Abstract
Chronic phase–to–blast crisis transition in chronic myelogenous leukemia (CML) is associated with differentiation arrest and down-regulation of C/EBPα, a transcription factor essential for granulocyte differentiation. Patients with CML in blast crisis (CML-BC) became rapidly resistant to therapy with the breakpoint cluster region–Abelson murine leukemia (BCR/ABL) kinase inhibitor imatinib (STI571) because of mutations in the kinase domain that interfere with drug binding. We show here that the restoration of C/EBPα activity in STI571-sensitive or -resistant 32D-BCR/ABL cells induced granulocyte differentiation, inhibited proliferation in vitro and in mice, and suppressed leukemogenesis. Moreover, activation of C/EBPα eradicated leukemia in 4 of 10 and in 6 of 7 mice injected with STI571-sensitive or -resistant 32D-BCR/ABL cells, respectively. Differentiation induction and proliferation inhibition were required for optimal suppression of leukemogenesis, as indicated by the effects of p42 C/EBPα, which were more potent than those of K298E C/EBPα, a mutant defective in DNA binding and transcription activation that failed to induce granulocyte differentiation. Activation of C/EBPα in blast cells from 4 patients with CML-BC, including one resistant to STI571 and BMS-354825 and carrying the T315I Abl kinase domain mutation, also induced granulocyte differentiation. Thus, these data indicate that C/EBPα has potent antileukemia effects even in cells resistant to ATP-binding competitive tyrosine kinase inhibitors, and they portend the development of anti-leukemia therapies that rely on C/EBPα activation.
Introduction
Chronic myelogenous leukemia (CML) is a clonal disorder arising from neoplastic transformation of hematopoietic stem/progenitor cells.1 The typical course of CML involves progression from a protracted chronic phase marked by the accumulation of apparently normal neutrophils to a rapidly fatal blast crisis characterized by clonal expansion of differentiation-arrested myeloid or lymphoid precursor.2-4 CML is consistently associated with an acquired genetic abnormality, the Philadelphia chromosome, a shortened chromosome 22 resulting from a reciprocal translocation of the long arms of chromosomes 9 and 22.5,6 This translocation generates the breakpoint cluster region–Abelson murine leukemia (BCR/ABL) fusion gene, which is translated in the p210 or, less frequently, the p230 oncoprotein.7-9
Expression of p210 BCR/ABL is necessary and sufficient for the transformation of hematopoietic cells and for disease maintenance, as demonstrated by in vitro assays, leukemogenesis in mice, and the antileukemia effect of the BCR/ABL kinase inhibitor STI571 (imatinib mesylate [Gleevec]; Novartis, Basel, Switzerland).10-14
The mechanisms responsible for chronic phase–to–blast crisis transition remain poorly understood. A plausible model predicts that increased BCR/ABL expression during disease progression15-17 promotes secondary genetic and epigenetic changes essential for the expansion of clones with increasingly malignant characteristics.17,18 The BCR/ABL tyrosine kinase inhibitor imatinib is the first-line treatment for patients with CML.19 Most patients with newly diagnosed chronic-phase CML (CML-CP) treated with imatinib achieve durable responses,14,20 but treatment is less effective in the accelerated and blast-crisis phases of the disease.21 A small percentage of patients with CML-CP and most with advanced-phase disease have relapses on imatinib therapy.22
The most common mechanism of resistance involves specific point mutations in the kinase domain of BCR/ABL that interfere with STI571 binding.23-26 Amplification of the BCR/ABL gene and BCR/ABL-independent mechanisms of resistance have also been reported.26-28
Hematopoietic cell differentiation, which is defective in CML in blast crisis (CML-BC), is regulated by lineage-specific transcription factors, suggesting that the differentiation arrest of CML-BC cells depends, in part, on their altered expression/activity. The transcription factor CCAAT/enhancer-binding protein α (C/EBPα) induces differentiation and inhibits proliferation of many cell types, including myeloid cells.29 Within hematopoietic cells, C/EBPα is expressed by granulocyte progenitors and precursors, but not by monocytes.30,31 Ectopic expression of C/EBPα in bipotential myeloid progenitors induces granulopoiesis and blocks monocytic differentiation,32 and loss of C/EBPα results in mice that retain monocytes but not mature granulocytes.33,34 A model of conditional knockout of C/EBPα has further demonstrated the critical role of C/EBPα in the transition of common myeloid progenitors into granulocyte-monocyte precursors.35
The induction of granulocyte differentiation by C/EBPα is thought to depend on transcription activation,36-38 but the direct interaction of C/EBPα with other proteins also has a profound influence on its function. For example, C/EBPα interacts directly with the cyclin-dependent kinases CDK2 and CDK4 and prevents the assembly of functional CDK complexes that impede cell cycle progression,39 but the CDK2/CDK4 interaction domain of C/EBPα, which is located between amino acids 175 and 188, is not required for C/EBPα regulation of granulocyte differentiation, which depends on its transcriptional activation function.40 C/EBPα also interacts with and represses E2F, a key transcriptional regulator of genes involved in cell cycle progression, an effect possibly involved in C/EBPα-dependent cell cycle arrest and induction of differentiation.41-43
Alteration of C/EBPα function is a common feature of leukemia cells.44 C/EBPα mutations have been reported in approximately 10% of patients with AML,45,46 and C/EBPα expression is transcriptionally repressed in samples from patients with t(8;21) AML1-ETO–positive leukemia47 and is down-modulated in cell lines expressing the FLT3-ITD protein, the inv16 fusion protein, and the AML1-MDS1-EVI1 fusion gene.48-50
In BCR/ABL-expressing cell lines and in marrow cells from patients with CML-BC, but not with CML-CP, C/EBPα expression is suppressed,51,52 suggesting that C/EBPα down-regulation is important in the progression of CML.18,51
Given that ectopic expression of C/EBPα in BCR/ABL-expressing cell lines can restore granulocyte differentiation,51,53 we sought to determine whether sustained C/EBPα expression could permanently suppress leukemogenesis in mice injected with BCR/ABL-expressing cells and whether it would also exert its antileukemia effect in cells carrying STI571-resistant mutant BCR/ABL.
We report here that ectopic expression/activity of C/EBPα induced cell cycle arrest and morphologic, immunophenotypic, and molecular features of granulocyte differentiation in cells expressing wild-type or STI571-resistant mutant BCR/ABL. Similarly, conditional activation of C/EBPα suppressed leukemogenesis in mice, whether it depended on wild-type or mutant BCR/ABL. C/EBPα expression in CML-BC cells carrying the wild-type BCR/ABL or the T315I mutant rapidly induced neutrophilic differentiation, suggesting that therapeutic strategies relying on C/EBPα activation may bypass STI571 resistance.
Materials and methods
Plasmids
ΔuORF/Δspacer C/EBPα-HA (p42 C/EBPα), K298E C/EBPα-HA, and Δ177-191 C/EBPα-HA were previously described.40 Plasmids p42 C/EBPα-ERTAM, K298E C/EBPα-ERTAM, and Δ177-191 C/EBPα-ERTAM were generated by polymerase chain reaction (PCR) as follows: the ligand-binding domain of the murine estrogen receptor (ER) was amplified by reverse transcription–PCR (RT-PCR) from 32Dc13 RNA and was point mutated (Gly525Arg) with the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The PCR product was then reamplified with an upstream oligomer containing a 5′-flapping BamHI site and a downstream oligomer containing a 3′-flapping EcoRI site. p42 C/EBPα, K298E C/EBPα, and Δ177-191 C/EBPα were amplified by PCR from the respective plasmids with an upstream primer containing a 5′-flapping XhoI site and a downstream primer containing a 3′-flapping BamHI site after a mutated C/EBPα stop codon. The p42 C/EBPα or K298E C/EBPα or Δ177-191 C/EBPα and ERTAM PCR products were directionally cloned into the XhoI/EcoRI-digested MigR1 vector. Each plasmid was sequenced to verify the presence of the expected mutations. The plasmid pBabe Puro Kα-ERTAM was a kind gift of A. D. Friedman (Johns Hopkins University, Baltimore, MD).
Cell cultures and retroviral infections
32D-BCR/ABL and derivative cell lines were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, and 10% WEHI-conditioned medium as a source of IL-3. For assays requiring the inhibition of BCR/ABL kinase activity, cells were cultured in IL-3–containing medium supplemented (2 μM) or not with the ABL-kinase inhibitor STI571 (Novartis).
STI571-resistant 32D-BCR/ABL cell lines were established by exposing 32D-BCR/ABL cells to increasing concentrations of STI571 (0.1-2 μM). After selection, RNA from resistant cells was extracted, the cABL kinase domain region was amplified by RT-PCR, and the PCR product was sequenced to assess the presence of mutations.
For retroviral infections, Phoenix cells (kind gift of G. P. Nolan, Stanford University School of Medicine, Stanford, CA) were transiently transfected with the indicated plasmids. The infectious supernatant was collected 48 hours later and was used to infect (a 48-hour procedure) 32D-BCR/ABL–expressing cells. Twenty-four hours later, infected cells were sorted (EPICS Profile Analyzer; Coulter, Hialeah, FL) for green florescent protein (GFP) expression and were kept in culture as described.40 Images were visualized using an Olympus CK2 microscope with a 40 ×/0.65 numeric aperture objective, and were photographed using an Olympus SC35 type 12 camera (Olympus, Melville, NY). JPEG images were viewed using Adobe Photoshop (Adobe Systems, San Jose, CA), and contrast adjustments were made.
Cell proliferation and differentiation assays
For proliferation and differentiation assays, 32D-BCR/ABL–cells transduced with wild-type or mutant C/EBPα-ERTAM were washed with phosphate-buffered saline (PBS) and treated with 4-hydroxytamoxifen (4-HT, 100 nM; Sigma, St Louis, MO). Viable cells were counted by trypan blue exclusion. Differentiation was monitored by May-Grünwald/Giemsa staining and by detection of the differentiation-related marker Gr-1 with a specific phycoerythrin (PE)–conjugated mouse monoclonal antibody (PharMingen, San Diego, CA). A PE-conjugated rat IgG2b isotype immunoglobulin (PharMingen) was also used as control for specific staining.
Western and Northern blot analyses
For Western blotting, cells were lysed (2 × 105 cells/20 μL) in Laemmli buffer, and proteins of interest were detected with anti-C/EBPα (14AA) polyclonal antibody raised against the C-terminus of C/EBPα (sc-61; Santa Cruz Biotechnology, Santa Cruz, CA), anti-C/EBPα (15C8) monoclonal antibody raised against amino acids 1 to 14 of mouse C/EBPα (ab 15047; Novus Biologicals, Littleton, CO), anti–G-CSFR (M20) polyclonal antibody (sc-694; Santa Cruz Biotechnology) with PY20H anti-PTyr/HRP (p11265; BD Transduction Laboratories, Lexington, KY) or with anti-GRB2 monoclonal antibody (610112, BD Transduction Laboratories).
For Northern blot analysis, total RNA of untreated or 4-HT–treated cells was extracted using Tri-Reagent (Molecular Research Center, Cincinnati, OH), fractionated (10 μg/lane) onto denaturing 1% agarose/6.6% formaldehyde gels, transferred onto Hybond-nylon membrane (Amersham Pharmacia Biotechnologies, Piscataway, NJ) and hybridized to a P32-labeled 470 bp PstI fragment of murine myeloperoxidase (MPO) from plasmid puc19MPO.54 RNA levels 28S and 18S were monitored as control for equal loading.
Mice
Sublethally irradiated (4.5 Gy) 4- to 6-week-old C3H/HeJ (Jackson Laboratories, Bar Harbor, ME) male mice were injected intravenously through the lateral tail vein with 32D-BCR/ABL cells transduced with p42 C/EBPα-ERTAM or K298E C/EBPα-ERTAM or with 32D-BCR/ABLT315I STI571-resistant cells transduced with Δ177-191 C/EBPα-ERTAM. Each mouse received 105 cells.
Tamoxifen treatment
4-HT was dissolved in ethanol at 100 mg/mL, then diluted in autoclaved sunflower seed oil (Sigma) at 10 mg/mL. Each mouse was injected intraperitoneally with 1 mg 4-HT on consecutive days.
Analysis of disease progression with acute and chronic activation of C/EBPα
For acute activation of C/EBPα, sublethally irradiated C3H/HeJ male mice were injected intravenously (105 cells/mouse) with 32D-BCR/ABL cells expressing p42 C/EBPα-ERTAM or K298E C/EBPα-ERTAM and with STI571-resistant 32D BCR/ABLT315I cells expressing Δ177-191 C/EBPα-ERTAM. Mice were monitored for leukemia progression with the use of peripheral blood taken retro-orbitally. After erythrocyte lysis, cells were plated in methylcellulose (1000 or 500 per plate) in the absence of cytokines, and colony formation was monitored after 5 days. When peripheral blood leukemia cell percentages reached 10% to 20%, mice were divided into 2 groups, 1 treated with 3 mg 4-HT injected intraperitoneally and the other treated with sunflower seed oil only. At the end of treatment, mice were killed, and GFP-positive bone marrow and spleen cells were used for colony-formation assays for the detection of Gr-1 marker expression and for DNA content analysis.
For chronic activation of C/EBPα, sublethally irradiated C3H/HeJ mice were injected intravenously with C/EBPα-ERTAM–expressing 32D-BCR/ABL (wild-type or mutant) cells (105 cells/mouse). Forty-eight hours later, mice were divided into 2 groups, 1 treated with 1 mg 4-HT, the other with sunflower seed oil, and both injected intraperitoneally on consecutive days for 15 days. Mice were killed at the end of the injections, and bone marrow and spleen cells were used for morphologic examination (May-Grünwald/Giemsa staining) and for assessment of leukemic load by flow cytometry and methylcellulose colony formation assays of GFP-positive cells.
Mice survival
Sublethally irradiated C3H/HeJ male mice were injected intravenously with transduced 32D-BCR/ABL, p42 C/EBPα-ERTAM cells (20 mice, 105 cells/mouse), K298E C/EBPα-ERTAM cells (14 mice, 105 cells/mouse), or Δ177-191 C/EBPα 32D-BCR/ABLT315I cells (14 mice, 105 cells/mouse). Forty-eight hours later, mice were divided in 2 groups, 1 treated with 1 mg 4-HT (intraperitoneally on consecutive days for 15 days) and the other with sunflower seed oil only. After the last injection, mice were monitored for survival.
Detection of the T315I mutation in CML-BC cells
Total RNA from peripheral blood blast cells of a patient with STI571- and BMS-354825–resistant CML-BC was used to generate a BCR/ABL-specific RT-PCR product including nucleotides corresponding to the Abl kinase domain with the use of a 5′ BCR primer (5′-TGAAACTCCAGACTGTCCACA-3′) and a downstream c-Abl 3′ primer (5′-CTGGATTCCTGGAACATTGTTT-3′). Sequences of the PCR product revealed the presence of a T-to-C substitution generating the T315I mutation.
Differentiation assay of CML-BC cells
Bone marrow cells from a patient with CML-BC with 20% blasts carrying a double Ph chromosome were depleted of lineage-positive cells and enriched for CD34+ cells using the Stem Span protocol (Stem Cell Technology, Vancouver, BC, Canada). Cells were cultured for 24 hours in the presence of IL-6, Flt3 ligand, SCF, and IL-3, retrovirally transduced with wild-type or mutant C/EBPα for 48 hours, selected for GFP positivity, and assessed for granulocyte differentiation (May-Grünwald/Giemsa staining) in the presence of 2 ng/mL human recombinant IL-3 or 25 ng/mL human recombinant G-CSF (R&D Systems, Minneapolis, MN).
Peripheral blood blast cells (greater than 90%) from 3 other patients with CML-BC (2 of whom carried the T315I mutation) were expanded for 7 days in the presence of IL-6, Flt3 ligand, SCF, and IL-3 and were retrovirally transduced with the MigRI empty vector or with p42 C/EBPα for 48 hours, selected for GFP positivity, and assessed for granulocyte differentiation in the presence of 25 ng/mL human recombinant G-CSF.
CML patient samples, obtained from the Division of Medical Oncology, Thomas Jefferson Medical College, and the Department of Hematology, M. D. Anderson Hospital, were used with approval by the Institutional Review Board (IRB) of Thomas Jefferson Medical College.
Results
STI571-dependent granulocyte differentiation of 32D-BCR/ABL cells is blocked by an inducible C/EBP transcription repressor
Myeloid precursor 32Dc13 cells are dependent on IL-3 for proliferation and undergo differentiation on treatment with G-CSF.55 Upon expression of BCR/ABL, these cells become growth factor independent and fail to differentiate, a phenotype associated with the down-modulation of C/EBPα.51,52 Ectopic expression of C/EBPα in 32D-BCR/ABL cells can overcome the block in differentiation by allowing the cells to respond to G-CSF.51 Treatment with the BCR/ABL kinase inhibitor STI571 leads to increased C/EBPα expression and reverses, in part, the block in granulocyte differentiation induced by BCR/ABL,52 suggesting that STI571-dependent activation of C/EBPα expression and the induction of differentiation in STI571-responsive cells are functionally linked.
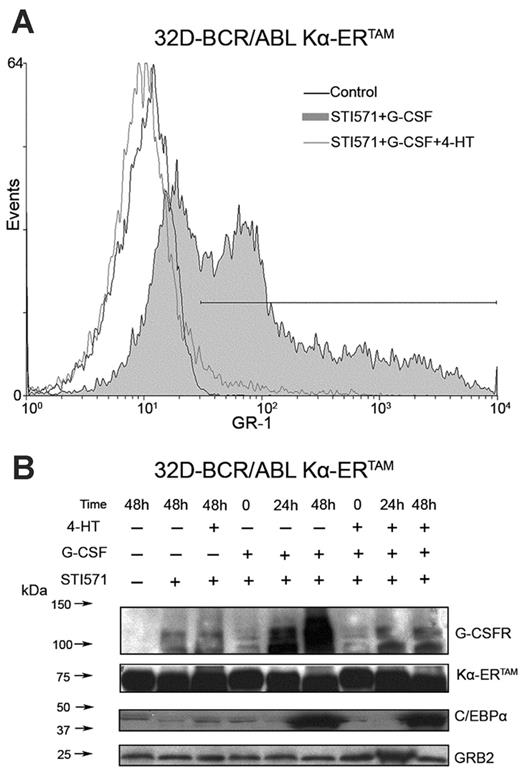
To assess the requirement of C/EBP activity in STI571-induced differentiation of 32D-BCR/ABL cells, a fusion protein consisting of the 89-amino acid Kruppel-associated box (KRAB) transrepression domain (K), the C/EBPα DNA–binding domain(α), and the 4-HT–responsive murine ER ligand–binding domain, Kα-ERTAM was expressed in 32D-BCR/ABL cells. Upon 4-HT treatment of these cells, STI571-dependent G-CSF–induced granulocyte differentiation was suppressed, as assessed by monitoring of the levels of the granulocyte surface marker Gr-1 (Figure 1A). Western blot analysis confirmed that C/EBPα expression was induced by G-CSF and STI571 treatment and showed that activation of the transrepressor Kα-ERTAM had no effect on C/EBPα expression but inhibited its activity,as indicated by the suppression of G-CSFR induction (Figure 1B).
Induction of differentiation in STI571-treated 32D-BCR/ABL cells is dependent on C/EBP activity. Gr-1 expression at 48 hours (A) and C/EBPα and G-CSFR levels at the indicated times (B) in STI571-treated 32D-BCR/ABL cells upon activation of Kα-ERTAM, a C/EBP transcription repressor. Levels of Kα-ERTAM were detected with anti-C/EBPα (14AA) raised against the C-terminus; endogenous C/EBPα was detected with anti-C/EBPα15C8 raised against amino acids 1 to 14 of mouse C/EBPα. GRB2 levels were measured as control of equal loading.
Induction of differentiation in STI571-treated 32D-BCR/ABL cells is dependent on C/EBP activity. Gr-1 expression at 48 hours (A) and C/EBPα and G-CSFR levels at the indicated times (B) in STI571-treated 32D-BCR/ABL cells upon activation of Kα-ERTAM, a C/EBP transcription repressor. Levels of Kα-ERTAM were detected with anti-C/EBPα (14AA) raised against the C-terminus; endogenous C/EBPα was detected with anti-C/EBPα15C8 raised against amino acids 1 to 14 of mouse C/EBPα. GRB2 levels were measured as control of equal loading.
The induction of C/EBPα in STI571-treated 32D-BCR/ABL cells and the demonstration that the STI571 induction of differentiation markers is suppressed by functional inhibition of C/EBP-regulated transcription suggest that ectopic expression/activity of C/EBPα might suppress the leukemogenic potential of STI571-sensitive and -resistant BCR/ABL-expressing cells.
Effect of wild-type and mutant C/EBPα on proliferation and differentiation of STI571-sensitive and -resistant 32D-BCR/ABL cells
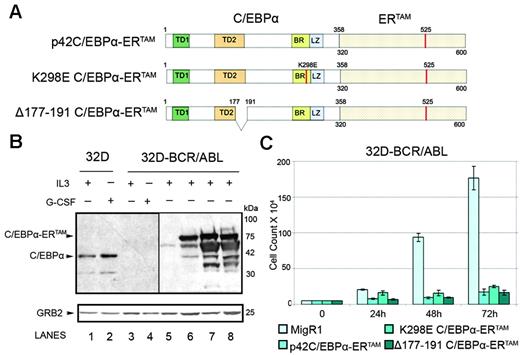
To assess the effects of ectopic C/EBPα expression in cells transformed by BCR/ABL and to identify the domains necessary for its activity, 32D-BCR/ABL cells were transduced with retroviruses expressing only GFP (the MigRI empty vector); p42 C/EBPα-ERTAM, a 4-HT–inducible form of wild-type C/EBPα; K298E C/EBPα-ERTAM, a 4-HT–inducible form of a basic region/DNA-binding domain mutant that maintains the alpha helical structure but is deficient in DNA binding40,56 ; and Δ177-191 C/EBPα-ERTAM, a 4-HT–inducible internally deleted mutant that fails to interact with CDK2/CDK439 (Figure 2A). Immunoblots of 32D-BCR/ABL cell lysates confirmed that the C/EBPα fusion proteins were equally expressed (Figure 2B, lanes 6-8) and that their levels were more abundant in endogenous protein in untreated and G-CSF–treated (3 days) parental and BCR/ABL-expressing 32Dcl3 cells (Figure 2B, lanes 1-4). 4-HT–dependent activation of p42 or mutant C/EBPα-ERTAM in 32D-BCR/ABL caused a marked decrease in cell number (Figure 2C).
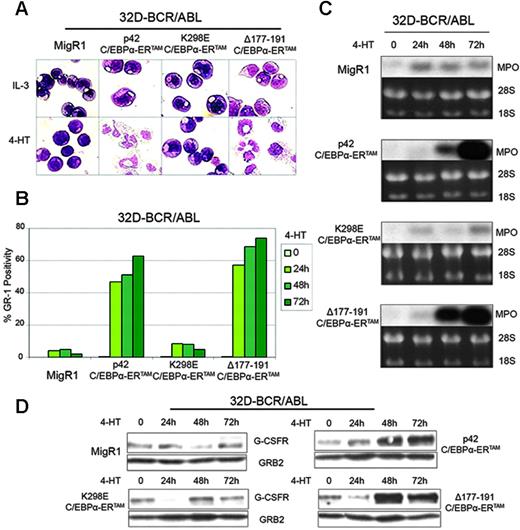
Cell morphology was assessed daily by May-Grünwald/Giemsa staining of cytospins (Figure 3A). Activation of p42 C/EBPα-ERTAM and Δ177-191 C/EBPα-ERTAM led to the rapid appearance of many cells with nuclear segmentation and of terminally differentiated neutrophils; by contrast, activation of K298E C/EBPα-ERTAM did not induce differentiation (Figure 3A). Moreover, activation of p42 C/EBPα-ERTAM and Δ177-191 C/EBPα-ERTAM, but not of K298E C/EBPα-ERTAM, led to increased expression of Gr-1, a marker of granulocyte differentiation (Figure 3B).
Total RNA and whole cell lysates of 4-HT–treated 32D-BCR/ABL cells expressing the various chimeric proteins were prepared daily for 3 days. Total RNA was subjected to Northern blotting for MPO expression. 4-HT treatment had no effect on MPO expression in cells transduced with the MigRI vector or with K298E C/EBPα-ERTAM, but MPO mRNA levels markedly increased by day 2 in cells expressing p42 C/EBPα-ERTAM and Δ177-191 C/EBPα-ERTAM (Figure 3C).
Effects of C/EBPα on 32D-BCR/ABL cells numbers. (A) Schematic diagram of wild-type and mutant C/EBPα-ERTAM chimeric proteins. (B) Levels of endogenous C/EBPα and ectopic C/EBPα-ERTAM in 32Dcl3 (lanes 1-2), in 32D-BCR/ABL cells (lanes 3-4), or in 32D-BCR/ABL cells transduced with the MigRI empty vector (lane 5) or wild-type or mutant C/EBPα-ERTAM (lanes 6-8). The anti-C/EBPα blot was exposed for 2 minutes (lanes 1-4) or 10 seconds (lanes 5-8). (C) Cell counts of 4-HT–treated MigRI- or C/EBPα-ERTAM–transduced 32D-BCR/ABL cells. Values represent mean ± SD of 3 different experiments.
Effects of C/EBPα on 32D-BCR/ABL cells numbers. (A) Schematic diagram of wild-type and mutant C/EBPα-ERTAM chimeric proteins. (B) Levels of endogenous C/EBPα and ectopic C/EBPα-ERTAM in 32Dcl3 (lanes 1-2), in 32D-BCR/ABL cells (lanes 3-4), or in 32D-BCR/ABL cells transduced with the MigRI empty vector (lane 5) or wild-type or mutant C/EBPα-ERTAM (lanes 6-8). The anti-C/EBPα blot was exposed for 2 minutes (lanes 1-4) or 10 seconds (lanes 5-8). (C) Cell counts of 4-HT–treated MigRI- or C/EBPα-ERTAM–transduced 32D-BCR/ABL cells. Values represent mean ± SD of 3 different experiments.
G-CSFR protein levels were also markedly increased in cells expressing p42 C/EBPα-ERTAM and Δ177-191 C/EBPα -ERTAM, but not in cells transduced with the MigRI vector or with K298E C/EBPα-ERTAM (Figure 3D). Thus, C/EBPα activation in 32D-BCR/ABL–expressing cells promotes granulocyte differentiation and mimics, in part, the effects of STI571. C/EBPα-dependent differentiation, but not cell cycle arrest, requires DNA binding and transactivation ability (as demonstrated by the effects of the K298E C/EBPα mutant), whereas the CDK2/CDK4-binding domain is not required for C/EBPα-dependent differentiation or cell cycle arrest.
To assess whether activation of C/EBPα can overcome the block in granulocyte differentiation of STI571-resistant cells, we exposed parental and Δ177-191 C/EBPα-ERTAM–expressing 32D-BCR/ABL cells to increasing concentrations (0.05-2 μM) of STI571. When the selection was complete, total RNA was extracted from resistant cells, and the segment encoding the c-Abl kinase domain was amplified by RT-PCR and sequenced to assess the presence of mutations. The Y253H mutation was identified in 32D-BCR/ABL cells, whereas the T315I mutation was found in Δ177-191 C/EBPα-ERTAM–expressing 32D-BCR/ABL cells.
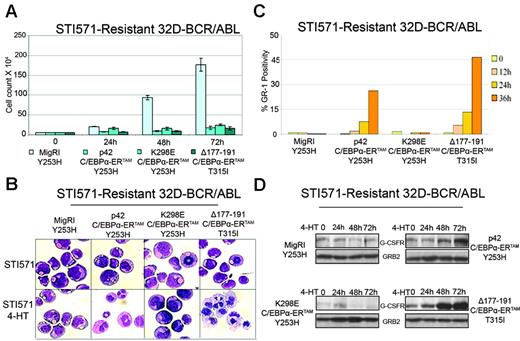
To confirm that the mechanism of resistance was BCR/ABL dependent, the 2 cell lines and control STI571-sensitive cells were treated with 2 μM STI571, and cell lysates were assessed for levels of tyrosine-phosphorylated BCR/ABL by antiphosphotyrosine Western blotting. As expected, levels of tyrosine-phosphorylated proteins (including BCR/ABL) were markedly down-modulated in STI571-sensitive cells but not in the STI571-resistant cell lines (not shown). STI571-resistant 32D-BCR/ABL cells were then retrovirally transduced with the MigRI empty vector, p42 C/EBPα-ERTAM,or K298E C/EBPα-ERTAM to determine whether C/EBPα activation bypasses STI571 resistance.
Compared with MigRI-transduced cells, activation of p42 or mutant C/EBPα caused a marked decrease in the number of 32D-BCR/ABL cells carrying the Y253H or the T315I mutation (Figure 4A). Cell morphology was assessed daily by May-Grünwald/Giemsa staining of cytospins (Figure 4B). After 3 days of 4-HT treatment, most p42 C/EBPα-ERTAM and Δ177-191 C/EBPα-ERTAM–expressing cells were morphologically differentiated, as indicated by the appearance of segmented nuclei typical of mature neutrophils; by contrast, features of morphologic differentiation were not detected in cultures of MigRI-transduced cells or of cells expressing the K298E mutant (Figure 4B). Activation of p42 and Δ177-191 C/EBPα-ERTAM, but not K298E C/EBPα-ERTAM, led to increased levels of Gr-1 (Figure 4C) and G-CSFR (Figure 4D).
Effects of C/EBPα on differentiation of 32D-BCR/ABL cells. Morphology (A), Gr-1 levels (B), MPO (C), and G-CSFR expression (D) in 4-HT–treated 32D-BCR/ABL cells retrovirally transduced with MigRI or C/EBPα-ERTAM.
Effects of C/EBPα on differentiation of 32D-BCR/ABL cells. Morphology (A), Gr-1 levels (B), MPO (C), and G-CSFR expression (D) in 4-HT–treated 32D-BCR/ABL cells retrovirally transduced with MigRI or C/EBPα-ERTAM.
CEBPα activation suppresses in vivo leukemogenesis of STI571-sensitive and -resistant 32D-BCR/ABL cells
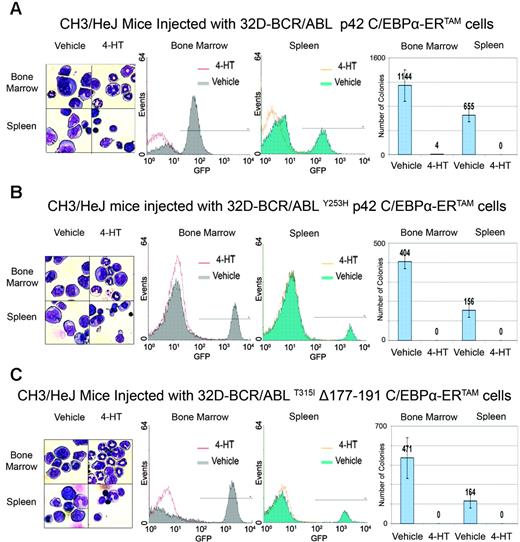
To assess whether C/EBPα can suppress BCR/ABL-dependent leukemogenesis in mice, 32D-BCR/ABL cells, 32D-BCR/ABLY253H cells expressing p42 C/EBPα-ERTAM, and 32D-BCR/ABLT315I cells expressing Δ177-191 C/EBPα-ERTAM were injected intravenously (105 cells/mouse) into sublethally irradiated syngeneic mice; 2 days later, mice were treated with vehicle or 4-HT (1 mg/d for 15 consecutive days). At the end of the treatment, mice were killed and cells were harvested from bone marrow and spleen. May-Grünwald/Giemsa staining of bone marrow cell suspensions of 4-HT–treated mice revealed the presence of normal myeloid and nonmyeloid cells, suggesting that the bone marrow was not infiltrated by BCR/ABL-transformed cells. By contrast, samples of vehicle-treated mice showed various proportions of blastlike cells characterized by high nuclear-cytoplasmic ratios and round nuclei (Figure 5A-C, left panel). Bone marrow samples of vehicle-treated mice displayed a high proportion of GFP-positive cells (32D-BCR/ABL–injected mice, 70% ± 1.16%, n = 2; 32D-BCR/ABLY253H–injected mice, 16.5% ± 2.0%, n = 2; 32D-BCR/ABLT315I–injected mice, 21.4% ± 4.5%, n = 2), whereas in 4-HT–treated mice, GFP positivity of marrow cells was negligible (32D-BCR/ABL–injected mice, 1.04% ± 1.6%, n = 3; 32D-BCR/ABLY253H–injected mice, 0.14% ± 0.3%, n = 2; 32D-BCR/ABLT315I–injected mice, 0.94% ± 0.2%, n = 3) (Figure 5A-C, middle panels). The clonogenic potential of bone marrow cells was evaluated by methylcellulose assays performed in the absence of cytokines. Five days after plating, cells of vehicle-treated mice formed numerous colonies consistent with the presence of growth factor–independent BCR/ABL-expressing cells; by contrast, few colonies formed from bone marrow cells of 4-HT–treated mice injected with 32D-BCR/ABL p42 C/EBPα-ERTAM cells (Figure 5A, right panel), and no colonies developed from bone marrow of 4-HT–treated mice injected with STI571-resistant 32D-BCR/ABL cells (Figure 5B-C, right panels).
Effects of C/EBPα on STI571-resistant 32D-BCR/ABL cells. Cell counts (A), morphology (B), Gr-1 levels (C), and G-CSFR expression (D) in STI571-resistant 32D-BCR/ABL cells upon 4-HT activation of C/EBPα-ERTAM. Cell count values represent mean ± SD of 3 different experiments.
Effects of C/EBPα on STI571-resistant 32D-BCR/ABL cells. Cell counts (A), morphology (B), Gr-1 levels (C), and G-CSFR expression (D) in STI571-resistant 32D-BCR/ABL cells upon 4-HT activation of C/EBPα-ERTAM. Cell count values represent mean ± SD of 3 different experiments.
The same analysis was performed on splenocytes harvested from vehicle or 4-HT–treated mice. Spleens of control mice were markedly larger than those of 4-HT–treated mice, suggesting they were infiltrated by BCR/ABL-transformed cells. Microscopic examination of spleen cell suspensions from 4-HT–treated mice showed that most cells were lymphocytes, whereas the splenocyte suspension from vehicle-treated mice was enriched in cells with a blastlike morphology (Figure 5A-C, left panel). Fluorescence-activated cell sorter (FACS) analysis displayed a cohort of GFP-positive cells in control mice (32D-BACR/ABL–injected mice, 23.4% ± 2.8%, n = 2; 32D-BCR/ABLY253H–injected mice: 5% ± 1.0%, n = 2; 32D-BCR/ABLT315I–injected mice, 4.7% ± 1.1%, n = 3), whereas no GFP-positive cells were detected in the spleen suspensions of 4-HT–treated mice (Figure 5A-C, middle panels). Colony-formation assays further demonstrated the presence of BCR/ABL-expressing cells in spleen cell suspensions of vehicle-treated, but not of 4-HT–treated, mice (Figure 5A-C, right panels). Because the leukemia load in mice injected with 32D-BCR/ABLY253H and 32D-BCR/ABLT315I cells was lower than that in mice injected with 32D-BCR/ABL cells, clonogenic assays were also performed by seeding splenocytes and bone marrow cells from mice injected with STI571-resistant 32D-BCR/ABL cells at 19 000 cells/plate, a number chosen to approximate the GFP positivity of cell suspensions from mice injected with STI571-sensitive 32D-BCR/ABL cells. No colonies formed from samples of 4-HT–treated mice (not shown), indicating that C/EBPα is equally effective in STI571-sensitive and -resistant cells.
In vivo effects of C/EBPα activation in STI571-sensitive and -resistant 32D-BCR/ABL cells. Mice were injected with C/EBPα-ERTAM-expressing 32D-BCR/ABL cells (A), C/EBPα-ERTAM–expressing 32D-BCR/ABLY253H cells (B), or Δ177-191 C/EBPα-ERTAM 32D-BCR/ABLT315I cells (C) and were treated with vehicle alone or with 4-HT for 15 consecutive days. (left panels) Morphology, (middle panels) GFP positivity, and (right panels) colony formation of bone marrow and spleen cells isolated at the end of treatment. Colony number values represent mean ± SD of 3 different experiments.
In vivo effects of C/EBPα activation in STI571-sensitive and -resistant 32D-BCR/ABL cells. Mice were injected with C/EBPα-ERTAM-expressing 32D-BCR/ABL cells (A), C/EBPα-ERTAM–expressing 32D-BCR/ABLY253H cells (B), or Δ177-191 C/EBPα-ERTAM 32D-BCR/ABLT315I cells (C) and were treated with vehicle alone or with 4-HT for 15 consecutive days. (left panels) Morphology, (middle panels) GFP positivity, and (right panels) colony formation of bone marrow and spleen cells isolated at the end of treatment. Colony number values represent mean ± SD of 3 different experiments.
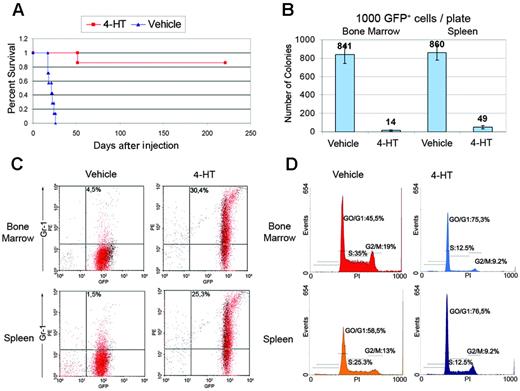
Because the T315I BCR/ABL mutant is completely insensitive to STI571 and other ATP-binding competitive inhibitors of the BCR/ABL kinase, including BMS-354825,57-59 we further tested the antileukemia effect of C/EBPα in 32D-BCR/ABLT315I cells. Thus, mice (7 per group) injected with 105 Δ177-191 C/EBPα-ERTAM 32D-BCR/ABLT315I cells were treated 2 days later with vehicle only or with 1 mg/d 4-HT for 15 consecutive days and then were assessed for survival. Untreated mice were all dead 25 days after the injection of leukemia cells; by contrast, only 1 mouse in the 4-HT–treated group died of leukemia (51 days after injection), whereas the remaining 6 were alive and without signs of disease 225 days after injection (Figure 6A). The effect of C/EBPα was also assessed in mice in which bone marrow and spleen were heavily infiltrated by leukemia cells. Thus, 32D-BCR/ABLT315I cells expressing Δ177-191 C/EBPα-ERTAM were injected intravenously in sublethally irradiated mice and were allowed to develop leukemia.
Effect of C/EBPα activation on mice injected with 32D-BCR/ABLT315Icells. (A) Survival of mice injected with Δ177-191 C/EBPα-ERTAM 32D-BCR/ABLT315I cells (105 cells/mouse) and treated 2 days later with vehicle only or with 4-HT (1 mg/d for 15 consecutive days). (B) Methylcellulose colony formation assays, (C) Gr-1 immunostaining, and (D) DNA content of GFP-positive cells isolated from bone marrow and spleen of mice injected with Δ177-191 C/EBPα-ERTAM 32D-BCR/ABL cells (105 cells/mouse) and treated 14 days later with vehicle only or with 4-HT (1 mg/12 hours, for 3 injections). Colony number values represent mean ± SD of 3 different experiments.
Effect of C/EBPα activation on mice injected with 32D-BCR/ABLT315Icells. (A) Survival of mice injected with Δ177-191 C/EBPα-ERTAM 32D-BCR/ABLT315I cells (105 cells/mouse) and treated 2 days later with vehicle only or with 4-HT (1 mg/d for 15 consecutive days). (B) Methylcellulose colony formation assays, (C) Gr-1 immunostaining, and (D) DNA content of GFP-positive cells isolated from bone marrow and spleen of mice injected with Δ177-191 C/EBPα-ERTAM 32D-BCR/ABL cells (105 cells/mouse) and treated 14 days later with vehicle only or with 4-HT (1 mg/12 hours, for 3 injections). Colony number values represent mean ± SD of 3 different experiments.
When leukemia cell percentages reached 15% to 20%, mice were treated with vehicle alone or with 1 mg 4-HT every 12 hours for a total of 3 injections. At 36 hours, mice were killed, leukemia cells in bone marrow and spleen were sorted by GFP positivity, and their proliferation potential and differentiation were tested. Compared with GFP-positive cells of vehicle-treated mice, activation of Δ177-191 C/EBPα-ERTAM by 4-HT induced a marked decrease in colony formation (841 ± 101 vs 14 ± 7 from bone marrow GFP-positive cells; 860 ± 79 vs 49 ± 17 from spleen GFP-positive cells) (Figure 6B) but led to an increased number of differentiating cells expressing the Gr-1 antigen (4.5% ± 3.6% vs 30.4% ± 7.9% in bone marrow and 1.5% ± 0.4% vs 25.3% ± 0.4% in spleen) (Figure 6C).
Compared with vehicle treatment only, activation of Δ177-191 C/EBPα-ERTAM in 32D-BCR/ABLT315I cells injected in mice led to a dramatic G0/G1 arrest of GFP-positive cells in bone marrow (vehicle-treated mice: G0/G1 phase, 45.4% ± 2.25%; S phase, 35% ± 2.0%; G2/M phase, 19% ± 1.1%; 4-HT–treated mice: G0/G1 phase, 75.3% ± 11.4%; S phase: 12.5% ± 6.4%; G2/M phase: 9.2% ± 4.0%) and spleen (vehicle-treated mice: G0/G1 phase, 58.5% ± 0.28%; S phase, 25.3% ± 1.0%; G2/M phase, 13% ± 0.4%; 4-HT–treated mice: G0/G1 phase, 76.5% ± 11.4%; S phase, 12.5% ± 6.0%; G2/M phase, 9.2% ± 4.0%) (Figure 6D). Thus, the activation of functional C/EBPα induced differentiation, suppressed the proliferation of 32D-BCR/ABLT315I cells injected in mice, and enhanced the survival of leukemic mice.
Differentiation induction and proliferation inhibition are both required for the antileukemia effect of C/EBPα
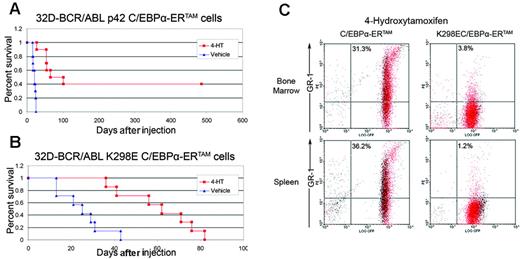
We further assessed whether C/EBPα suppression of in vivo leukemogenesis was dependent on both proliferation inhibition and differentiation induction. Thus, 105 32D-BCR/ABL cells expressing p42 C/EBPα-ERTAM or K298E C/EBPα-ERTAM were injected into sublethally irradiated syngeneic mice and, 2 days later, were treated with vehicle only or 1 mg/d 4-HT for 15 consecutive days. Activation of K298E C/EBPα-ERTAM prolonged the survival of leukemic mice (treated: mean, 60.5 days; median, 62 days; n = 7; untreated: mean, 25 days; median, 25 days; n = 7; difference between means, 35.5 days; P = .001; difference between medians, 37 days; P = .001, Wilcoxon 2-sample exact test). However, the activation of p42 C/EBPα-ERTAM had a more potent effect (treated: mean, 230 days; median, 83 days; n = 10; untreated: mean, 20.2 days; median, 21 days; n = 10; difference between means, 209.8 days; P < .001; difference between medians, 62 days; P < .001, Wilcoxon 2-sample exact test).
Notably, 4 of 10 treated mice were still alive more than 15 months after the last injection, indicating that they were cured of their disease (Figure 7A-B). On comparing the effect of the 2 C/EBPα proteins, the differentiation-inducing function of p42 C/EBPα, but not of K298E C/EBPα, may explain the more potent antileukemia effect of the former. Thus, we tested cell cycle inhibition and differentiation induction by p42 C/EBPα and K298E C/EBPα in 32D-BCR/ABL injected in mice.
Leukemia was allowed to develop in mice injected with 32D-BCR/ABL cells expressing p42 or K298E C/EBPα-ERTAM. When percentages of leukemia cells in the peripheral blood were 15% to 20%, mice were treated with vehicle only or with 1 mg/d 4-HT for 3 days. On day 4, mice were killed, bone marrow and splenocytes were harvested, GFP-positive cells were sorted, and Gr-1 positivity and DNA content were evaluated. 4-HT activation of p42 C/EBPα-ERTAM led to markedly increased Gr-1 expression in bone marrow (31.3% ± 11.7% [n = 2] vs 4.5% ± 3.6% [n = 2] in vehicle-treated mice) and spleen (36.2% ± 4.6 [n = 2] vs 1.5% ± 0.4 [n = 2] in vehicle-treated mice) GFP-positive cells (Figure 7C). By contrast, 4-HT activation of K298E C/EBPα-ERTAM did not induce an increase of Gr-1 levels in bone marrow (3.8% ± 1.1%) or spleen (1.2% ± 0.5%) GFP-positive cells (Figure 7C). DNA content analysis of bone marrow and spleen GFP-positive cells from mice injected with p42 or K298E C/EBPα-ERTAM–expressing 32D-BCR/ABL cells revealed that 4-HT induced in both increased numbers of G0/G1 cells (bone marrow: G0/G1, 57.2% ± 2.0% and 59.2% ± 2.2%, n = 2, respectively; spleen: G0/G1, 64.25% ± 1.0% and 63.5% ± 2.1%, n = 2, respectively) compared with treatment with vehicle (bone marrow: G0/G1, 45.4% ± 2.3%, n = 2; spleen: G0/G1, 57.5% ± 0.5%, n = 2). Thus, the more potent effect of p42 C/EBPα-ERTAM in suppressing leukemogenesis by 32D-BCR/ABL cells likely depends on its ability to induce the differentiation of leukemia cells in mice.
Effect of C/EBPα activation on survival of mice injected with 32D-BCR/ABL cells and on in vivo differentiation of 32D-BCR/ABL cells. (A) Survival of mice (n = 10) injected with p42 C/EBPα-ERTAM 32D-BCR/ABL cells (105/mouse) and left untreated or treated with 4-HT (1 mg/d for 15 consecutive days). (B) Survival of mice (n = 7) injected with K298E C/EBPα-ERTAM 32D-BCR/ABL cells (105/mouse) and left untreated or treated with 4-HT (1 mg/d for 15 consecutive days). (C) Gr-1 levels in GFP-positive 32D-BCR/ABL cells expressing p42 C/EBPα-ERTAM or K298E C/EBPα-ERTAM and treated with 4-HT.
Effect of C/EBPα activation on survival of mice injected with 32D-BCR/ABL cells and on in vivo differentiation of 32D-BCR/ABL cells. (A) Survival of mice (n = 10) injected with p42 C/EBPα-ERTAM 32D-BCR/ABL cells (105/mouse) and left untreated or treated with 4-HT (1 mg/d for 15 consecutive days). (B) Survival of mice (n = 7) injected with K298E C/EBPα-ERTAM 32D-BCR/ABL cells (105/mouse) and left untreated or treated with 4-HT (1 mg/d for 15 consecutive days). (C) Gr-1 levels in GFP-positive 32D-BCR/ABL cells expressing p42 C/EBPα-ERTAM or K298E C/EBPα-ERTAM and treated with 4-HT.
Ectopic expression of p42 C/EBPα induces differentiation of CML-BC cells
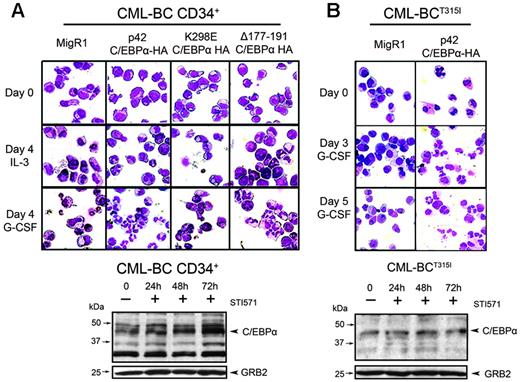
The effects of C/EBPα were also investigated in human CML-BC cells. Thus, wild-type or mutant C/EBPα was retrovirally transduced in CD34+ cells of a patient with CML-BC with 20% blasts carrying a double Ph chromosome and no mutations in the BCR/ABL kinase domain and GFP-positive cells assessed for granulocyte differentiation in the presence or in the absence of G-CSF. In the presence of G-CSF, differentiation was essentially complete by day 4 (Figure 8A, upper panel, second and fourth rows), whereas in the absence of G-CSF the process was slower and most p42 C/EBPα and Δ177-191 C/EBPα–expressing cells underwent granulocyte differentiation by day 7 (not shown). By contrast, few MigRI-transduced cells or cells expressing the K298E C/EBPα mutant showed morphologic features of differentiation (Figure 8A, first and third rows). Of note, C/EBPα expression was low in bone marrow mononuclear cells but increased 3- to 4-fold after STI571 treatment (Figure 8A, lower panel). Peripheral blood blast cells of another CML-BC patient without mutations in the BCR/ABL kinase domain were also induced to differentiate by ectopic expression of p42 C/EBPα (not shown).
The ability of C/EBPα to induce granulocyte differentiation of CML-BC cells was also tested with the use of peripheral blood blasts from a patient with CML-BC resistant to STI571 and to BMS-354825 and, predictably, carrying the T315I mutation. Blast cells from this patient with CML-BCT315I were retrovirally transduced with MigRI or wild-type p42 C/EBPα for 48 hours, selected for GFP positivity, and assessed for granulocyte differentiation. In the presence of G-CSF, most p42 C/EBPα-expressing cells underwent granulocyte differentiation by day 5; by contrast, few MigRI-transduced cells showed features of differentiation in the presence of G-CSF (Figure 8B, upper panel). C/EBPα expression was low in untreated cells and, as expected, did not increase upon STI571 treatment (Figure 8B, lower panel). Peripheral blood blast cells of another patient with CML-BC carrying the T315I mutation were also induced to differentiate by ectopic expression of p42 C/EBPα (not shown).
Effect of C/EBPα in CML-BC cells. (Top panel) Induction of granulocyte differentiation by constitutive expression of C/EBPα in CML-BC cells. Bone marrow CD34+ cells (A) or peripheral blood blasts (B) from 2 CML-BC patients were retrovirally transduced with the MigRI empty vector or with wild-type or mutant C/EBPα. After sorting, GFP-positive cells were cultured in the presence of IL-3 or G-CSF and were assessed (May-Grünwald/Giemsa staining of cytospins) for morphologic differentiation. (Bottom panel) Western blot analysis shows C/EBPα expression in untreated and STI571-treated cells of the 2 patients with CML-BC. GRB-2 levels were measured as control of equal loading.
Effect of C/EBPα in CML-BC cells. (Top panel) Induction of granulocyte differentiation by constitutive expression of C/EBPα in CML-BC cells. Bone marrow CD34+ cells (A) or peripheral blood blasts (B) from 2 CML-BC patients were retrovirally transduced with the MigRI empty vector or with wild-type or mutant C/EBPα. After sorting, GFP-positive cells were cultured in the presence of IL-3 or G-CSF and were assessed (May-Grünwald/Giemsa staining of cytospins) for morphologic differentiation. (Bottom panel) Western blot analysis shows C/EBPα expression in untreated and STI571-treated cells of the 2 patients with CML-BC. GRB-2 levels were measured as control of equal loading.
Discussion
CML therapy had been revolutionized by the development of STI571, the first ATP-binding competitive inhibitor of the BCR/ABL kinase.24 STI571 is highly effective in patients with CML-CP; BCR/ABL transcripts remain detectable in a cohort of STI571-treated CML-CP patients,60 but the long-term consequences of this finding are unclear. Resistance to STI571 develops in most patients with advanced-stage CML and in a few with CML-CP, often as a consequence of mutations in the BCR/ABL kinase domain.26,61 Given that the development of STI571 resistance reduces the therapeutic options in CML patients, it is important to identify molecular targets of STI571 that may mimic its effects. When activated, some of these targets might bypass STI571 resistance, raising the possibility of novel therapeutic strategies that use effector molecules functioning downstream of the drug target. One such target of STI571 is the transcription factor C/EBPα because its expression is repressed by BCR/ABL and is restored by BCR/ABL kinase inhibition. Enhanced C/EBPα expression after STI571 treatment of BCR/ABL-expressing cells is likely to be important for the drug's effects because markers of granulocyte differentiation are not induced by STI571 in cells conditionally expressing a C/EBP transcription repressor (Figure 1).
Although this repressor is also expected to inhibit C/EBPβ- and C/EBPϵ-dependent transcription, these effects are likely to be less important than those on C/EBPα because C/EBPϵ is induced late during myeloid differentiation52 and C/EBPβ is less potent than C/EBPα in inducing the differentiation of BCR/ABL-expressing cells.62 Thus, we assessed the antileukemia effects of C/EBPα using STI571-sensitive and -resistant BCR/ABL-expressing cells in vitro and in mice in which C/EBPα was conditionally activated.
For these studies, we tested the effect of wild-type p42 C/EBPα and of 2 mutants, K298E and Δ177-191 C/EBPα, that are deficient in DNA-binding/transcription activation and CDK2/CDK4 interaction, respectively. In vitro, p42 C/EBPα and both mutants all induced a marked decrease in cell number, indicating that neither transcription activation nor CDK2/CDK4 interaction is required for the cell cycle effects. By contrast, the induction of granulocyte differentiation by C/EBPα depended on its transcription activation function but not on CDK2/CDK4 interaction. Together, these findings are essentially identical to those in normal cells.40
A previous study failed to show differentiation of 32D-BCR/ABL cells conditionally expressing p42 C/EBPα-ER.63 We do not have a simple explanation for the discrepancy in our findings. Perhaps cells transduced with the C/EBPα-ER retrovirus express less chimeric protein than cells transduced with the C/EBPα-ERTAM retrovirus because of negative selection possibly attributed to the presence of estrogens in the culture media. This may select cells expressing reduced levels of C/EBPα-ER, which may be sufficient for the cell cycle effects but not for differentiation induction.
In vivo, conditional activation of p42 or K298E C/EBPα markedly prolonged the survival of leukemic mice. However, p42 C/EBPα was more effective because 4 of 10 leukemic mice were cured of their disease. Combined with the potent in vivo effects of Δ177-191 C/EBPα (Figure 6), an explanation for these differences probably rests in the ability of the transcription activation-competent p42 C/EBPα and Δ177-191 C/EBPα to induce cell cycle arrest and differentiation of BCR/ABL-expressing cells in mice compared with the DNA binding and transcription activation–deficient K298E mutant, which failed to induce differentiation (Figure 7C).
Activation of C/EBPα-induced differentiation caused cell cycle arrest and suppressed leukemogenesis of STI571-resistant 32D-BCR/ABL cells. Of the 2 mutants tested here, the Y253H mutation renders the BCR/ABL kinase activity more than 100-fold less inhibitable,64 whereas the T315I mutation renders BCR/ABL essentially insensitive to STI571.64 Of interest, C/EBPα activation completely suppressed leukemia induced by 32D-BCR/ABLT315I cells in 6 of 7 mice (Figure 6).
The antileukemia effect of C/EBPα was also evident in mice treated with 4-HT 15 days after the injection of STI571-resistant 32D-BCR/ABLT315I cells, when bone marrow and spleen were heavily infiltrated by BCR/ABL-transformed cells (Figure 6). Activation of C/EBPα led very rapidly to the inhibition of proliferation (methylcellulose assays and DNA content analysis) and the induction of differentiation (Gr-1 positivity), a finding that would support the therapeutic potential of C/EBPα-inducing agents in CML-BC patients characterized by high disease burden and resistance to tyrosine kinase inhibitors.
Of greater interest, C/EBPα expression in the blast cells of 4 patients with CML-BC led to the rapid induction of granulocyte differentiation. In particular, one of the patients was resistant to both STI571 and the dual Src/Abl kinase inhibitor BMS-354825, which blocks the activity of all BCR/ABL mutants tested except T315I.58 Indeed, all (or most) blast cells of this patient carried the T315I mutation, as indicated by sequence analysis of a BCR/ABL-specific RT-PCR product encompassing the kinase domain.
We previously reported that the expression of C/EBPα is repressed by BCR/ABL at the translational level by interaction of the RNA binding protein hnRNPE2 with a C-rich region in the 5′ UTR of c/ebpβ mRNA.51 In this study, we used C/EBPα retroviruses lacking the 5′ UTR to prevent BCR/ABL suppression and to maximize the biologic effects of the protein. Indeed, the biologic effects of the 5′ UTR-containing C/EBPα retrovirus were less potent (data not shown). This suggests that optimal strategies for C/EBPα-based therapies would require use of the protein itself or of small molecules disrupting the hnRNPE2-c/ebpα mRNA interaction.
In conclusion, this proof-of-principle study demonstrates the potential usefulness of therapeutic strategies restoring C/EBPα expression in CML-BC and, perhaps, in other types of acute myelogenous leukemia in which C/EBPα activity is disrupted.
Prepublished online as Blood First Edition Paper, May 2, 2006; DOI 10.1182/blood-2006-01-011833.
Supported in part by National Institutes of Health grants CA 95111 and PO1 78890 (B.C.). G.F.-A. was supported in part by a fellowship from the A. Serra Foundation for Cancer Research. C.G. was supported in part by a fellowship from the Italian-American Foundation for Cancer Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Cathy Tomastik for editorial assistance and Edward Pequignot for help with statistical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal