The stem cell leukemia gene SCL, also known as TAL-1, encodes a basic helix-loop-helix transcription factor expressed in erythroid, myeloid, megakaryocytic, and hematopoietic stem cells. To be able to make use of the unique tissue-restricted and spatio-temporal expression pattern of the SCL gene, we have generated a knock-in mouse line containing the tTA-2S tetracycline transactivator under the control of SCL regulatory elements. Analysis of this mouse using different tetracycline-dependent reporter strains demonstrated that switchable transgene expression was restricted to erythrocytes, megakaryocytes, granulocytes, and, importantly, to the c-kit-expressing and lineage-negative cell fraction of the bone marrow. In addition, conditional transgene activation also was detected in a very minor population of endothelial cells and in the kidney. However, no activation of the reporter transgene was found in the brain of adult mice. These findings suggested that the expression of tetracycline-responsive reporter genes recapitulated the known endogenous expression pattern of SCL. Our data therefore demonstrate that exogenously inducible and reversible expression of selected transgenes in myeloid, megakaryocytic, erythroid, and c-kit-expressing lineage-negative bone marrow cells can be directed through SCL regulatory elements. The SCL knock-in mouse presented here represents a powerful tool for studying normal and malignant hematopoiesis in vivo.
Introduction
The basic helix-loop-helix transcription factor stem cell leukemia (SCL) (also known as TAL-1 or TCL5) was originally identified by virtue of a chromosomal translocation associated with acute human lymphoblastic leukemia.1-3 In addition to its involvement in leukemia, loss-of-function studies in mice demonstrated an essential role of SCL for the specification of mesoderm to primitive and definitive blood cell formation (reviewed in Begley and Green4 and Lecuyer and Hoang5 ). The absolute requirement for SCL expression during early embryonic development has led to the view that SCL acts as a master regulator of blood cell formation.6 Furthermore, conditional gene targeting of SCL in adult mice not only has revealed a regulatory function of SCL in both erythropoiesis and megakaryopoiesis,7-9 but also has suggested that SCL function is not required for self-renewal or long-term repopulation capacity of hematopoietic stem cells (HSCs). Within blood cell lineages, SCL expression has been reported in granulocytic, erythroid, megakaryocytic, and HSC/progenitor populations.4,5
Human and murine SCL genes are transcribed from 3 distinct lineage-specific promoters leading to a complex pattern of differentially spliced transcripts.10-16 DNase I hypersensitivity mapping, restriction endonuclease accessibility assays, and functional in vitro experiments revealed several enhancer and silencer elements within the SCL genomic locus.17 In addition, reporter mice were used to identify distinct regulatory elements of the SCL locus responsible for directing expression to specific subdomains of the endogenous SCL expression pattern.18-24 Complementary studies examining the expression of a lacZ reporter knocked into exon III of the SCL gene locus provided evidence that SCL regulatory elements can direct expression of the lacZ transgene to progenitors of lymphoid, erythroid, and myeloid lineages.25 Analysis of SCL lacZ knock-in embryos further revealed expression of the reporter gene in parts of the central nervous system, the vascular endothelium, and in primitive and definitive blood cells.26 These findings, together with the loss-of-function data, suggest that SCL regulatory elements are active in HSCs and blood progenitors and that this activity is selectively maintained during ontogeny in myeloid, erythroid, megakaryocytic, and HSCs/progenitors but extinguished in all other mature blood cell lineages.
To be able to reversibly express transgenes in SCL-positive blood cells, we have made use of the tetracycline regulatory system.27 Tetracycline-mediated control of transgenes has become an excellent strategy for studying gene function in mice (Gossen and Bujard28 and Bockamp et al29 ). Since transgene expression in these animals is exclusively dependent on the administration/absence of tetracycline or tetracycline derivatives,30 the function of any gene product can be studied during selected developmental windows or at critical stages of disease. Furthermore, inducible expression of toxic genes can be used to ablate selected cell populations in vivo, allowing direct studies of the function of the targeted cells and the creation of conditional disease models.31 The unique experimental potential of tet on/off mouse models for approaching crucial questions about normal and malignant blood cell development is illustrated by numerous reports investigating the in vivo function of conditionally expressed transgenes.32-41 In these reports, the combination of a tissue-specific effector with a responder mouse was used to express selected genes in a tetracycline-controlled fashion.
For studying the etiology of hematologic malignancies and, in particular, leukemias, the ability to control gene function in vivo is a major advantage, since reversible induction can reveal whether transgene expression is needed for initiation, progression, maintenance, or remission of the disease. In addition, for several leukemias, distinct oncogenes or leukemia-associated factors have been reported to be already expressed in HSCs or blood cell progenitors.42,43 This observation, together with the obvious similarity between stem cells and cancer cells, has led to the emerging concept of the leukemic stem cell.44,45 Research focusing on the role of leukemic stem cells would therefore greatly benefit from mouse models allowing the reversible induction of oncogenes and/or leukemia-associated factors in HSCs or blood cell progenitors.
To be able to reversibly target the expression of transgenes to SCL-positive cells, we have generated an SCL tTA-2S knock-in mouse. Detailed analysis of this mouse demonstrated that in hematopoietic tissues tetracycline-mediated transgene expression was completely restricted to myeloid, megakaryocytic, and erythroid cells, and, most importantly, to c-kit-expressing lineage-negative cells of the bone marrow. In addition, conditional transgene expression also was found in a very minor fraction of platelet endothelial cell adhesion molecule 1 (PECAM-1)-expressing endothelial cells and in a subset of cells in the kidney. However, no induction of transgenes was detected in histologic brain sections. These findings suggest that the SCL tTA-2S knock-in mouse recapitulates the known endogenous expression pattern of SCL. The SCL knock-in mouse presented here therefore represents an excellent model for studying controlled gene expression in SCL-positive blood cells and, most importantly, to conditionally direct expression of selected gene products to c-kit+/lin- hematopoietic cells of the bone marrow.
Materials and methods
Construction of the targeting vector
The murine genomic SCL locus was obtained by screening a 129/Sv lambda phage library. A 4.2-kb fragment upstream of SCL exon V was used as the 5′ homology arm and an 8.1-kb fragment downstream of the unique XbaI site in exon VI as the 3′ homology arm and cloned into pGem11 ZF+ (Promega, Madison, WI). All ATG codons of exon IV and the first ATG codon in exon V were changed to GGG codons, thus preventing translational initiation from these sites. The unique Not I recognition site in exon V was used for insertion of the tTA-2S transactivator,46 followed by the bovine growth hormone polyA signal and a loxP-flanked neomycin-resistant cassette under the control of the Herpes simples virus TK promoter (Figure 1A). All modified sequences were confirmed by sequence analysis.
Animals
The W9.5 embryonic stem (ES) cell line47 was electroporated with the linearized targeting vector. G-418-resistant single clones containing the correctly recombined locus were injected into blastocysts and transferred into pseudopregnant mothers following standard procedures.48 Successful germ-line transmission and correct integration was confirmed by Southern blotting using an 800-bp fragment upstream of SCL exon Ia as a 5′ outside probe and a 1025-bp polymerase chain reaction (PCR) fragment as an inside probe to confirm correct integration. The 800-bp 5′ probe was excised by Hind III digestion of the -2000 SCL Ia pGL-2 plasmid,13 and the 3′ probe was generated by PCR using oligonucleotide 5′-CCTCAGAAGCTGTCACTGTGTC-3′ as a forward and oligonucleotide 5′-TTGCTCAGGGACTTTACTGTCAG-3′ as a reverse primer. For in vivo excision of the neomycin-resistant cassette, germ-line-transmitting SCL-TA-2S knock-in mice were crossed to the SYCP-Cre deleter line.49 Successful excision of the cassette was confirmed by using a 3-primer PCR approach with the oligonucleotides 5′-TGGCCAAGTTACTCAATGACC-3′ and 5′-GGAAGTATCAGCTCGACCAA-3′ as forward primers and the 5′-GGATGGATCAACATGGACCT-3′ oligonucleotide as reverse primer.
Genotyping of mice
For genotyping of the SCL-tTA-2S knock-in mouse primers 5′-CCCTGCTCGATGCCCTGGC-3′ and 5′-AGGAAGGCAGGTTCGGCTCC-3′ were used. The LC-1 mouse was typed using primers 5′-CCGTACACCAAAATTTGCCTGC-3′ and 5′-GAACATCTTCAGGTTCTGCGGG-3′. The EGFP-lacZ tetracycline-responsive responder mouse was typed using primers 5′-CTCAAGTTCATCTGCACCACC-3′ and 5′-CGTTCTTCTGCTTGTCGGCC-3′.
Luciferase assays
Organs from adult mice were dissected, extracted, and assayed for luciferase activity as described.53 Luciferase activity was normalized against the amount of 10 μg protein. A linear relationship between light units and volume was confirmed in all experiments. Luciferase values in the presence and without doxycycline (DOX) were obtained in each case from at least 3 different animals producing a similar pattern of activity.
Collagenase treatment
Dissected tissues were digested at 37°C for 40 minutes in phosphate buffered saline (PBS) (pH 7.4) containing 0.5 μg/mL collagenase together with 50 units DNase I per mL (both Sigma, St Louis, MO) and subsequently subjected to fluorescence activated cell sorting (FACS) analysis.
FACS analysis and cell sorting
Lineage contribution of EGFP-marked blood cells was analyzed with a 4-color-equipped FACSCalibur (Becton Dickinson [BD], San Jose, CA) by co-staining with phycoerythrin (PE)-conjugated antibodies against CD11b, CD19, Gr-1, TER119 (BD), CD3, CD11c, DX5 (Caltag, Burlingame, CA), CD23 (Southern Biotech, Birmingham, AL) or with purified antibodies against CD41 (BD) detected with anti-rat-PE (Caltag). Collagenase-treated suspensions of peripheral organs were simultaneously incubated with an endothelial-specific PECAM-1 rat monoclonal antibody (CD31, BD) and a mix of TER119/CD45 antibodies (BD). Prior to staining, the samples (not the samples stained with secondary reagents) were blocked with PBS supplemented with 5% rat serum for 10 minutes. Dead cells were excluded from analysis via 7AAD staining (BD). Detection levels over background were confirmed for the PECAM-1 antibody in parallel control experiments using a rat PE-conjugated IgG 2A isotype control antibody (BD). The stem cell fraction was defined by lin-PE- and c-kit+APC (CD117, BD) staining. Data were analyzed using the CellQuest Pro software (BD). In all cases the lineage contribution of EGFP-expressing cells was determined in 3 independent experiments, analyzing each time a minimum of 5 × 105 cells.
Preparative FACS sorting of lin- c-kit+ cells was performed using a FACS Vantage SE Turbo (BD). Lin+ cells were first depleted from the femoral mononuclear population using a magnetic affinity lineage depletion kit (MACS, Miltenyi Biotech, Auburn, CA). The lineage-depleted fraction was then stained with c-kit-APC antibody and the c-kit+ population sorted simultaneously into EGFP+ and EGFP- fractions. Because of the small number of lin- c-kit+ cells available, the EGFP sort gates were preset using mononuclear cells from DOX-treated and untreated mice.
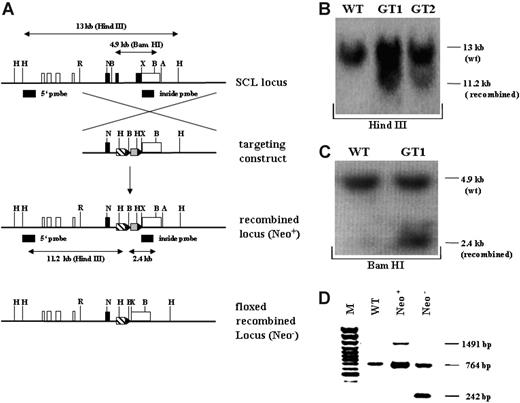
Targeting strategy and confirmation of the recombined SCL genomic locus. (A) Schematic overview of the targeting strategy. In the upper representation the SCL wild-type genomic locus is shown. Coding exons (IV, V, and VI) are depicted as black, and noncoding exons (Ia, Ib, IIb, III, and part of VI) are depicted as white boxes. The targeting construct is shown below the SCL genomic locus, consisting of 2 homology arms, the tTA-2S coding sequence (striped box), and the floxed neomycin-resistant selection cassette (gray box). In the targeting construct all ATG codons in exon IV and the first ATG in exon V were changed to GGG codons. LoxP Cre-recombinase recognition sites flanking the neomycin cassette are indicated as black triangles. Below the targeting construct the recombined mutant SCL locus is shown still containing the neomycin cassette (Neo+). At the bottom of the representation the recombined SCL locus is depicted after excision of the neomycin cassette (Neo-). H indicates Hind III; R, EcoRI; N, Not I; X, XbaI;A, ApaI, and B, BamHI. (B) 5′ confirmation of the recombined SCL locus by Southern blotting using a specific outside probe. Digestion with Hind III of wild-type (WT) DNA gives rise to a 13-kb fragment, whereas the correctly recombined locus will result in a smaller 11.2-kb fragment (GT1 and GT2, germ-line-transmitting mouse founder line 1 and 2). (C) 3′ confirmation of the recombined SCL locus by Southern blotting. BamHI digestion of genomic DNA followed by hybridization with an inside probe produces a 4.9-kb fragment for the wild-type allele (WT) and a 2.4-kb fragment for the mutant knock-in allele (GT1). (D) In vivo excision of the neomycin-resistant cassette. PCR was used to verify the excision of the neomycin-resistant cassette from the germ-line of the SCL tTA-2S knock-in mouse. The recombined SCL locus still containing the cassette will produce a 1491-bp amplification product (Neo+). After excision of the neomycin cassette the same primers will amplify a 242-bp fragment (Neo-). The 764-bp amplification product is specific for the SCL wild-type allele.
Targeting strategy and confirmation of the recombined SCL genomic locus. (A) Schematic overview of the targeting strategy. In the upper representation the SCL wild-type genomic locus is shown. Coding exons (IV, V, and VI) are depicted as black, and noncoding exons (Ia, Ib, IIb, III, and part of VI) are depicted as white boxes. The targeting construct is shown below the SCL genomic locus, consisting of 2 homology arms, the tTA-2S coding sequence (striped box), and the floxed neomycin-resistant selection cassette (gray box). In the targeting construct all ATG codons in exon IV and the first ATG in exon V were changed to GGG codons. LoxP Cre-recombinase recognition sites flanking the neomycin cassette are indicated as black triangles. Below the targeting construct the recombined mutant SCL locus is shown still containing the neomycin cassette (Neo+). At the bottom of the representation the recombined SCL locus is depicted after excision of the neomycin cassette (Neo-). H indicates Hind III; R, EcoRI; N, Not I; X, XbaI;A, ApaI, and B, BamHI. (B) 5′ confirmation of the recombined SCL locus by Southern blotting using a specific outside probe. Digestion with Hind III of wild-type (WT) DNA gives rise to a 13-kb fragment, whereas the correctly recombined locus will result in a smaller 11.2-kb fragment (GT1 and GT2, germ-line-transmitting mouse founder line 1 and 2). (C) 3′ confirmation of the recombined SCL locus by Southern blotting. BamHI digestion of genomic DNA followed by hybridization with an inside probe produces a 4.9-kb fragment for the wild-type allele (WT) and a 2.4-kb fragment for the mutant knock-in allele (GT1). (D) In vivo excision of the neomycin-resistant cassette. PCR was used to verify the excision of the neomycin-resistant cassette from the germ-line of the SCL tTA-2S knock-in mouse. The recombined SCL locus still containing the cassette will produce a 1491-bp amplification product (Neo+). After excision of the neomycin cassette the same primers will amplify a 242-bp fragment (Neo-). The 764-bp amplification product is specific for the SCL wild-type allele.
CAFC assay
The cobblestone area-forming cell (CAFC) assay was performed essentially as described.54,55 Briefly, the lin- c-kit+ EGFP+, lin- c-kit+ EGFP-, and the whole mononuclear cell populations were counted, then titrated through serial dilutions onto established OP-9 stromal feeder layers, each cell concentration being represented by 20 independent wells. Cultures were fed by refreshing half of the medium weekly. All wells were scored for the presence of cobblestone areas (groups of 5 or more hematopoietic cells growing underneath the stromal layer) at day 14 and day 35 of culture, and the frequency of CAFCs calculated using Poisson statistics.
Controlled expression of transgenes
To exogenously switch the expression of luciferase, EGFP, and β-galactosidase in tTA-2S-SCL/LC-1 or tTA-2S-SCL/EGFP-lacZ tetracycline-responsive mice, animals were either provided with normal drinking water (reporter gene expression on) or fed a solution of 7.5 mg DOX (Sigma)/mL water containing 1% sucrose (reporter gene expression off).
Immunofluorescence and X-gal staining
Mice were killed by cervical neck dislocation and organs snap frozen in isopenthane. Cryostat sections (5-12 μm) were fixed in 100% acetone at 4°C for 1 hour, air dried, and stained for β-galactosidase by washing twice in PBS (pH 7.4), followed by overnight incubation at 37°C in X-gal solution (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 1 mg/mL X-gal in PBS). To visualize endothelial cells, sections were incubated with a purified rat anti-mouse CD31 monoclonal antibody against PECAM-1 (BD), followed by a second biotin-conjugated goat anti-rat Ig-specific polyclonal antibody (BD) using the Renaissance TSA fluorescence system (Perkin Elmer Life Sciences, Shelton, CT). Images were captured using a color view digital camera running on an Olympus BX50 WI microscope (Olympus, Hamburg, Germany) and a 20×/0.50 numeric aperture objective. Images were captured using a Color View 12 digital charge-coupled device (CCD) camera (Olympus). Images were digitalized using the analySIS software package 3.1 (Soft Image Systems, Münster, Germany) and imported into Adobe Photoshop 4.0 (Adobe Systems, San Jose, CA). In all cases, electronic adjustments were applied to the whole image.
β-Galactosidase expression and Cre expression in the brains of mice were analyzed as described.51
Results
Generation of the SCL tTA-2S knock-in mouse
To conditionally express transgenes under the control of SCL regulatory elements, gene targeting was used to insert the coding sequence for the tTA-2S transactivator46 into exon V of the SCL gene locus. We selected insertion of tTA-2S into exon V to ensure that all known SCL regulatory elements were present in the recombined locus.12-23,56,57 Figure 1A shows a schematic representation of the targeting strategy. Correct homologous recombination in ES cells and germ-line transmission was confirmed by Southern blotting (Figure 1B,C). Consistent with the introduction of 2 novel HindIII sites in the recombined locus, an 11.2-kb band was detected in addition to the 13-kb wild-type band after digestion of genomic DNA from the germ-line-transmitting founder animals and hybridization with the 5′ outside probe (Figure 1B). Similarly, correct 3′ recombination was confirmed by BamHI digestion of genomic DNA, followed by Southern hybridization with an inside probe. As shown in Figure 1C in the germ-line-transmitting founder GT1, the expected 2.4 kb was detected in addition to the 4.9-kb wild-type specific band (see also the schematic representation of the expected fragments in Figure 1A). Correct recombination was further confirmed for the overlap between the 3′ targeting arm and the adjacent genomic SCL locus using 2 additional probes (data not shown). Taken together, Southern blot analysis of the germ-line-transmitting founder GT1 demonstrated correct homologous recombination into the SCL locus.
To completely exclude unwanted transcriptional interference effects from the TK promoter governing the expression of the neomycin-resistant cassette, this cassette was removed from the recombined SCL locus by in vivo excision using the SYCP-Credeleter mouse line.49 Successful excision of the floxed neomycin-resistant cassette was confirmed by PCR. As shown in Figure 1D, removal of the floxed cassette resulted in a 242-bp PCR product (lane Neo-). By contrast, the recombined locus still containing the neomycin-resistant cassette produced a 1491-bp PCR product (lane Neo+). A 764-bp product specific for the wild-type SCL locus was detected both in wild-type (lane WT) and rearranged mice (lanes Neo+ and Neo-), indicating the presence of at least one SCL wild-type allele. For all subsequent experiments heterozygous SCL-tTA-2S mice lacking the neomycin-resistant cassette were used (homozygous SCL-tTA-2S knock-in mice were embryonic lethal, data not shown).
Tissue-specific expression of transgenes with the SCL tTA-2S knock-in mouse is completely dependent on DOX
The schematic representation in Figure 2A illustrates the DOX-dependent regulatory strategy used here. As shown in Figure 2A, in the presence of DOX the tTA-2S transactivator does not bind to the tetO binding sequence and, thus, transgene expression is not initiated. Conversely, in the absence of DOX tTA-2S homodimers will bind to the tetO sequence upstream of the cytomegalovirus (CMV) minimal promoter, resulting in transcriptional activation of the luciferase transgene.
SCL expression in the adult is mainly restricted to hematopoietic tissues.4,5 In addition, the presence of a small number of SCL-positive cells also has been reported for the adult kidney.58 To evaluate if the SCL-tTA-2S effector mouse also will direct conditional expression of transgenes to these cells, SCL-tTA-2S knock-in effector mice were crossed to the LC-1 reporter mouse line.50 In this mouse the luciferase gene is under the control of a tetracycline-responsive promoter element. As expected, extracts prepared from different organs of bitransgenic SCL-tTA-2S/LC-1 mice, kept in the presence of DOX, did not show luciferase activity (bottom bar graph +DOX in Figure 2B, luciferase off). By contrast, high levels of luciferase activity were detected in bone marrow and spleen of bitransgenic littermates that were never exposed to DOX (upper bar graph -DOX in Figure 2B, luciferase on). In addition, lower luciferase activity was found in the thymus of induced animals. Interestingly, extracts prepared from brain, heart, kidney, liver, lung, tongue, esophagus, and pancreas also exhibited luciferase activity over background, suggesting the presence of tTA-2S-expressing cells in these tissues. No substantial luciferase activity was detectable in the salivary gland, the stomach, the small and large intestine, the muscle, or the lymph nodes. These results demonstrated that the SCL-tTA-2S effector mouse induced reporter gene activity in adult hematopoietic tissues and that this expression was strictly dependent on DOX (compare luciferase activity between bitransgenic mice in the presence and absence of DOX in Figure 2B). The observed high levels of luciferase activity in bone marrow and spleen were expected, as SCL is known to be expressed in these tissues. The low luciferase activity in the thymus is probably explained by the presence of a minor population of CD8/CD4 double-negative and/or positive thymocytes or other cells of hematopoietic origin. Whether the somewhat unexpected luciferase activity in brain, heart, liver, lung, tongue, esophagus, and pancreas represented organ-specific activation of the reporter gene or was the result of tTA-2S expressing circulating blood and/or endothelial cells could not be addressed at this point. Finally, the detected luciferase activity in the kidney was in line with the published expression of SCL in this organ.58
Tissue-specific induction of the luciferase transgene is completely DOX-dependent. (A) Schematic representation of the tetracycline regulatory system. Restriction endonuclease recognition sites are as in Figure 1. DOX indicates doxycycline; tTA-2S, tetracycline-dependent transactivator; tetO, DNA-binding consensus for tTA-2S homodimers; pCMV, human cytomegalovirus minimal promoter; pA, polyA signal. (B) Luciferase activity expressed as relative light units (RLU) per microgram of protein extract was determined for different organs as indicated. The top bar graph shows luciferase activities of double heterozygous SCL-tTA-2S/LC-1 mice in the absence of DOX (-DOX, luciferase on). The bottom bar graph represents luciferase values obtained from double-transgenic SCL-tTA-2S/LC-1 mice that were kept from conception onwards in the presence of DOX (+DOX, luciferase off). The luciferase values in each graph are shown for a single bitransgenic mouse. A similar pattern of activity was obtained also in 2 additional independent experiments using different mice.
Tissue-specific induction of the luciferase transgene is completely DOX-dependent. (A) Schematic representation of the tetracycline regulatory system. Restriction endonuclease recognition sites are as in Figure 1. DOX indicates doxycycline; tTA-2S, tetracycline-dependent transactivator; tetO, DNA-binding consensus for tTA-2S homodimers; pCMV, human cytomegalovirus minimal promoter; pA, polyA signal. (B) Luciferase activity expressed as relative light units (RLU) per microgram of protein extract was determined for different organs as indicated. The top bar graph shows luciferase activities of double heterozygous SCL-tTA-2S/LC-1 mice in the absence of DOX (-DOX, luciferase on). The bottom bar graph represents luciferase values obtained from double-transgenic SCL-tTA-2S/LC-1 mice that were kept from conception onwards in the presence of DOX (+DOX, luciferase off). The luciferase values in each graph are shown for a single bitransgenic mouse. A similar pattern of activity was obtained also in 2 additional independent experiments using different mice.
Histologic and flow-cytometric analysis of transgene induction in peripheral organs
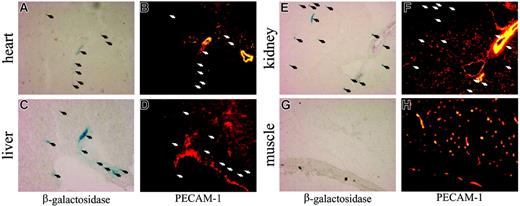
In the adult, SCL is restricted to hematopoietic cells and the kidney.4,5,58 In addition, expression of endogenous SCL in endothelial cells has been described for the early embryo, the vasculature of tumors, and the lining of newly arising blood vessels but is absent in quiescent adult vasculature.59-63 Intriguingly, lysates obtained from SCL-tTA-2S/LC-1 mice exhibited luciferase activity in heart, liver, lung, tongue, esophagus, and pancreas (Figure 2B). To clarify, if transgene induction in the SCL-tTA-2S knock-in mouse was due to endogenous organ-specific expression or reflected the presence of circulating blood cells and/or resident endothelial cells, SCL tTA-2S knock-in mice were mated to EGFP-lacZ tetracycline-responsive reporter mice.51 The resulting bitransgenic SCL tTA-2S/EGFP-lacZ mice were either kept from conception onwards in the presence of DOX (reporter gene off) or kept on normal drinking water (reporter gene on). At the age of 6 to 8 weeks organs from these mice were subjected to histologic analysis. As shown in the left panel of Figure 3, heart, liver, and kidney of bitransgenic SCL tTA-2S/EGFP-lacZ mice harbored blue β-galactosidase-expressing cells, consistent with the previously detected luciferase activity in these organs. No β-galactosidase activity was detected in bitransgenic animals permanently kept in the presence of DOX (data not shown) or in muscle (Figure 3G). Immunofluorescence analysis for the endothelial-specific PECAM-1 marker further revealed that β-galactosidase-expressing cells typically did not colocalize with PECAM-1-positive endothelial populations (Figure 3, right panel). These results indicated that in the analyzed organs transgene expression was in general not directed to endothelial cells.
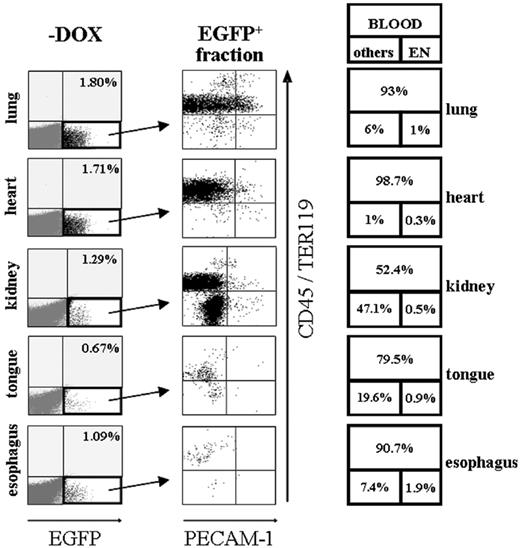
To analyze transgene-expressing cells of different organs more precisely, dissected tissues from induced and noninduced SCL-tTA-2S/EGFP-lacZ bitransgenic mice were treated with collagenase and the resulting cell suspensions examined by FACS. Major advantages of this strategy are that large numbers of cells can be tested and that each individual cell can be simultaneously analyzed for the presence of several different tissue-specific markers. First, we wanted to determine the overall percentage of transgene-expressing cells in lung, heart, kidney, tongue, and esophagus. The result of this analysis is shown in Figure 4 and demonstrated that lung, heart, kidney, tongue, and esophagus of noninduced bi-transgenic animals did not contain any EGFP+ cells (data not shown). Consistent with the previously detected luciferase activity, a small fraction of EGFP-expressing cells was present in lung (1.8%), heart (1.71%), kidney (1.29%), tongue (0.67%), and esophagus (1.09%) of induced animals (Figure 4, -DOX).
To distinguish whether conditionally induced EGFP-expressing cells were organ specific or represented migrating blood cells and/or rare tTA-2S-expressing endothelial cells, EGFP+ cells were tested for co-expression of the endothelial marker PECAM-1 together with CD45 and TER119 pan-hematopoietic markers. The result of these experiments is shown in the central panel of Figure 4 and indicates that in lung, heart, esophagus, and tongue the majority of EGFP+ cells were of hematopoietic origin (CD45+/TER119+ cells contained in the 2 upper quadrants of each organ plot). In the boxes on the right of Figure 4 the percentage of EGFP-expressing cells falling either into the category blood (CD45+/TER119+, large upper box) or endothelium (exclusively PECAM-1 expressing, bottom right box) and other cell types (CD45-/TER119- and PECAM-1-, bottom left box) is indicated for each organ. Even though a significant proportion of EGFP+ cells of the kidney expressed hematopoietic markers (52.4%), a major population of kidney cells lacked expression of both the endothelial PECAM-1 marker and the pan-hematopoietic combination of CD45/TER119 surface antigens (47.1%). The presence of a significant population of EGPF-expressing cells lacking blood and endothelial markers suggests that in renal tissues tTA-2S is expressed in a kidney-specific fashion. This observation is in line with the preciously described presence of SCL-expressing cells in the kidney.58 Finally, in all analyzed peripheral organs very few EGFP+ cells exclusively expressed the PECAM-1 endothelial marker (bottom right quadrant of each plot). This suggested that conditional transgene expression also was directed to very rare endothelial cells. This finding was further supported by control experiments using an isotype antibody instead of PECAM-1. In several control experiments the absolute percentage of PECAM-1 single-positive cells was in all cases higher than the percentages detected with the matched isotype antibody (Figure S2, available at the Blood website; see the Supplemental Figures link at the top of the online article). For this reason we conclude that a very minor population of all PECAM-1+ cells did express the tTA-2S transactivator. It is most likely that these cells represented newly forming or regenerating vasculature known to express SCL.59-63
DOX-induced expression of β-galactosidase in peripheral organs of SCL-tTA-2S/EGFP-lacZ double-transgenic mice does not generally colocalize to vascular endothelium. Representative sections from (A) heart, (C) liver, (E) kidney, and (G) muscle of double-transgenic mice were analyzed for the presence of β-galactosidase-expressing cells (left panel). Vascular endothelium was identified by immunofluorescence using a monoclonal antibody against murine PECAM-1 (B, D, F, and H, right panel). The location of β-galactosidase-expressing cells is indicated by arrows.
DOX-induced expression of β-galactosidase in peripheral organs of SCL-tTA-2S/EGFP-lacZ double-transgenic mice does not generally colocalize to vascular endothelium. Representative sections from (A) heart, (C) liver, (E) kidney, and (G) muscle of double-transgenic mice were analyzed for the presence of β-galactosidase-expressing cells (left panel). Vascular endothelium was identified by immunofluorescence using a monoclonal antibody against murine PECAM-1 (B, D, F, and H, right panel). The location of β-galactosidase-expressing cells is indicated by arrows.
Induction of EGFP in peripheral organs of SCL-tTA-2S/EGFP-lacZ double-transgenic mice is primarily restricted to hematopoietic cells and a subset of organ-specific cells in the kidney. Representative FACS profiles of collagenase-digested tissues from lung, heart, kidney, tongue, and esophagus are shown. Left panel (-DOX): Induced organs of bitransgenic mice do contain a small fraction of EGFP+ cells (bottom right quadrant). Percentages of EGFP-expressing cells are shown in the top right quadrant. Central panel: The EGFP+ fraction of cells from the left panel of organ plots (indicated by an arrow) was used for plotting CD45/TER119 pan-hematopoietic markers (y-axis) against the PECAM-1 endothelial marker (x-axis). Right panel: Percentages of EGFP+ hematopoietic cells are shown in the large top box and percentages of endothelial cells in the bottom right box. The percentage of EGFP-expressing cells lacking blood and endothelial markers is indicated in bottom box on the left. Note the substantial increase of EGFP-expressing double-negative CD45-/TER119-and PECAM-1- cells in the kidney.
Induction of EGFP in peripheral organs of SCL-tTA-2S/EGFP-lacZ double-transgenic mice is primarily restricted to hematopoietic cells and a subset of organ-specific cells in the kidney. Representative FACS profiles of collagenase-digested tissues from lung, heart, kidney, tongue, and esophagus are shown. Left panel (-DOX): Induced organs of bitransgenic mice do contain a small fraction of EGFP+ cells (bottom right quadrant). Percentages of EGFP-expressing cells are shown in the top right quadrant. Central panel: The EGFP+ fraction of cells from the left panel of organ plots (indicated by an arrow) was used for plotting CD45/TER119 pan-hematopoietic markers (y-axis) against the PECAM-1 endothelial marker (x-axis). Right panel: Percentages of EGFP+ hematopoietic cells are shown in the large top box and percentages of endothelial cells in the bottom right box. The percentage of EGFP-expressing cells lacking blood and endothelial markers is indicated in bottom box on the left. Note the substantial increase of EGFP-expressing double-negative CD45-/TER119-and PECAM-1- cells in the kidney.
In conclusion, our data suggest that in lung, heart, tongue, and esophagus, expression of tTA-2S was almost completely restricted to hematopoietic cells. In the kidney the majority of EGFP-expressing cells were either hematopoietic or organ specific.
SCL regulatory elements target induction of EGFP to red blood cells, megakaryocytes, granulocytes, and the c-kit+/lin- population of the bone marrow
Next, we wanted to determine in which hematopoietic lineages the SCL-tTA-2S effector mouse could induce expression of conditional transgenes. For this purpose reporter mice carrying the EGFP coding region under the control of a tetracycline-inducible promoter51 were mated to the SCL-tTA-2S effector mouse line. In the resulting bitransgenic animals hematopoietic organs were analyzed for the presence of EGFP+ cells by FACS. As shown in Figure 5, hematopoietic organs from bitransgenic effector/reporter mice permanently kept in the presence of DOX did not contain any EGFP+ cells (right panel +DOX, EGFP off). By contrast, bitransgenic mice without DOX contained a fraction of EGFP+ cells in spleen (1.3%), bone marrow (1.72%), thymus (0.03%), and lymph nodes (0.13%). These results indicated that induction of the EGFP reporter gene in these mice was strictly dependent on DOX and that expression of EGFP occurred only in a subset of cells.
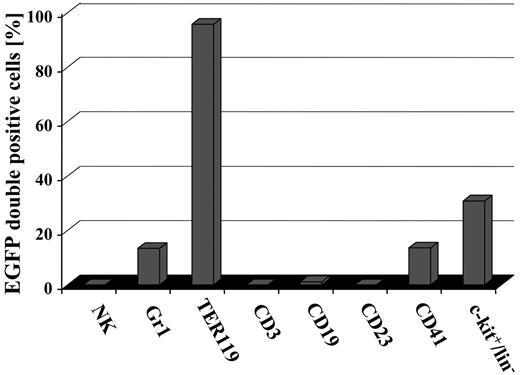
To investigate more precisely whether conditional induction of EGFP was tissue restricted to certain blood cell types or whether all hematopoietic lineages contained EGFP-expressing cells, distinct hematopoietic cell types were analyzed for the presence of EGFP. As shown in Figure 6, no EGFP-positive DX5+ NK-cells, CD3+ T-lymphoid cells, a very minor fraction of CD19+ cells, no CD23+ mature B cells, activated macrophages, eosinophils, and follicular dendritic cells were detected in hematopoietic organs of induced bitransgenic mice. Indeed, as no EGFP+ cells expressed CD23, the very minor fraction of CD19-expressing EGFP+ cells might represent early myelomonocytic cells and/or immature B cells. By contrast, in the same animals EGFP+ cells were detected in Gr1+ granulocytes, TER119+ erythrocytes, CD41+ megakaryocytes, and the c-kit/lin- fraction.
To further evaluate the presence of HSCs/progenitor cells within the EGFP-expressing c-kit+/lin- population, limiting dilution cobblestone area-forming cell (CAFC) assays were performed. CAFC assays are providing a generally accepted in vitro readout of both primitive and progenitor HSCs in mice.54,55,64 Cobblestone areas apparent after 14 days accurately measure spleen colony-forming units (CFU-S) day 12, and those present after 35 days of culture contain long-term HSC repopulating activity.54,55,64 To investigate if the EGFP-expressing population of bone marrow cells did contain CAFC activity, lin-/c-kit+ EGFP+, lin-/c-kit+ EGFP-, and, as a negative control, mononuclear bone marrow cells of induced SCL-tTA-2S/EGFP-lacZ mice were preparatively sorted and tested for their CAFC activities. As expected the mononuclear fraction of bone marrow cells essentially contained no CAFCs (Table 1, MNC). In contrast, day 14 and day 35 CAFCs were generated from the lin-/c-kit+ EGFP-expressing fraction, indicating the presence of progenitors/HSCs proficient to generate early and late CAFCs (Table1). Furthermore, the lin-/c-kit+ EGFP-negative fraction also contained CAFC activity. The presence of CAFC activity in both the lin-/c-kit+ EGFP-expressing and EGFP-negative fraction is not surprising, since SCL is not homogeneously expressed in hematopoietic progenitors/HSCs.65,66 However, the generation of day 14 and day 35 CAFC with the EGFP-expressing lin-/c-kit+ fraction suggests that the SCL-tTA-2S knock-in mouse line directs expression of EGFP to a subset of progenitors/HSCs.
EGFP-expressing c-kit+/lin– cells from the bone marrow of induced SCL-tTA-2S/EGFP-lacZ mice contain early and late CAFC activity
. | Experiment 1, CAFCs per 104 cells . | . | Experiment 2, CAFCs per 104 cells . | . | ||
|---|---|---|---|---|---|---|
| Tested cell population . | Day 14 . | Day 35 . | Day 14 . | Day 35 . | ||
| MNCs | 1.2 (0.8-1.7) | 0.25 (0.2-0.3) | 1 (0.6-1.3) | 0.2 (0.1-0.3) | ||
| lin c-kit+ EGFP+ | 72 (41-102) | 15 (4-26) | 18 (9-27) | 6 (2-11) | ||
| lin– c-kit+ EGFP– | 38 (24-52) | 5 (3-7) | 35 (22-47) | 10 (5-14) | ||
. | Experiment 1, CAFCs per 104 cells . | . | Experiment 2, CAFCs per 104 cells . | . | ||
|---|---|---|---|---|---|---|
| Tested cell population . | Day 14 . | Day 35 . | Day 14 . | Day 35 . | ||
| MNCs | 1.2 (0.8-1.7) | 0.25 (0.2-0.3) | 1 (0.6-1.3) | 0.2 (0.1-0.3) | ||
| lin c-kit+ EGFP+ | 72 (41-102) | 15 (4-26) | 18 (9-27) | 6 (2-11) | ||
| lin– c-kit+ EGFP– | 38 (24-52) | 5 (3-7) | 35 (22-47) | 10 (5-14) | ||
Bone marrow cells were isolated and cultured on OP-9 cells for limiting dilution analysis of CAFC activity as described in “Materials and methods.” Mean CAFC frequencies scored at day 14 and day 35 are shown for 2 independent experiments using, in total, 4 different mice. Numbers in parentheses indicate the range of the 95% confidence limit
MNCs indicates mononuclear cells
Expression of EGFP in hematopoietic organs is dependent on DOX. FACS analysis of adult spleen, bone marrow, thymus, and lymph nodes from double-transgenic effector/responder mice, demonstrating that the induction of EGFP was strictly dependent on DOX. Note the lack of EGFP+ cells in the FACS plots on the right where EGFP expression was inhibited by DOX. The percentage of EGFP-positive cells in each organ is indicated in the top right quadrant.
Expression of EGFP in hematopoietic organs is dependent on DOX. FACS analysis of adult spleen, bone marrow, thymus, and lymph nodes from double-transgenic effector/responder mice, demonstrating that the induction of EGFP was strictly dependent on DOX. Note the lack of EGFP+ cells in the FACS plots on the right where EGFP expression was inhibited by DOX. The percentage of EGFP-positive cells in each organ is indicated in the top right quadrant.
Induction of EGFP expression in SCL-tTA-2S/EGFP-lacZ double-transgenic mice is restricted to granulocytes, red blood cells, megakaryocytes, and c-kit+/lin- cells of the bone marrow. The presence of EGFP+ cells in DX5+ NK cells, Gr1+ myeloid cells, TER119+ red blood cells, CD3+ T-lymphoid cells, CD19+ cells, CD41+ megakaryocytes, CD23 mature B cells, activated macrophages, eosinophils, follicular dendritic cells, and the bone marrow lin-/c-kit+ population was determined by FACS.
Induction of EGFP expression in SCL-tTA-2S/EGFP-lacZ double-transgenic mice is restricted to granulocytes, red blood cells, megakaryocytes, and c-kit+/lin- cells of the bone marrow. The presence of EGFP+ cells in DX5+ NK cells, Gr1+ myeloid cells, TER119+ red blood cells, CD3+ T-lymphoid cells, CD19+ cells, CD41+ megakaryocytes, CD23 mature B cells, activated macrophages, eosinophils, follicular dendritic cells, and the bone marrow lin-/c-kit+ population was determined by FACS.
Taken together, our results show that the SCL-tTA-2S knock-in line exclusively targeted EGFP expression to a subset of hematopoietic lineages, namely, erythrocytes, megakaryocytes, granulocytes, and also to c-kit+/lin- bone marrow cells. These findings suggest that conditional targeting of the EGFP transgene recapitulated the reported lineage-restricted expression pattern of SCL in adult blood.
Analysis of transgene induction in the brain
Expression of SCL has been reported in V2b interneurons of the developing embryo.67-69 In addition, in a recent report it was shown that SCL plays a critical role for the initial specification of primitive neural precursors to astrocytes.69 However, SCL mRNA is not expressed in the brain of postnatal mice.70 Using the EGFP-lacZ and the tetO-Cre responder mouse lines,51,52 functional tTA-2S activity could not be detected in coronal sections through the entire brain of induced SCL-tTA-2S mice. The lack of Cre-recombinase expression in SCL-tTA-2S/tetO-Cre mice (data not shown) and the absence of detectable β-galactosidase activity in induced SCL-tTA-2S/lacZ-EGFP mice (compare induced and noninduced sections in Figure S1) indicated that tTA-2S expression in the brain was either absent or too low to drive the expression of the indicator transgenes. We conclude, therefore, that the SCL-tTA-2S effector mouse is not suitable for robust expression of transgenes in the adult brain.
Discussion
The aim of this study was to generate a conditional mouse model that recapitulates the unique spatio-temporal and lineage-restricted expression pattern of the SCL gene. In particular, we wished to generate a mouse line allowing reversible targeting of transgene expression to HSCs and blood progenitors. Such a conditional SCL effector mouse would be an invaluable experimental tool for approaching fundamental issues concerning normal and malignant hematopoiesis.
The basic helix-loop-helix transcription factor SCL is one of the very few genes known to be expressed both in embryonic and adult HSCs.4,5 This unique expression pattern suggests that SCL regulatory elements could be used to direct conditional expression to HSCs and blood cell progenitors. Radomska and colleagues36 had previously used the human CD34 locus to direct tetracycline-controlled expression of heterologous transgenes to HSCs and early progenitors. In this mouse inducible transgene expression was reported for endothelial and early blood cell progenitors. In a similar fashion elements from the 3′ SCL enhancer were used to direct DOX-inducible expression of transgenes to hematopoietic tissues and HSCs.41 However, in this study, only lung, intestine, and hematopoietic organs were analyzed for DOX-dependent transgene induction. For this reason it is not clear to what extent conditional expression was exclusively restricted to hematopoietic tissues and the lung but was absent from other organs. Interestingly, when this effector mouse was used to express the BCR-ABL oncogene, a chronic myeloid leukemia (CML)-like disease was induced.41 However, since overexpression of SCL under the control of the 3′ SCL enhancer led only to a partial rescue of the lethal SCL knock-out phenotype, it is to be assumed that the 3′ enhancer is not sufficient for recapitulating the endogenous SCL expression pattern.23 Here, we report the generation of a tTA-2S knock-in mouse line that mirrors the known expression pattern of SCL in the adult.
Transcriptional regulation of the murine SCL gene has been extensively studied in vitro and in vivo.12-23,56 Based on this information, we reasoned that inserting the tTA-2S coding sequence into exon V of the SCL locus would ensure the conservation of critical regulatory elements and result in a faithful recapitulation of the endogenous SCL expression pattern by tTA-2S. The capacity and tissue specificity of the SCL-tTA-2S effector mouse line was tested using luciferase, lacZ, and EGFP tetracycline-dependent reporter mice. In a first series of experiments the LC-1 luciferase responder line50 was used to determine in which organs the expression of the luciferase transgene was induced. Since luciferase is known to be a sensitive reporter, low levels of transgene induction should be detectable. These experiments demonstrated high and strictly DOX-dependent transgene induction in bone marrow and spleen and intermediate levels in brain, heart, kidney, liver, lung, tongue, esophagus, pancreas, and thymus (Figure 2B). The intermediate induction of luciferase activity in these organs was somewhat unexpected, as SCL expression in the adult had been reported only for hematopoietic tissues and the kidney.4,5,58 However, given that the analyzed organs were not perfused prior to dissection, we could not exclude the possibility that the measured luciferase activities were due to tTA-2S-expressing, circulating blood, and/or resident endothelial cells. To address this question and to visualize transgene expressing cells in situ, sections from kidney, muscle, liver, and heart of induced bitransgenic SCL-tTA-2S/EGFP-lacZ mice were stained for β-galactosidase activity. Inspection of these sections revealed the presence of lacZ-expressing blue cells in kidney, liver, and heart, but not in the muscle (Figure 3). Subsequent staining of these sections with the PECAM-1 endothelial-specific marker further revealed no obvious general co-localization of tTA-2S and PECAM-1-expressing cells (Figure 3, right panel). Therefore, the histologic analysis suggested that tTA-2S was not expressed in the majority of endothelial cells of these organs. To further clarify the origin of tTA-2S-expressing cells in peripheral organs and to permit analysis of large numbers of individual cells, we used FACS. As the EGFP-lacZ responder mice will simultaneously express EGFP and lacZ upon induction,51 kidney, heart, lung, esophagus, and tongue tissues were subjected to collagenase digestion followed by FACS analysis. These experiments showed that all analyzed tissues contained a fraction of cells expressing EGFP, thus confirming the previously measured luciferase activities in these organs (Figure 4). In addition, examination of EGFP-expressing cells using blood- and endothelial-specific markers revealed that the majority of the analyzed cells were of hematopoietic origin and that only a minor subset represented endothelial or other cell types that were not analyzed further. Moreover, the kidney contained a significant proportion of EGFP-expressing cells lacking both blood and endothelial markers, directly suggesting that these cells were organ-specific (Figure 4). This finding is in line with a recent report showing the expression of SCL in the kidney.58 To determine if adult brain tissues were targeted by the SCL-tTA-2S knock-in mouse, β-galactosidase induction of neuronal tissues also was determined in SCL-tTA-2S/EGFP-lacZ mice. No difference between induced and noninduced brain tissues was seen in these mice, demonstrating that SCL regulatory elements did not direct transgene induction to the brain. Taken together, histologic and flow cytometric analyses suggested that the observed induction of transgenes closely mirrored the known expression pattern of SCL.
Finally, the specificity of tTA-2S-mediated transgene expression in mature blood cells and c-kit-expressing lineage-negative cells was determined. In a previously published report of mice harboring a lacZ reporter gene in exon III of the SCL locus, lacZ expression was confined to HSCs, blood cell progenitors, and red blood cells.25 These findings contrast with the endogenous SCL expression pattern and also with the induced transgene expression pattern observed here, which also included megakaryocytes and granulocytes. However, the differences between these 2 SCL knock-in lines are most likely explained by differences in the design of the targeting strategy (lack of the third SCL promoter and actively transcribing neomycin gene in the case of the lacZ knock-in line). Most notably, the SCL-tTA-2S knock-in mouse directed expression of inducible transgenes to c-kit+/lin- bone marrow cells known to contain blood progenitors/HSCs. Furthermore, measurement of CAFC frequencies from tTA-2S-targeted EGFP-expressing c-kit+/lin- bone marrow cells demonstrated the presence of day 14 CAFCs and day 35 CAFCs, which are an accepted in vitro correlate for CFU-S and bone marrow repopulating stem cell activity (Table1). The ability of EGFP+/c-kit+/lin- cells from the bone marrow to generate day 35 CAFCs thus strongly suggests that the SCL-tTA-2S knock-in mouse is suitable for conditional expression of transgenes in adult HSCs/progenitors.
Taken together, our data show that the SCL-tTA-2S knock-in mouse model will direct conditional DOX-dependent expression of transgenes within blood to erythrocytes, megakaryocytes, granulocytes, and, most importantly, to c-kit+/lin- cells of the bone marrow. This expression profile therefore represents a recapitulation of the known endogenous SCL expression pattern. It is to be expected that the mouse presented here will be a valuable tool for raising fundamental questions about normal and malignant blood cell development.
Prepublished online as Blood First Edition Paper, May 4, 2006; DOI 10.1182/blood-2005-12-012104.
Supported by the European Union (E.B.), the Deutsche Forschungsgemeinschaft (E.B., L.E.), the Stiftung Rheinland-Pfalz für Innovation (E.B.), the Mainz-Forschungsfonds (MAIFOR) program from the Johannes Gutenberg-Universität Mainz (E.B.), and the Deutsche Krebshilfe (E.B.).
E.B. and C.A. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank H. Bujard for the LC-1 reporter mouse line and the tTA-2S transactivator cDNA. In addition, we are grateful to J. Mann, who gave us the W9.5 ES cell line. We also would like to thank the animal technicians of the Mainz animal house for excellent assistance and mouse care, and the Interdisciplinary Center for Clinical Research (IZKF) Core Unit of Fluorescence Technology in Leipzig for preparative cell sorting. Finally, we would like to acknowledge Annette Herold for her excellent technical assistance in the preparation and analysis of brain sections.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal