PLB cells by 1,25(OH)2D3 alone (D) or with addition of 10 μM H-89 (D+H89) every day during 4 days of differentiation. Undifferentiated cells are shown by the unlabeled plot overlapping with D+H89 treatment. (C-D) Induction of FcγRIIA expression by dbcAMP. Immunofluorescent analysis of FcγRIIA expression in PLB cells (histogram C) and PLB-D cells (histogram D) at 1 day of differentiation by 0.3 mM dbcAMP. (E) The effect of PKA inhibitor on FcγRII expression induced by dbcAMP. Induction of differentiation of PLB cells by dbcAMP alone or with addition of 10 μM H-89 (dbcAMP+H89) every day during 4 days of differentiation. (F) Kinetics of FcγRII protein expression (detected by immunofluorescent analysis) and of RT-PCR FcγRIIA mRNA (insert) induced by dbcAMP in PLB cells. (G) Immunofluorescent analysis of CR3 protein expression at 1 day and 3 days of differentiation induced by dbcAMP in PLB cells. In all experiments, results are representative of 3 experiments. The left plots are the negative controls (N) and unlabeled plots represent undifferentiated cells.
Involvement of PKA in induction of FcγRII expression during differentiation. (A) EP4 mRNA: representative RT-PCR analysis of EP4 mRNA during 4 days of differentiation of PLB cells or PLB-D cells by 1,25(OH)2D3. Amplification of β-actin was done as a control. Three other experiments showed similar results. (B) The effect of PKA inhibitor on FcγRIIA expression detected by immunofluorescent analysis induced by 1,25(OH)2D3. Induction of differentiation of PLB cells by 1,25(OH)2D3 alone (D) or with addition of 10 μM H-89 (D+H89) every day during 4 days of differentiation. Undifferentiated cells are shown by the unlabeled plot overlapping with D+H89 treatment. (C-D) Induction of FcγRIIA expression by dbcAMP. Immunofluorescent analysis of FcγRIIA expression in PLB cells (histogram C) and PLB-D cells (histogram D) at 1 day of differentiation by 0.3 mM dbcAMP. (E) The effect of PKA inhibitor on FcγRII expression induced by dbcAMP. Induction of differentiation of PLB cells by dbcAMP alone or with addition of 10 μM H-89 (dbcAMP+H89) every day during 4 days of differentiation. (F) Kinetics of FcγRII protein expression (detected by immunofluorescent analysis) and of RT-PCR FcγRIIA mRNA (insert) induced by dbcAMP in PLB cells. (G) Immunofluorescent analysis of CR3 protein expression at 1 day and 3 days of differentiation induced by dbcAMP in PLB cells. In all experiments, results are representative of 3 experiments. The left plots are the negative controls (N) and unlabeled plots represent undifferentiated cells.
Discussion
FcγRIIA expressed on neutrophils and monocytes has a fundamental role in combating bacterial infections. FcγRIIA contains immunoreceptor tyrosine-activating motif, mediating positive signaling, resulting in internalization of immune complexes and initiation of inflammatory responses.34 Elevated expression of FcγRIIA was detected on monocytes of patients with inflammatory diseases such as rheumatoid arthritis.35 Therefore understanding the mechanism that regulates the induction of the FcγRIIA surface protein is of great clinical significance. The results of the present study demonstrate that cPLA2 has a central role in induction of FcγRIIA expression, during differentiation of PLB cells, because in its absence (in PLB-D cells) FcγRIIA is not acquired during differentiation. This requirement is specific for the induction of FcγRIIA as other Fcγ and complement receptors are expressed normally in differentiated PLB-D cells (Table 1). Consistent with the known physiologic role of this receptor, the diminished FcγRIIA surface expression in differentiated PLB-D cells resulted in a significant reduction in the rate of phagocytosis of opsonized particles (Figure 2E), which is mediated by the Fc and complement receptors. The participation of cPLA2 in the induction of FcγRIIA is not restricted to differentiation with 1,25(OH)2D3 toward the monocytic phenotype, because PLB-D cells differentiating with RA or IFN-γ toward the granulocyte or monocyte lineage, respectively, also did not express FcγRIIA. We have recently identified 3 types of secreted PLA2 (type II, V, and X) in PLB cells and in PLB-D cells whose levels did not change after differentiation,25 indicating that these PLA2s cannot replace cPLA2 for the induction of FcγRIIA.
Our findings show that PGE2 but not AA induces FcγRIIA expression during differentiation of PLB cells because the presence of the COX inhibitor, indomethacin, inhibited during differentiation its expression (Figure 2B), whereas AA has been shown to regulate NADPH oxidase activity.22 Maximal levels of PGE2 were secreted from parent PLB cells at day 2 of differentiation (Figure 2B insert), which coincided with the appearance of FcγRIIA mRNA (Figure 1B). Treatment with the indomethacin inhibited both the release of PGE2 (Figure 2B insert) and the expression of FcγRIIA (Figure 2B) in differentiating PLB cells. Furthermore, addition of PGE2 during differentiation of PLB-D cells, which do not produce any PGE2,25 significantly restored the expression of FcγRIIA (Figure 2A). The absolute requirement of PGE2 for induction of FcγRIIA is not restricted to the PLB cell line only and was observed also during differentiation of HL-60 cells (Figure 2C). Furthermore, addition of indomethacin to cultured monocytes, which already express high levels of FcγRIIA surface protein, caused a marked reduction in its expression (Figure 2D).
Expression of specific types of EP receptors appears to be the mechanism by which different cell types carry out various physiologic eicosanoid functions.36 Four subtypes of PGE2 receptors have been identified and characterized: EP1 receptor, which causes influx of Ca2+ and activation of protein kinase C (PKC); EP2 and EP4 receptors, which activate adenylate cyclase thereby increasing cellular cAMP levels, which in turn activates PKA; and EP3 receptor, which signals primarily through an inhibitory G protein to decrease intracellular cAMP levels.37 The expression of EP4 after differentiation (Figure 3A), the inhibition of FcγRIIA induction during differentiation of PLB cells by a specific PKA inhibitor (Figure 3B), and the induction of FcγRIIA expression by a cAMP analog, which directly activates PKA (Figure 3C-D), suggest that the PKA pathway participates in signaling FcγRIIA induction. Early and potent induction by dbcAMP was restricted only to the FcγRIIA (Figure 3F) and not to induction of the whole differentiation process, as analyzed by the expression of C3 receptor (Figure 3G), which further supports the direct role of PKA in induction of FcγRIIA expression. In undifferentiated cells, PGE2 alone was unable to induce FcγRIIA expression, because of the absence of the EP4 receptor (Figure 3A). Only after the induction of EP4 by 1,25(OH)2D3, did PGE2 induce the up-regulation of FcγRIIA expression (Figure 2A).
Several sequence elements that may mediate the regulation of FcγRIIA transcription have been identified within the 5′-flanking region,31 including potential binding sites for GATA-1, and elements that mediate response to glucocorticoids (GRE), cytokines (CK-1), interferon (IRE), phorbol ester (AP-1), and vitamin D. The response elements GATA-1, GATA-2, and NF-Y have been shown to participate in transcriptional regulation of FcγRIIA in megakaryocytic cells but not myelomonocytic cells.38 The results of the present study are the first to show that the promoter of the FcγRIIA gene contains a functional CRE-binding domain (in contrast to FcγRI or FcγRIII promoters) and that CREB is a key regulator in mediating activation of FcγRIIA gene transcription in myeloid lineages (myelomonocytic and myelogranulocytic cells). The nuclear CREB protein is phosphorylated on Ser133 and interacts with CRE (Figure 4B) during differentiation of PLB cells with 1,25(OH)2D3. CREB phosphorylation and CREB-CRE interaction were detected after 2 days of differentiation, which coincided with both the maximal secretion of PGE2 and the appearance of the EP4 receptor (Figure 3A) thus enabling PKA activation by PGE2. CREB-CRE interaction could not be detected during differentiation of PLB-D cells or during differentiation of parent PLB cells in the presence of the COX inhibitor, indomethacin (Figure 4C), indicating the role of cPLA2 and the production of PGE2 in CREB activation for FcγRIIA gene transcription. Our results are in agreement with the known role of CREB stimulation by PKA in cAMP-mediated activation of gene transcription.39 PKA catalytic subunits translocate to the nucleus where they phosphorylate CREB family members.40 Phosphorylation of CREB on Ser133 has been shown to be required for CREB-induced gene transcription.41 The expression of CD14 in mouse macrophages have also been shown to be regulated by PGE2 via cAMP-dependent PKA.42 However, for CD14 expression PKA caused stimulation of the AP-1 transcription factor.
Involvement of CREB (Ser133) in the induction of FcγRII transcription. (A) Representative Western blot analysis of phospho-CREB (Ser133) or total CREB in nuclear extracts from PLB cells, PLB-D cells, or PLB-D cells with daily addition of 12 μM PGE2 during 4 days of differentiation by 1,25(OH)2D3. The intensity of each band of phospho-CREB was divided by the intensity of each band of CREB after quantification by densitometry scanning. (B) Representative EMSA for DNA-binding activity of CREB. Nuclear extracts isolated from PLB cells during 4 days of differentiation by 1,25(OH)2D3 were incubated with 32P-labeled probe containing CREB consensus sequence from FcγRIIA promoter. For competitive inhibition assay, 50-fold molar excess of unlabeled CREB antibodies against CREB or labeled CREB mutant probe was added. (C) EMSA for DNA-binding activity of CREB was not detected during differentiation by 1,25(OH)2D3 in PLB-D cells and in parent PLB cells in the presence of 30 μM indomethacin. DNA-protein complexes were analyzed on a 7% nondenaturing polyacrylamide gel. Three other experiments showed similar results.
Involvement of CREB (Ser133) in the induction of FcγRII transcription. (A) Representative Western blot analysis of phospho-CREB (Ser133) or total CREB in nuclear extracts from PLB cells, PLB-D cells, or PLB-D cells with daily addition of 12 μM PGE2 during 4 days of differentiation by 1,25(OH)2D3. The intensity of each band of phospho-CREB was divided by the intensity of each band of CREB after quantification by densitometry scanning. (B) Representative EMSA for DNA-binding activity of CREB. Nuclear extracts isolated from PLB cells during 4 days of differentiation by 1,25(OH)2D3 were incubated with 32P-labeled probe containing CREB consensus sequence from FcγRIIA promoter. For competitive inhibition assay, 50-fold molar excess of unlabeled CREB antibodies against CREB or labeled CREB mutant probe was added. (C) EMSA for DNA-binding activity of CREB was not detected during differentiation by 1,25(OH)2D3 in PLB-D cells and in parent PLB cells in the presence of 30 μM indomethacin. DNA-protein complexes were analyzed on a 7% nondenaturing polyacrylamide gel. Three other experiments showed similar results.
CREB has been shown to confer a response with or without cooperating factors. For example, in PC12 cells in the absence of the SRE-SRF complex, CREB phosphorylation was not sufficient to activate c-fos transcription in response to growth factors.43,44 On the other hand, in primary cortical neurons, brain-derived neurotrophic factor stimulated activation of CREB-dependent transcription of c-fos.44,45 This promoter contains the consensus sequence for CREB binding but not the consensus sequences of transcription factors reported to work in cooperation with CREB such as SRE, AP1, CBP, or EBP.46,47 Whether and which transcription factors are working in cooperation with CREB/CRE interaction are still not known. A recent study48 has demonstrated that addition of PGE2 to chondrocytes transfected with a luciferase reporter driven by a promoter containing only 4 copies of cAMP response element (CRE-Luc) was sufficient for a robust induction of luciferase activity.
Activation of CRE in PLB cells. PLB cells were transiently transfected with pGL3-ΔFcγRIIA-Luc reporter plasmid or pGL3-Luc as described in “Materials and methods,” and treated for 16 hours with 10 mM dbcAMP in the absence or presence of 10 μM H-89. Reporter activity is expressed as fold increase in luciferase activity, which was standardized to Renilla luciferase expression used as an internal control. Relative luciferase units ± SE (bars) of 3 independent experiments, each done in triplicate.
Activation of CRE in PLB cells. PLB cells were transiently transfected with pGL3-ΔFcγRIIA-Luc reporter plasmid or pGL3-Luc as described in “Materials and methods,” and treated for 16 hours with 10 mM dbcAMP in the absence or presence of 10 μM H-89. Reporter activity is expressed as fold increase in luciferase activity, which was standardized to Renilla luciferase expression used as an internal control. Relative luciferase units ± SE (bars) of 3 independent experiments, each done in triplicate.
Activation of MAP kinases during differentiation of PLB and PLB-D cells by 1,25(OH)2D3. Kinetics of JNK, ERK, or p38 MAP kinase phosphorylation were analyzed by immunoblot analysis with phospho-specific antibodies (p-JNK, p-ERK, or p-p38) in PLB and PLB-D cells during differentiation by 1,25(OH)2D3. The levels of JNK, ERK, or p38 MAP kinase in each sample were evaluated by immunoblotting anti-JNK, anti-ERK, or anti-p38 MAP kinase antibodies, respectively. Results are representative of 3 experiments.
Activation of MAP kinases during differentiation of PLB and PLB-D cells by 1,25(OH)2D3. Kinetics of JNK, ERK, or p38 MAP kinase phosphorylation were analyzed by immunoblot analysis with phospho-specific antibodies (p-JNK, p-ERK, or p-p38) in PLB and PLB-D cells during differentiation by 1,25(OH)2D3. The levels of JNK, ERK, or p38 MAP kinase in each sample were evaluated by immunoblotting anti-JNK, anti-ERK, or anti-p38 MAP kinase antibodies, respectively. Results are representative of 3 experiments.
Although CREB was originally identified as a target of the cAMP signaling pathway, studies on immediate-early gene activation33 revealed that CREB is also a target of other signaling pathways activated by a diverse array of stimuli. For example, growth factors can induce CREB phosphorylation via activation of different MAP kinases.49-52 Activation of ERKs causes stimulation of RSKs, which then translocate to the nucleus and phosphorylate CREB. Alternatively, ERKs or p38 MAP kinase, by themselves, can also translocate into the nucleus to activate the kinase MSK1, which phosphorylates CREB.51 Our findings (Figure 6) show a similar pattern of activation of the 3 MAP kinases, ERK, p38 MAP kinase, and JNK, during differentiation of PLB cells containing cPLA2 and PLB-D cells lacking cPLA2 with 1,25(OH)2D3. Yet FcγRIIA was not expressed in differentiated PLB-D cells, indicating that the MAP kinases are not involved in CREB phosphorylation or in FcγRIIA gene transcription signaling.
In conclusion, the present study demonstrates, as summarized in Figure 7, that induction of differentiation causes among other processes, a release of AA by cPLA2, which is then converted to PGE2 and released from the cells. At day 2 of differentiation there is a significant release of PGE2 that coincides with the expression of the EP4 receptor. EP4 is not expressed in undifferentiated PLB cells and its induction during differentiation is independent of cPLA2, because it is also developed in differentiating PLB-D cells. Thus, we suggest that 2 independent synchronized processes participate in up-regulation of FcγRIIA expression: the production and release of PGE2 and the induction of EP4 receptor. The activation of EP4 by PGE2 results in the activation of PKA, CREB phosphorylation, and CREB-CRE interaction leading to the induction of FcγRIIA gene transcription.
Proposed schematic signaling pathway from the activation of cPLA2 to the FcγRIIA protein surface expression, during differentiation in PLB cells. The scheme is based on the present study demonstrating that during differentiation of PLB cells PGE2 is synthesized from AA released by cPLA2. PGE2 is released from the cells and through the EP4 receptor exerts its effect on PKA activation, CREB phosphorylation, and CREB-CRE binding activates FcγRIIA gene and protein expression.
Proposed schematic signaling pathway from the activation of cPLA2 to the FcγRIIA protein surface expression, during differentiation in PLB cells. The scheme is based on the present study demonstrating that during differentiation of PLB cells PGE2 is synthesized from AA released by cPLA2. PGE2 is released from the cells and through the EP4 receptor exerts its effect on PKA activation, CREB phosphorylation, and CREB-CRE binding activates FcγRIIA gene and protein expression.
Prepublished online as Blood First Edition Paper, May 18, 2006; DOI 10.1182/blood-2006-05-021881.
Supported by a grant from the Israel Sciences Foundation founded by the Israel Academy of Sciences and Humanities 438/03.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal