Serum amyloid A (SAA) is the major acute-phase protein in man and most mammals. Recently we demonstrated that SAA binds to many Gram-negative bacteria including Escherichia coli and Pseudomonas aeruginosa through outer membrane protein A (OmpA) family members. Therefore we investigated whether SAA altered the response of innate phagocytic cells to bacteria. Both the percentage of neutrophils containing E coli and the number of bacteria per neutrophil were greatly increased by SAA opsonization, equivalent to the increase seen for serum opsonization. In contrast, no change was seen for Streptococcus pneumoniae, a bacteria that did not bind SAA. Neutrophil reactive oxygen intermediate production in response to bacteria was also increased by opsonization with SAA. SAA opsonization also increased phagocytosis of E coli by peripheral blood mononuclear cell-derived macrophages. These macrophages showed strong enhancement of TNF-α and IL-10 production in response to SAA-opsonized E coli and P aeruginosa. SAA did not enhance responses in the presence of bacteria to which it did not bind. These effects of SAA occur at normal concentrations consistent with SAA binding properties and a role in innate recognition. SAA therefore represents a novel innate recognition protein for Gram-negative bacteria.
Introduction
During systemic inflammation the most notable change in protein synthesis involves hepatic synthesis of a set of acute-phase proteins whose functions, where identified, are in homeostasis or protection from the cause of the inflammation. Two of these proteins, human C-reactive protein (CRP) and serum amyloid A (SAA) protein, show massive increases in concentration within 24 hours in response to systemic inflammation. Both can be used as indicators of severity of inflammation, although currently CRP is the main clinical marker. The SAA protein family comprises up to 4 members with no disulphide bonds and for which no structure has yet been elucidated and no clear function ascribed.1 The 2 major acute-phase forms (SAA1 and SAA2) are approximately 12 kDa and are synthesized largely by hepatocytes. During severe inflammation, acute-phase SAA concentrations in plasma may increase from 1 to 5 μg/mL to approach 1 mg/mL1 and at such time can comprise more than 2% of total hepatic protein synthesis.2 A third gene product, SAA3, is synthesized largely in extrahepatic sites and is also induced by inflammatory cytokines such as interleukin-6 and interleukin-1. A constitutive form (SAA4) has an 8-amino acid insert and comprises the most abundant serum SAA form in healthy individuals. In the absence of inflammation, major sites of synthesis of SAA include the epithelium.3 The SAA protein family is found in mammals and other vertebrates such as marsupials and fish1 but more anciently in the sea cucumber, an echinoderm, where it is expressed in the coelomic epithelium and is induced in response to lipopolysaccharide (LPS).4 SAA isoforms associate with high-density lipoprotein (HDL), in particular subfraction 3,5 but studies on alteration of HDL properties by SAA have yet to demonstrate a convincing role for SAA. Alternative studies have examined direct effects of SAA on a number of immune cells. For instance, SAA has been reported to cause chemotaxis of several cell types6-8 but this response does not occur in the presence of HDL,6,7 leaving it uncertain if this occurs in vivo. The receptor that causes the chemotactic activity was identified as FPRL1, which is a low-affinity receptor for fMLP and binds to lipoxin A4.6 This receptor has also been identified as being responsible for other activities such as neutrophil IL-8 release or intracellular calcium mobilization.8 In addition, the induction of matrix metalloproteinases has been described for rabbit and human SAA isoforms,9,10 and induction of secretory phospholipase A2 has been reported11 as well as other neutrophil activatory responses.12 SAA may interact with a number of other potential receptors including scavenger receptor-B113,14 as well as Tanis15 and receptor for advanced glycation end products.16 A variety of cell types including platelets and lymphocytes have been reported to bind SAA.17,18 Other SAA ligands include certain extracellular matrix glycoproteins,18,19 heparin, and heparan sulfate.20
Recently we demonstrated that SAA binds to a range of Gram-negative bacteria including Escherichia coli, Klebsiella pneumoniae, Shigella flexneri, Vibrio cholerae, and Pseudomonas aeruginosa but not Burkholderia cepacia or Gram-positive organisms such as Streptococcus pneumoniae and Staphylococcus aureus. Since SAA bound to a ligand in bacterial lysates we were able to fractionate the bacteria and identify the major ligand as outer membrane protein A (OmpA) in E coli.21 SAA bound to bacteria when the source of SAA was normal or acute-phase serum and the amount of SAA required for half-maximal binding was at or below the normal serum concentration.21 Since the OmpA/OprF family protein is found across almost all Gram-negative bacteria, it could represent a potential pathogen-associated molecular pattern (PAMP) with SAA having the role of a pattern-recognition protein. In this report we investigate this hypothesis and demonstrate that indeed SAA does act as an opsonin for macrophages and neutrophils. In addition to promoting phagocytosis, SAA opsonization also enhanced neutrophil respiratory burst and macrophage TNF-α and IL-10 production. SAA depletion studies showed that in normal human serum in the presence of a humoral response, SAA was a minor contributor to total opsonic activity. This report establishes SAA as an innate immune protein.
Patients, materials, and methods
Bacteria
Clinical S pneumoniae strain 3 was kindly provided by Dr S. Gillespie (Department of Microbiology, Royal Free Hospital, London, United Kingdom); P aeruginosa laboratory strains 7 and 7/1 and clinical-derived strains 18S and CF003 were provided by Dr T. Pitt (Central Public Health Laboratory, Colindale, London, United Kingdom). B cepacia was provided by P. Donachie (Microbiology, London School of Hygiene and Tropical Medicine [LSHTM], London, United Kingdom). E coli, P aeruginosa, and B cepacia were cultured in brain heart infusion (BHI) broth at 37°C for 18 hours until stationary phase. S pneumoniae was cultured on blood agar plates in an atmosphere containing 10% (vol/vol) CO2 at 37°C. Bacteria were washed and harvested at 400g for 10 minutes. E coli BL21 (DE3; Novagen, Nottingham, United Kingdom) was either transformed by an enhanced green fluorescent protein (pEGFP) vector (BD Clontech, Oxford, United Kingdom) or E coli or S pneumoniae were labeled with 10 μM PKH67 cell marker (Sigma, Poole, United Kingdom). For certain experiments, bacteria at 109 colony-forming units (cfu's)/mL were γ-irradiated with approximately 1.4 × 107 Gy in a 131GAMMACELL 1000 ELITE (Nordion International, Toronto, ON, Canada) for 4 hours. Organisms were plated onto blood agar plates or cultured into BHI broth and incubated overnight at 37°C to confirm they were incapable of growth after irradiation.
SAA and binding studies
SAA in this study was the isoform SAA1, purified as described22 from plasma derived from patients undergoing plasmapheresis as part of treatment using hydrophobic interaction chromatography (HIC) on octyl-Sepharose, gel filtration, and anion exchange chromatography, with the modification that the first stage of purification on octyl-Sepharose 4B was performed in the presence of 10 mM EDTA. SAA was radiolabeled with carrier-free [125I] using the N-bromo-succinimide method to specific activities between 70 to 120 kBq/μg. SAA was freshly dialyzed from 4 M urea into distilled water overnight before incorporation into experimental media in all studies. All studies used purified SAA1 except where stated, and this preparation was greater than 95% monomeric by gel filtration. Recombinant SAA (rSAA; Peprotech, London, United Kingdom), which corresponds to SAA1.1, with the exception of an addition of methionine at the N-terminal and substitution of aspartic acid for asparagine at position 60 and histidine for arginine at position 71 (both substitutions occurring in the SAA2 isoform), was reconstituted as directed by the suppliers.
Normal serum (2 mL) was collected from healthy donors and depleted of SAA by passage through an affinity-purified goat polyclonal anti-SAA column (5 mL). SAA in depleted and nondepleted serum was measured by a sandwich enzyme-linked immunosorbent assay (ELISA) using coating monoclonal antibody (clone 115) and detection biotinylated monoclonal antibody (clone 607; Anogen, Mississauga, ON, Canada) according to the specified instructions. Alternatively, normal serum (2 mL) was passed through an octyl-Sepharose (HIC) column (4 mL; Amersham Bioscience, Slough, United Kingdom) and SAA was measured similarly. Sera depleted using antibody contained SAA at 100 ng/mL and sera depleted using HIC contained less than 50 ng/mL.
Live bacteria (108 cfu's) were incubated with 0.01 to 5 μg/mL SAA in PBS at various dilutions at 4°C for 1 hour followed by washing 3 times with PBS followed by addition of 2 μg/mL mouse anti-human SAA (clone 513; Anogen) or isotype control for 1 hour at 4°C. Cells were washed 3 times and 40 μg/mL f(ab′)2 goat anti-mouse (IgG) FITC (Sigma) was added for 1 hour at 4°C. Bacteria were washed 3 times and fixed with 4% (wt/vol) paraformaldehyde (PFA) for 30 minutes at 4°C and analyzed by FACSCalibur and Cell Quest software (BD Biosciences, Oxford, United Kingdom).
Macrophage and neutrophil isolation
For neutrophil isolation, acid citrate dextrose anticoagulated blood from healthy donors under ethical consent was transferred to a tube containing dextran solution: 3% (wt/vol) dextran T500 (Pharmacia, Milton Keynes, United Kingdom) in 0.9% (wt/vol) NaCl in a 1:2 ratio (dextran-blood). The leukocyte-enriched supernatant was carefully layered onto 15 mL of Histopaque (1.077g/mL) and centrifuged for 30 minutes at 400g at 4°C to remove peripheral blood mononuclear cells (PBMCs). Distilled water was added for 30 seconds to eliminate contaminant red blood cells. Greater than 95% were confirmed as polymorphonuclear cells and viability was greater than 97%.
Macrophages were obtained from buffy coats (North Thames Blood Transfusion Service) and mononuclear cells separated on Histopaque (1.077g/mL). Monocytes from PBMCs were allowed to adhere for 2 hours in serum-free medium and then washed before incubation for 6 days in RPMI supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10% FCS (Gibco, Invitrogen, Paisley, United Kingdom). Adherent monocyte-derived macrophages were removed by cell dissociation buffer (Gibco) and counted before use in phagocytosis assays.
Respiratory burst (intracellular hydrogen peroxide) assay
Isolated neutrophils were resuspended at 2 × 106 cells/mL in CD hybridoma medium (Gmbco) with E coli or S pneumoniae (multiplicity of infection [MOI] 30:1) in the absence or presence of SAA (1, 10, 50 μg/mL) diluted in PBS for 20 minutes at 37°C. Dihydrorhodamine-1,2,3 (DHR; Calbiochem, Nottingham, United Kingdom) was added to a final concentration of 30 μM and incubated with cells for a further 10 minutes. The reaction was stopped with PBS containing 0.02% (wt/vol) EDTA and washed 3 times with PBS. The cells were fixed with 2% (wt/vol) PFA at 4°C. Cells were acquired by FACSCalibur and analyzed by Cell Quest software.
Binding and phagocytosis assay and confocal microscopy
Monocyte-derived macrophages or neutrophils (2 × 106 cells/mL) in RPMI 1640 supplemented with 2 mM l-glutamine and 0.2% (wt/vol) BSA were either incubated at 37°C for an hour (30 min for neutrophils) with E coli-GFP or PKH67-labeled bacteria (MOI 50:1 for macrophages and 30:1 for neutrophils) in the absence or presence of SAA (1, 10, 50 μg/mL) or 20% (vol/vol) normal human serum (NHS). To determine if the increase was due to phagocytosis, cells were pretreated with cytochalasin D (10 μg/mL) for 30 minutes at 37°C and then incubated with various stimuli. The cells were washed 3 times with PBS before fixing with 2% (wt/vol) PFA and analyzed by fluorescence-activated cell sorter (FACS). For depletion experiments, serum preparations were used at 33% of original serum concentration.
For confocal microscopy, macrophages (2 × 105 cells/well) on glass coverslips in 24-well plates were loaded with 200 nM LysoTracker red DND-99 probe (543 nm; Molecular Probes, Eugene, OR) by incubating at 37°C for 1 hour. E coli-GFP (MOI 50:1) was added in the absence or presence of SAA for 1 hour at 37°C. The cells were washed twice, reloaded with LysoTracker probe, fixed, washed once, and quenched with 50 mM ammonium chloride for 45 minutes at room temperature (rt). The cells were washed once and permeabilized and blocked with 0.1% (wt/vol) saponin, 1.5% (vol/vol) normal goat serum in PBS for 45 minutes at rt. Cells were then incubated with Phalloidin-BODIPY (633 nm; 1:100 dilution; Molecular Probes) at rt for 45 minutes followed by washing with PBS (× 4). The coverslips were air dried, mounted on glass slides, and viewed by confocal microscopy (LSM510; Zeiss, Oberkochen, Germany). Bacteria per cell and number of cells with internalized bacteria were counted in a blind manner. Real-time live confocal microscopy was performed on neutrophils (2 × 106 cells/mL) loaded with LysoTracker dye for 1 hour at 37°C in RPMI with additives in 4-well chamber slides (Nunc, SLS, Nottingham, United Kingdom). E coli-GFP (MOI 10:1) was added to SAA (10 μg/mL) and neutrophils at 37°C in an atmosphere of 5% CO2 (vol/vol) and images were acquired over 30 minutes by confocal microscope.
Cytokine production by monocyte-derived macrophages
After 6 days, monocyte-derived macrophages (3 × 105/well) in 96-well plates were incubated with γ-irradiated bacteria (MOI 50:1) in the absence or presence of SAA (1, 10, 50 μg/mL) and SAA alone. Supernatants were harvested at 16 hours and IL-10 and TNF-α levels were determined using antibody pairs according to the manufacturer's guidelines (BD Pharmingen, Oxford, United Kingdom). The limit of sensitivity for TNF-α was 50 pg/mL and for IL-10 was 30 pg/mL.
Results
SAA opsonizes Gram-negative bacteria for phagocytosis by neutrophils
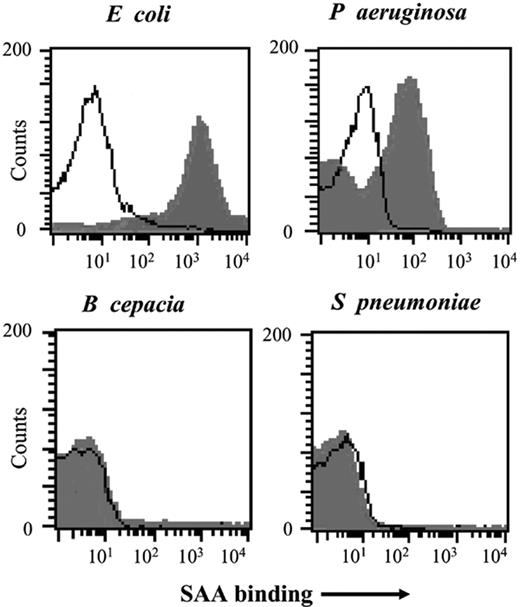
Previous studies demonstrated that bacteria bound SAA1 whether it was offered as a component of HDL in normal or acute-phase serum, in purified HDL, or when purified as described previously.21 SAA binding to bacteria for use in this study was examined using radiolabeled SAA and ligand immunoblotting (data not shown) or unlabeled SAA followed by detection using FACS analysis (Figure 1). Titration of SAA revealed maximal binding at about 1 μg/mL for E coli and 5 μg/mL for P aeruginosa. In contrast, no detectable binding was seen to the Gram-positive organisms S pneumoniae (virulent serotype 3 and unencapsulated strain R36A) or S aureus or to certain Gram-negative organisms such as B cepacia (Figure 1). S pneumoniae and B cepacia were selected as SAA nonbinding controls for functional assays.
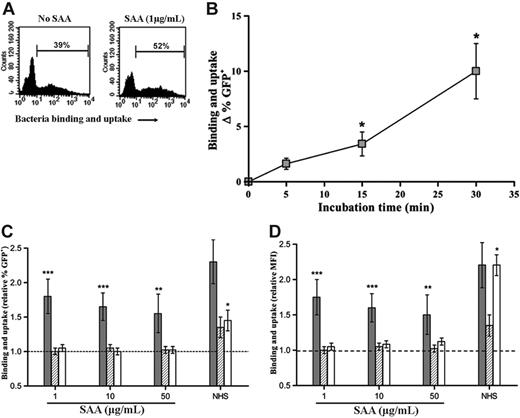
We initially examined neutrophil uptake of SAA-opsonized or untreated E coli-GFP (transformed with pEGFP). E coli-GFP bound [125I]-SAA to the same extent as untransformed bacteria (data not shown). SAA was added at concentrations similar to that found in normal plasma (1 μg/mL), at concentrations equivalent to the upper limit of normal (10 μg/mL), or at an acute-phase concentration (50 μg/mL). SAA at each concentration increased association (binding and uptake) of bacteria with neutrophils as measured by FACS. That this increase was caused by phagocytosis of bacteria was confirmed by cytochalasin D treatment, which prevented the increase in uptake of bacteria either by SAA or NHS. Increasing SAA concentrations above 1 μg/mL did not further enhance responses (Figure 2) consistent with maximal binding at this concentration. Internalization was also confirmed by real-time live confocal microscopy with E coli-GFP in the absence or presence of SAA (data not shown). The time course of uptake was examined and it was found that 30 minutes was an appropriate time to examine responses, although a significant increase could be seen as early as 15 minutes (Figure 2B). This uptake was due to an increase in both the number of neutrophils that internalized bacteria (Figure 2C) and the average number of bacteria per phagocytic cell (Figure 2D). The increase in uptake seen with SAA was of the same order of magnitude as that seen with human serum. In contrast, S pneumoniae that was effectively opsonized with NHS did not show any change in uptake with added SAA at any concentration. Thus SAA alone was not directly activating the neutrophil but only increasing phagocytosis of bacteria to which it was bound.
SAA binds to E coli and P aeruginosa but not B cepacia or S pneumoniae. Live E coli, P aeruginosa (7/1), S pneumoniae, and B cepacia (all at 108 cfu's) were incubated with SAA (5 μg/mL; filled histogram) or without (open histogram) and binding was examined by FACS following incubation with monoclonal anti-SAA and FITC-labeled secondary antibody. Data show analysis of 30 000 events.
SAA binds to E coli and P aeruginosa but not B cepacia or S pneumoniae. Live E coli, P aeruginosa (7/1), S pneumoniae, and B cepacia (all at 108 cfu's) were incubated with SAA (5 μg/mL; filled histogram) or without (open histogram) and binding was examined by FACS following incubation with monoclonal anti-SAA and FITC-labeled secondary antibody. Data show analysis of 30 000 events.
SAA opsonization of bacteria increases the neutrophil intracellular reactive oxygen intermediate response
Neutrophil intracellular reactive oxygen intermediate production was determined by measuring rhodamine fluorescence as a result of activation of NADPH oxidase. SAA alone had no effect on neutrophil respiratory burst compared with neutrophils incubated with medium alone (Figure 3A). The time course of reactive oxygen intermediate production indicated that 20 minutes was a suitable incubation period for assay of reactive oxygen intermediate, although effects of SAA on response to bacteria could be observed as early as 10 minutes (Figure 3B). SAA addition at the same time as E coli addition increased the percentage of neutrophils producing intracellular reactive oxygen intermediates compared with bacteria alone (Figure 3C). Reactive oxygen intermediate production was increased when SAA was present at 1 μg/mL, and a further significant increase (P < .05) was seen when SAA was added at 10 μg/mL, although the higher SAA concentration of 50 μg/mL yielded no further increase. In contrast, SAA addition with S pneumoniae did not increase responses over that with the bacteria alone.
SAA opsonization increases phagocytosis of Gram-negative bacteria by macrophages
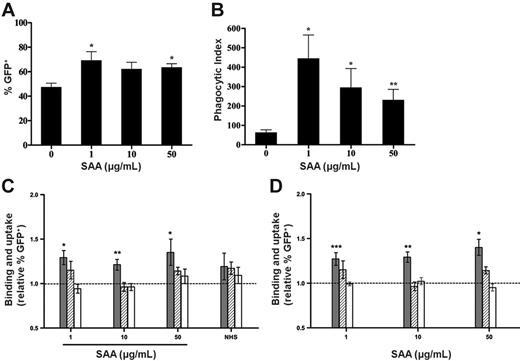
E coli-GFP association and phagocytosis by monocyte-derived macrophages was increased in the presence of SAA as demonstrated by counting macrophages associating with bacteria as well as internalized bacteria per macrophages by confocal microscopy (Figure 4A-B). Any apparent decrease in uptake at higher concentrations of SAA was not significant. The increase in phagocytosis of E coli-GFP by macrophages was also observed using FACS, while uptake of S pneumoniae was unaltered (Figure 4C). Cytochalasin D treatment reduced the increase in E coli association with macrophages following SAA addition, confirming internalization of bacteria. Previously we had determined that rSAA, which comprised SAA1 with 2 amino acid substitutions, was also able to bind to the same range of bacteria, although binding was weaker than observed for purified SAA1.21 rSAA at the same concentrations was able to cause a similar increase in phagocytosis of E coli but not S pneumoniae (Figure 4D). This confirmed that SAA does play a role as an opsonin. PKH67 as a fluorescent label for analysis of uptake of E coli by FACS produced results similar to that seen with E coli-GFP (data not shown). The failure of SAA to alter phagocytosis of S pneumoniae was consistent with SAA having an opsonic effect rather than directly activating macrophages.
SAA enhances neutrophil phagocytosis of E coli but not S pneumoniae. (A) Typical FACS profiles of E coli-GFP association with neutrophils in the absence or presence of SAA (1 μg/mL). Percentage of GFP+ cells within the region shown with bar were determined. (B) Time course of phagocytosis of E coli-GFP shows increases at approximately 15 minutes. Phagocytosis (difference in % GFP+ cells in the absence and presence of cytochalasin D) is expressed as Δ GFP+ in the presence of SAA (1 μg/mL) minus absence of SAA. (C-D) Binding and uptake of E coli-GFP ( ) by human neutrophils was increased in the presence of SAA at all concentrations tested. This response was due to phagocytosis, since no increase was seen when cytochalasin D treatment was included (
) by human neutrophils was increased in the presence of SAA at all concentrations tested. This response was due to phagocytosis, since no increase was seen when cytochalasin D treatment was included ( ). Phagocytosis of S pneumoniae-PKH67 (□) was not increased by SAA but was increased by NHS. (C) Relative ratio of percentage of GFP+/PKH67+ cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. (D) Relative ratio of mean fluorescence intensity (MFI) of cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 11 donors for E coli, 6 for E coli + cytochalasin D, and 4 for S pneumoniae. Statistical significance of samples with bacteria + SAA or NHS versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01; ***P < .001).
). Phagocytosis of S pneumoniae-PKH67 (□) was not increased by SAA but was increased by NHS. (C) Relative ratio of percentage of GFP+/PKH67+ cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. (D) Relative ratio of mean fluorescence intensity (MFI) of cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 11 donors for E coli, 6 for E coli + cytochalasin D, and 4 for S pneumoniae. Statistical significance of samples with bacteria + SAA or NHS versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01; ***P < .001).
SAA enhances neutrophil phagocytosis of E coli but not S pneumoniae. (A) Typical FACS profiles of E coli-GFP association with neutrophils in the absence or presence of SAA (1 μg/mL). Percentage of GFP+ cells within the region shown with bar were determined. (B) Time course of phagocytosis of E coli-GFP shows increases at approximately 15 minutes. Phagocytosis (difference in % GFP+ cells in the absence and presence of cytochalasin D) is expressed as Δ GFP+ in the presence of SAA (1 μg/mL) minus absence of SAA. (C-D) Binding and uptake of E coli-GFP ( ) by human neutrophils was increased in the presence of SAA at all concentrations tested. This response was due to phagocytosis, since no increase was seen when cytochalasin D treatment was included (
) by human neutrophils was increased in the presence of SAA at all concentrations tested. This response was due to phagocytosis, since no increase was seen when cytochalasin D treatment was included ( ). Phagocytosis of S pneumoniae-PKH67 (□) was not increased by SAA but was increased by NHS. (C) Relative ratio of percentage of GFP+/PKH67+ cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. (D) Relative ratio of mean fluorescence intensity (MFI) of cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 11 donors for E coli, 6 for E coli + cytochalasin D, and 4 for S pneumoniae. Statistical significance of samples with bacteria + SAA or NHS versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01; ***P < .001).
). Phagocytosis of S pneumoniae-PKH67 (□) was not increased by SAA but was increased by NHS. (C) Relative ratio of percentage of GFP+/PKH67+ cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. (D) Relative ratio of mean fluorescence intensity (MFI) of cells in region denoted by bar calculated from values obtained with bacteria + SAA or NHS over values obtained with bacteria alone. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 11 donors for E coli, 6 for E coli + cytochalasin D, and 4 for S pneumoniae. Statistical significance of samples with bacteria + SAA or NHS versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01; ***P < .001).
SAA opsonization enhances macrophage cytokine production
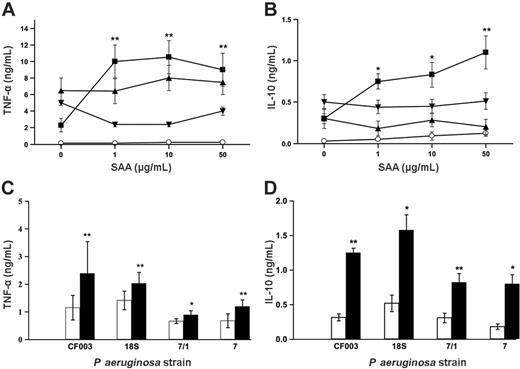
We investigated the ability of monocyte-derived macrophages to produce TNF-α and IL-10 in response to SAA-opsonized or untreated bacteria and SAA in the presence of bacteria that did not bind. All the concentrations of SAA induced a significant increase in both cytokines in response to E coli as shown in Figure 5A-B. In comparison, the control bacteria (S pneumoniae and B cepacia) to which SAA did not bind showed no increased responses. SAA alone generated no TNF-α response, although it did cause a minimal response in IL-10 release at higher concentrations. In order to show that this was a typical response to pathogenic bacteria, we investigated the response to treatment with 2 clinically derived strains of P aeruginosa (18S and CF003) as well as 2 laboratory strains (7 and 7/1). P aeruginosa alone induced very strong cytokine responses from macrophages. Therefore in order to test responses to SAA, donors were selected for low cytokine responses to bacteria alone. SAA addition 10 μg/mL to 107 cfu's/mL increased the macrophage release of cytokines above the response to bacteria alone. TNF-α and IL-10 showed significant increases in response to all of the strains of P aeruginosa (Figure 5C-D). Cytokine production in response to P aeruginosa at 108 cfu's/mL was also significantly increased by the addition of SAA (data not shown). Despite the fact that TNF-α and IL-10 are proinflammatory and anti-inflammatory, respectively, enhancement of both is typical of macrophage activation.
SAA increases neutrophil respiratory burst in response to E coli but not S pneumoniae. Activation of NADPH oxidase was determined by FACS analysis of rhodamine fluorescence (RHO+) in response to E coli or S pneumoniae in the absence or presence of SAA. (A) Typical FACS profiles of neutrophil respiratory burst when neutrophils are incubated with medium alone (control), SAA alone, E coli alone, or E coli + SAA (1 μg/mL). (B) Time course of reactive oxygen production expressed as percentage of RHO+ cells following incubation of neutrophils from a typical donor with E coli alone (▴) or E coli + SAA 1 μg/mL (▾), 10 μg/mL (▪), or 50 μg/mL (○). (C) Relative ratio of percentage of RHO+ cells in region denoted by bar calculated from values obtained with bacteria + SAA over values obtained with bacteria alone or values from SAA alone over values obtained with medium alone. SAA alone ( ) had no effect on activation of NADPH oxidase at any of the concentrations. SAA enhanced percentage of RHO+ neutrophils in response to E coli (▪) but not S pneumoniae (□). The dotted line represents unaltered respiratory burst in the absence of SAA. Data represent mean ± SEM of 10 donors for E coli and 8 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone, or bacteria with 1 μg/mL SAA versus bacteria with 10 μg/mL SAA, was calculated from raw percentage of RHO+ cells by Wilcoxon matched pair test (*P < .05).
) had no effect on activation of NADPH oxidase at any of the concentrations. SAA enhanced percentage of RHO+ neutrophils in response to E coli (▪) but not S pneumoniae (□). The dotted line represents unaltered respiratory burst in the absence of SAA. Data represent mean ± SEM of 10 donors for E coli and 8 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone, or bacteria with 1 μg/mL SAA versus bacteria with 10 μg/mL SAA, was calculated from raw percentage of RHO+ cells by Wilcoxon matched pair test (*P < .05).
SAA increases neutrophil respiratory burst in response to E coli but not S pneumoniae. Activation of NADPH oxidase was determined by FACS analysis of rhodamine fluorescence (RHO+) in response to E coli or S pneumoniae in the absence or presence of SAA. (A) Typical FACS profiles of neutrophil respiratory burst when neutrophils are incubated with medium alone (control), SAA alone, E coli alone, or E coli + SAA (1 μg/mL). (B) Time course of reactive oxygen production expressed as percentage of RHO+ cells following incubation of neutrophils from a typical donor with E coli alone (▴) or E coli + SAA 1 μg/mL (▾), 10 μg/mL (▪), or 50 μg/mL (○). (C) Relative ratio of percentage of RHO+ cells in region denoted by bar calculated from values obtained with bacteria + SAA over values obtained with bacteria alone or values from SAA alone over values obtained with medium alone. SAA alone ( ) had no effect on activation of NADPH oxidase at any of the concentrations. SAA enhanced percentage of RHO+ neutrophils in response to E coli (▪) but not S pneumoniae (□). The dotted line represents unaltered respiratory burst in the absence of SAA. Data represent mean ± SEM of 10 donors for E coli and 8 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone, or bacteria with 1 μg/mL SAA versus bacteria with 10 μg/mL SAA, was calculated from raw percentage of RHO+ cells by Wilcoxon matched pair test (*P < .05).
) had no effect on activation of NADPH oxidase at any of the concentrations. SAA enhanced percentage of RHO+ neutrophils in response to E coli (▪) but not S pneumoniae (□). The dotted line represents unaltered respiratory burst in the absence of SAA. Data represent mean ± SEM of 10 donors for E coli and 8 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone, or bacteria with 1 μg/mL SAA versus bacteria with 10 μg/mL SAA, was calculated from raw percentage of RHO+ cells by Wilcoxon matched pair test (*P < .05).
Monocyte-derived macrophages show increased phagocytosis of E coli-GFP in the presence of SAA. (A) E coli-GFP association and (B) phagocytosis were measured by confocal microscopy and counting of bacteria associated with or internalized by macrophages counterstained with LysoTracker and Phalloidin-BODIPY. Data represent (A) percentage of GFP+ macrophages associated to bacteria and (B) phagocytic index, which refers to the number of bacteria internalized per 100 macrophages. Data represent mean ± SEM of quadruplicates from 4 individual donors. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated by Wilcoxon matched pair test (*P < .05; **P < .01). (C-D) Binding and uptake or phagocytosis of E coli ( ) or E coli in the presence of cytochalasin D (
) or E coli in the presence of cytochalasin D ( ), or S pneumoniae (□) in the absence or presence of (C) purified SAA and (D) recombinant SAA (rSAA), was analyzed by FACS. Binding and uptake were expressed as the ratio of percentage of GFP+ macrophages in the presence of bacteria and SAA to that in the presence of bacteria alone. Increase in response to SAA was largely inhibited by the addition of cytochalasin D, implicating phagocytosis as the reason for increases in the presence of SAA. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 14 donors for E coli and 3 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01 ***P < .001).
), or S pneumoniae (□) in the absence or presence of (C) purified SAA and (D) recombinant SAA (rSAA), was analyzed by FACS. Binding and uptake were expressed as the ratio of percentage of GFP+ macrophages in the presence of bacteria and SAA to that in the presence of bacteria alone. Increase in response to SAA was largely inhibited by the addition of cytochalasin D, implicating phagocytosis as the reason for increases in the presence of SAA. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 14 donors for E coli and 3 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01 ***P < .001).
Monocyte-derived macrophages show increased phagocytosis of E coli-GFP in the presence of SAA. (A) E coli-GFP association and (B) phagocytosis were measured by confocal microscopy and counting of bacteria associated with or internalized by macrophages counterstained with LysoTracker and Phalloidin-BODIPY. Data represent (A) percentage of GFP+ macrophages associated to bacteria and (B) phagocytic index, which refers to the number of bacteria internalized per 100 macrophages. Data represent mean ± SEM of quadruplicates from 4 individual donors. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated by Wilcoxon matched pair test (*P < .05; **P < .01). (C-D) Binding and uptake or phagocytosis of E coli ( ) or E coli in the presence of cytochalasin D (
) or E coli in the presence of cytochalasin D ( ), or S pneumoniae (□) in the absence or presence of (C) purified SAA and (D) recombinant SAA (rSAA), was analyzed by FACS. Binding and uptake were expressed as the ratio of percentage of GFP+ macrophages in the presence of bacteria and SAA to that in the presence of bacteria alone. Increase in response to SAA was largely inhibited by the addition of cytochalasin D, implicating phagocytosis as the reason for increases in the presence of SAA. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 14 donors for E coli and 3 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01 ***P < .001).
), or S pneumoniae (□) in the absence or presence of (C) purified SAA and (D) recombinant SAA (rSAA), was analyzed by FACS. Binding and uptake were expressed as the ratio of percentage of GFP+ macrophages in the presence of bacteria and SAA to that in the presence of bacteria alone. Increase in response to SAA was largely inhibited by the addition of cytochalasin D, implicating phagocytosis as the reason for increases in the presence of SAA. The dotted line represents unaltered bacterial association in the absence of SAA. Data represent mean ± SEM of 14 donors for E coli and 3 for S pneumoniae. Statistical significance of samples with bacteria + SAA versus bacteria alone was calculated from raw percentage of GFP+ cells by Wilcoxon matched pair test (*P < .05; **P < .01 ***P < .001).
Depletion of SAA from human serum shows variable effect on phagocytosis
In order to address the question as to how much contribution SAA makes to the opsonic activity of serum, we chose to examine fresh normal human serum that had been depleted using 2 methods: first, an antibody to human SAA that would be expected to remove most of the HDL containing SAA1 and SAA2 but leave a considerable amount of HDL; and second, an HIC method that would remove all HDL and SAA including SAA4. This experiment therefore is examining the contribution of innate responses against a background of acquired immune response to E coli and, in particular, the known antibody response to OmpA, which is a major antigen of E coli and other Gram-negative bacteria.23,24 In Table 1 we show the amount of phagocytic activity for these SAA-depleted sera compared with the undepleted sera for cells from 2 individual donors as well as the mean for all 8 donors examined. SAA depletion for donors A and B led to a decrease in opsonic activity. Removal of SAA4 as well as SAA1, 2 (HIC SAA depletion) compared with depletion of only SAA1 and 2 (anti-SAA depletion) did not lead to further decrease in phagocytosis. Unexpectedly, donor C showed an increase in uptake following depletion with either method. Overall, despite the observation that most donors showed a decrease in uptake after SAA depletion, no significant decrease was seen. Thus innate SAA recognition does not always contribute significantly in the presence of an acquired response and/or other soluble innate opsonizing factors.
Effect of depletion of SAA from serum on opsonic activity
. | % GFP+ neutrophils . | . | . | ||
|---|---|---|---|---|---|
. | No depletion . | Anti-SAA depleted . | HIC depleted . | ||
| Donor A | 66 | 37 | 53 | ||
| Donor B | 16 | 11 | 12 | ||
| Donor C | 36 | 52 | 53 | ||
| Mean ± SEM, 8 donors | 43.0 ± 8.1 | 40.1 ± 6.2 | 40.8 ± 7.8 | ||
. | % GFP+ neutrophils . | . | . | ||
|---|---|---|---|---|---|
. | No depletion . | Anti-SAA depleted . | HIC depleted . | ||
| Donor A | 66 | 37 | 53 | ||
| Donor B | 16 | 11 | 12 | ||
| Donor C | 36 | 52 | 53 | ||
| Mean ± SEM, 8 donors | 43.0 ± 8.1 | 40.1 ± 6.2 | 40.8 ± 7.8 | ||
Normal serum or SAA-depleted normal serum was added to E coli–GFP and neutrophils for 30 minutes at 37°C, and phagocytosis was determined by FACS analysis of cells in the presence or absence of cytochalasin D. Phagocytosis was expressed as the difference between percentage of GFP+ cells with or without cytochalasin D
Discussion
In this report we have shown that SAA not only binds to many Gram-negative bacteria but, when it has done so, can induce a number of responses from both macrophages and neutrophils, which are responsible for rapid killing of invading bacterial pathogens. Phagocytosis is increased to an extent comparable with that seen by normal human serum that would be expected to have immunoglobulin, complement, and other factors capable of opsonizing the bacteria used here. This effect is therefore highly significant and equivalent to that seen with other opsonins, for instance CRP and S pneumoniae performed under identical conditions and reported elsewhere.25 It is in fact impressive that macrophages and neutrophils show a strong increase in uptake above the background, since even in the absence of other opsonins, macrophages have several receptors for phagocytosis of such common bacteria including scavenger receptors and C-type lectin receptors. We have shown that not only is phagocytosis increased for both cell types but neutrophils respond with increased production of reactive oxygen species consistent with their role in killing of bacteria. SAA opsonization would help in rapidly clearing bacteria by phagocytes, which is important primarily for an efficient response to combat infection. The bacteria used in this report represent only a few in the range of Gram-negative bacteria that bind SAA.21 Therefore it would be reasonable to predict similar uptake of other Gram-negative bacteria expressing the SAA ligand.
Monocyte-derived macrophages secrete increased amounts of cytokine in response to E coli and P aeruginosa in the presence of SAA. (A-B) Monocyte-derived macrophages were incubated with SAA alone (○) or γ-irradiated bacteria (at 107 cfu's/mL) in the absence or presence of SAA (1, 10, 50 μg/mL) for 16 hours and supernatants were assayed for (A) TNF-α and (B) IL-10. The bacteria used were E coli (▪), S pneumoniae (▾), and B cepacia (▴). Data represent mean ± SEM of 4 donors. Statistical significance between bacteria + SAA and bacteria alone was calculated by Student paired t test (*P < .05; **P < .01). (C-D) Monocyte-derived macrophages from donors with low cytokine responses to P aeruginosa alone were incubated with γ-irradiated P aeruginosa strains at 107 cfu's/mL in the absence (□) or presence (▪) of SAA (10 μg/mL) for 16 hours. Supernatants were assayed for (C) TNF-α and (D) IL-10. Data represent mean ± SEM of 5 donors. Statistical significance between bacteria + SAA and bacteria alone was calculated by Student paired t test (*P < .05; **P < .01).
Monocyte-derived macrophages secrete increased amounts of cytokine in response to E coli and P aeruginosa in the presence of SAA. (A-B) Monocyte-derived macrophages were incubated with SAA alone (○) or γ-irradiated bacteria (at 107 cfu's/mL) in the absence or presence of SAA (1, 10, 50 μg/mL) for 16 hours and supernatants were assayed for (A) TNF-α and (B) IL-10. The bacteria used were E coli (▪), S pneumoniae (▾), and B cepacia (▴). Data represent mean ± SEM of 4 donors. Statistical significance between bacteria + SAA and bacteria alone was calculated by Student paired t test (*P < .05; **P < .01). (C-D) Monocyte-derived macrophages from donors with low cytokine responses to P aeruginosa alone were incubated with γ-irradiated P aeruginosa strains at 107 cfu's/mL in the absence (□) or presence (▪) of SAA (10 μg/mL) for 16 hours. Supernatants were assayed for (C) TNF-α and (D) IL-10. Data represent mean ± SEM of 5 donors. Statistical significance between bacteria + SAA and bacteria alone was calculated by Student paired t test (*P < .05; **P < .01).
It has previously been shown that HDL does not alter the binding of SAA to bacteria21 and SAA binds when presented as serum, thus the response described here would be expected to occur in vivo. The fact that SAA1 works effectively at the concentrations found in normal serum without the need for acute-phase concentrations appears consistent for an innate protection mechanism that needs to be effective without a time lag for induction by inflammatory stimuli. We attempted to deplete fresh human serum of SAA in order to examine the contribution of SAA to total serum opsonic activity. SAA depletion led to a slight but not significant reduction in uptake overall, but when examined individually, responses were heterogeneous. This is not surprising because of a number of factors. First, OmpA is an immunodominant antigen23,24 and all samples that were available to us would be expected to have antibody to OmpA, thus effects seen will be due to the balance between SAA's ability to reduce antibody binding and its own positive opsonic effect. There are no data on the epitopes of OmpA that are bound by SAA and antibody, but for antibody at least the epitopes may be variable. Second, responses of individuals to SAA opsonization are themselves heterogeneous. Third, the depletion of SAA may presumably also remove other HDL components, such as LPS binding protein, which may affect responses. Depletion of SAA1 led to serum with 50 to 100 ng/mL remaining in normal serum, which would lead to reduced binding but not completely prevent it.21 HDL itself also has considerable ability to downregulate inflammatory responses, although the mechanism is not clear and acute-phase or normal HDL differ in not only relative amounts of associated proteins but also anti-inflammatory activity.26 Finally, it is unclear as yet whether other SAA isoforms bind in addition to SAA1. Our studies were restricted to SAA1 and a chimeric form of SAA1 (recombinant SAA), which contains some features of SAA2 as mentioned in “SAA and binding studies.” SAA2, which normally occurs at lower concentrations than SAA1, differs by only approximately 5 to 7 residues from SAA1, and all 4 isoforms have 2 regions of high homology in the central and C-terminal regions of the protein. We are as yet unsure whether SAA4 binds to bacteria and have yet to develop a system that allows the examination of this protein in cellular response assays due to difficulty in handling this protein. Thus to accurately assess the contribution of SAA to innate recognition, further studies need to be carried out in a model that examines a naive individual with no humoral response to the chosen bacteria.
Both macrophages and neutrophils were demonstrated to show altered states of activation following SAA-mediated phagocytosis. The time required for responses was shown to be as rapid as 15 minutes following addition of SAA and bacteria, showing that cells could respond rapidly to this stimulus. Macrophage responses examined were over a longer time course. Macrophages increased cytokine production consistent with their major role in driving immune responses. This suggests that the subsequent immune response to the bacteria will be altered by opsonization with SAA. The observed cellular activation may be due to direct activation of a receptor by SAA, or alternatively SAA-mediated opsonization may merely act to enhance phagocyte binding and uptake, with subsequent increased responses due to increased recognition of various bacterial PAMPs. None of the candidate receptors6,13-16 have yet been shown to cause both phagocytosis and activation of the macrophage and neutrophil responses described here. SAA can however be taken up by endocytosis through scavenger receptor-B1 (CLA-1 human orthologue) and has been shown to activate MAP kinase pathways known to be involved in macrophage activation.13,14 It should not be assumed that other activatory responses of neutrophils or macrophages will necessarily be induced by SAA-opsonized bacteria. For instance, we did not observe increased calcium signaling or IL-8 release from neutrophils, which are features of FPRL-1-mediated responses.
It was significant that normal SAA concentrations were sufficient to cause a strong response. This is expected because SAA binds maximally at such concentrations.21 In addition, if SAA is to have an innate function, then it would be needed during the first few hours of exposure to pathogen. Following an inflammatory stimulus, it takes approximately 24 hours for SAA to reach acute-phase levels because of the requirement for de novo synthesis of both cytokine inducer and acute-phase protein. The role of the acute-phase increase in SAA concentration may be to ensure adequate extravasation to tissue sites, although a separate unconnected function is equally possible. Although the most obvious site of immune protection would be in blood, it is clear that our observations also provide new potential meaning to previous reports concerning epithelial synthesis of SAA. These reports showed that SAA synthesis in the normal noninflamed state was largely by epithelial cells,3,27 particularly in the small and large intestine; that SAA was induced in mouse intestinal epithelium in response to normal mucosal enteric bacteria28 ; and that intestinal epithelial cell lines produced significant amounts of SAA during an acute-phase response.29 Our data indicate that this synthesis, at the site where there is need for protection against a range of Gramnegative bacteria, is likely to be significant. SAA is likely to have a role in maintenance of the balance of inflammation and immunity in the gut in the presence of commensal or pathogenic bacteria. SAA may also be of importance in milk where it has been shown to be present particularly following local inflammation. Not only is SAA1 synthesized but SAA3, which was thought to be a pseudogene, has now been reported to be expressed in human mammary epithelia.30
OmpA has been suggested to act as a PAMP that has been shown to have properties of an adjuvant carrier protein that activates through a gp96-like receptor.31-33 SAA might be expected to contribute to or alter these effects. SAA may also influence the observed interaction with OmpA of pathogenic E coli and the blood-brain endothelial layer.34 In addition, SAA may bind to vesicles containing OmpA released from Gram-negative bacteria into serum of sepsis patients.35,36
As yet no reports have identified a human deficiency of serum amyloid A, and its synthesis in response to inflammatory stimuli is only moderately blunted in malnutrition. As previously stated, there are several SAA genes and no information has yet been gained about the ability of these or mouse SAA isoforms to bind bacteria and/or act as an opsonin. It will be practical to attempt meaningful in vivo experiments only when binding and opsonic properties of murine SAA isoforms have been determined.
Thus SAA has the ability to opsonize Gram-negative bacteria and induce activatory and protective responses from macrophages and neutrophils. These are features of a pattern-recognition protein and thus we report that SAA is an innate pattern-recognition protein with specificity for the bacterial OmpA protein family.
Prepublished online as Blood First Edition Paper, May 25, 2006; DOI 10.1182/blood-2005-11-011932.
Supported in part by a Medical Research Council (MRC) studentship (C.S.).
R.H.-D. contributed to the SAA-bacterial binding experiments and cytokine production; C.S. contributed to the phagocytosis, respiratory burst, and cytokine experiments; and J.G.R. was responsible for funding and direction of the project.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal