Abstract
The E2F proteins are major regulators of the transcriptional program required to coordinate cell cycle progression and exit. In particular, E2f4 has been proposed to be the principal family member responsible for the regulation of cell cycle exit chiefly through its transcriptional repressive properties. We have previously shown that E2f4–/– mice display a marked macrocytic anemia implicating E2f4 in the regulation of erythropoiesis. However, these studies could not distinguish whether E2f4 was required for differentiation, survival, or proliferation control. Here, we describe a novel function for E2f4 in the promotion of erythroid proliferation. We show that loss of E2f4 results in an impaired expansion of the fetal erythroid compartment in vivo that is associated with impaired cell cycle progression and decreased erythroid proliferation. Consistent with these observations, cDNA microarray analysis reveals cell cycle control genes as one of the major class of genes down-regulated in E2f4–/– FLs, and we provide evidence that E2f4 may directly regulate the transcriptional expression of a number of these genes. We conclude that the macrocytic anemia of E2f4–/– mice results primarily from impaired cellular proliferation and that the major role of E2f4 in fetal erythropoiesis is to promote cell cycle progression and cellular proliferation.
Introduction
The E2F family is composed of transcription factors that are critical regulators of cell cycle entry, progression, and exit that connect extracellular cues to the cell cycle machinery.1,2 In addition to being the main transcriptional controllers of G1 to S phase progression, E2Fs have been shown to regulate the expression of genes required for DNA replication, DNA repair, cell cycle checkpoint activation and maintenance, mitosis, and apoptosis. Of importance, E2Fs have also recently been shown to regulate genes involved in differentiation and development1,3,4 and therefore provide a critical link between cell cycle and differentiation programs during development.
The E2F family comprises a group of at least 8 related E2F proteins that have been divided into distinct subsets, based on both structural and functional homology and their ability to bind to pRB family members, pRB, p107, and p130.1,2 E2F1, E2F2, and E2F3a comprise a group primarily associated with activation of transcription of E2F-responsive genes, can promote cell cycle progression, and bind exclusively to pRB.2,5 In contrast, E2F4 and E2F5 have been proposed to be primarily involved in transcriptional repression, which is achieved through the binding and recruitment of pRB family members and associated chromatin remodeling complexes to E2F-dependent promoters. As E2F4 can bind to all 3 pRB family members and forms the majority of cellular pRB family complexes, E2F4 has been proposed to be the major repressive E2F in vivo and to play a critical role in coordinating cell cycle exit.2,5 E2F6 and the divergent E2F7 and E2F8 proteins appear quite distinct from the other E2F subgroups in both structure and function and do not bind pRB family members. Current evidence suggests a role for these proteins primarily in transcriptional repression.6-12
Because of its proposed role as the major repressive E2F, our initial experiments focused on understanding the function of E2F4 in vivo. Our original analysis of E2f4–/– animals demonstrated a critical role for E2f4 in normal development. In particular, E2f4–/– animals display fetal macrocytic anemia and growth retardation detectable as early as E13.5 of gestation.13,14 The anemia of E2f4–/– mice is restricted to the fetal stage of development, although adult peripheral red blood cells remain macrocytic and display Howell-Jolly bodies (nuclear remnants of condensed DNA thought to be derived from either improper enucleation or from chromosomal aberrations). Transplant studies established the erythroid defects to be cell autonomous.13 Despite the anemia, the numbers of earliest erythroid progenitors (erythroid–blast-forming units [BFU-Es] and erythroid–colony-forming units [CFU-Es]) were elevated in E2f4–/– fetuses, indicating that the defects must occur at a later stage in erythroid differentiation.13 Together, these experiments indicate that E2f4 is critical for normal erythropoiesis in vivo. However, how E2f4 mediates its effects on erythropoiesis, whether through regulation of differentiation, survival, or proliferation, is not known.
To investigate the cellular function of E2f4 in erythropoiesis, we have conducted a detailed investigation of fetal erythroid development and production in E2f4–/– animals. Our data indicate that impaired proliferation is the underlying cause of the E2f4–/– animal macrocytic anemia and identifies E2f4 in a new role as a positive regulator of cell cycle progression and cellular proliferation in vivo.
Materials and methods
Timed matings and tissue harvesting
E2f4+/+ and E2f4–/– embryos13 were obtained at 12.5 and 15.5 days after coitus, and individual FLs were harvested and immediately snap frozen for protein or RNA extraction or alternatively resuspended in 2% fetal calf serum (FCS) in PBS by passing through a cell strainer for fluorescence-activated cell sorter (FACS) analysis. Genotyping of embryos derived from E2f4+/– matings was performed as previously described.13
Immunohistochemistry
Whole E12.5 and E15.5 embryos were formalin fixed and paraffin embedded. Sections were stained with anti-E2f4 primary antibody (LLF4.215 ), detected using the LSAB2 kit (DAKO, Carpinteria, CA) according to the manufacturer's specifications and counterstained with hematoxylin. To detect apoptosis, in situ TUNEL was performed on E15.5 embryo sections according to the manufacturer's specifications (In Situ Cell Death Detection Kit: Fluorescein; Roche, Indianapolis, IN), with the subsequent use of anti–fluorescein-POD (Roche) and DAB staining for detection by light microscopy.
Erythroid cultures, flow cytometry, and microscopy
Erythroid cultures were derived from E12.5 FL and grown essentially as previously described.16,17 After 3 to 4 days in expansion media, cultures were magnetic affinity column sorting (MACS) purified by depletion of Ter119-positive cells, and cells were resuspended at 1 × 106 cells/mL in differentiation media. Media were replenished every 24 hours and analysis was performed at 0, 12, 24, 48, and 72 hours. Flow cytometry and microscopy were conducted as previously described.17 Microscopy was performed using a Zeiss Axioskop 2 microscope (Carl Zeiss Microimaging, Thornwood, NJ) with a Zeiss Plan-Neofluar 40× or 5× objective (numerical aperture, 0.75). Digital images were obtained using a Diagnostic Instruments Spot RT Slider camera (Sterling Heights, MI) and analyzed with Spot Software Windows version 4.0.2 imaging software.
Western and gel-shift analysis
Cell cycle analysis
Propidium iodide (PI), DRAQ5, and in vitro BrdU labeling was performed as previously described.17 For PH3 and MPM2 analysis, cells were fixed in cold 95% ethanol, permeabilized in 0.25% Triton-X, and stained using anti-PH3 antibody (1:400; Upstate Biotech, Lake Placid, NY) or anti-MPM2 antibody (1:10; DAKO) as described.20 For in vivo BrdU analysis, 200 μL of 10 mg/mL BrdU was injected intraperitoneally into pregnant females, embryos were harvested 1 hour later, and FL single cell suspensions were isolated. Cells were stained with Ter119-PE and BrdU was detected according to the manufacturer's specifications (BrdU Flow Kit; BD Pharmingen, San Diego, CA) and analyzed by flow cytometry on a FACScalibur (Becton Dickinson, San Jose, CA).
Transcriptional analysis
See Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) for detailed description of cDNA microarray procedures, real-time polymerase chain reaction (PCR) primers, and chromatin immunoprecipitation (ChIP) protocol used.
Results
Expression of E2f4 protein and associated complexes during fetal erythropoiesis
To better understand how E2f4 may regulate erythropoiesis, we examined the expression of E2f4 protein during embryonic development and erythropoietic differentiation. Our analysis shows a distinctive expression pattern for E2f4 during development (Figure 1A). E2f4 is highly expressed in fetal liver (FL) at E12.5 through to E15.5 (Figure 1A; data not shown). At these developmental stages, the FL is the major site of definitive erythropoiesis and consists of 80% to 90% erythroid cells.21,22 Of note, the developmental timing of FL expression of E2f4 corresponds to the time of onset of fetal anemia.13 E2f4 was not detectable in other tissues at these stages except for very faint expression in the Rathke pouch (K.M.K. and P.O.H., unpublished observation, May 2001). High-power views of E12.5 FL cellular morphology (Figure 1A, lower left panel) indicate that E2f4 is present at most stages of definitive erythroid differentiation (ie, FL derived) but is not detectable in primitive (yolk-sac derived) nucleated erythroid cells. E2f4 exhibited both nuclear and cytoplasmic expression in FL erythroid cells with similar localization throughout different stages of differentiation of erythroid cells.
As the activity of E2f4 is modulated by the presence of pRB family members, we next examined the expression and composition of E2F/pRb complexes throughout fetal erythroid development using Western and gel-shift analyses. Markers of erythroid commitment (Ter119) and hematopoietic progenitors (c-kit) were used to isolate cell populations by flow cytometry corresponding to hematopoietic progenitors (H), proerythroblasts (P), and mature nucleated erythroblasts (M) (Figure 1B). Consistent with our immunohistochemical data, E2f4 was found to be present in all the FL populations, with some decreased expression in the most mature erythroblast fraction (Figure 1C). E2F2 and E2F3 appeared to be expressed in a differentiation-regulated pattern, and we did not detect significant levels of E2F1 or E2F5 (Figure 1C; data not shown). Similarly, there was no detectable expression of p130 in FL cells, and p107 was present in all stages of erythroid differentiation, with increased levels present in the most highly proliferative proerythroblast population (Figure 1C). pRb expression increased dramatically with erythroid differentiation, pRb being present in both hyperphosphorylated and hypophosphorylated forms (Figure 1C). To examine the nature of E2F/pRb family complexes in fetal erythoid cells, we performed gel-shift analysis on protein extracts of E12.5 FLs and sorted populations of E15.5 FLs (Figure 1D-E). At E12.5, antibodies specific to E2f4 but not to other E2Fs could supershift the majority of E2F DNA-binding complexes in both whole-cell extracts and nuclear extracts, indicating that E2f4 contributes to the majority of E2F DNA-binding activity in FL cells (Figure 1Di lanes 5 and 11; arrowheads denote supershifted complexes). This was further demonstrated by the loss of the majority of E2F DNA-binding activity in equivalent extracts from E12.5 E2f4–/– FLs (Figure 1Diii lanes 6-10). Of note, we have not detected any compensatory changes in other E2F family member expression using gel-shift, Western, or microarray analysis (Figure 1Diii; data not shown). Despite the expression of p107 in E12.5 FL cells, only a small amount of E2f4/p107 complexes is present in both whole-cell and nuclear extracts (Figure 1Dii lanes 4 and 9; monoclonal antibody to p107 supershifts complex, open arrow). This was further validated using an independent antibody to p107 (Figure 1Diii lane 4; polyclonal antibody to p107 destroys the slower migrating complex). Therefore, in extracts from E12.5 FL cells, E2f4 is predominantly found in “free” complexes (ie, in potentially transcriptionally active complexes). We also assessed E2F complex formation within the fractionated E15.5 FL populations (Figure 1E). Throughout each stage of differentiation, we observed that the major E2F complexes were “free” E2f4 complexes, with the remaining E2F DNA-binding activity corresponding to E2f4/p107 complexes (Figure 1E). Of interest, the proerythroblast population, which is the most highly cycling fetal erythroid population in vivo (Figure 1B), consistently showed the highest E2f4 DNA-binding activity (Figure 1E lanes 2, 5, and 8). Despite the high expression of pRb in these extracts, we did not detect DNA-bound pRb/E2f complexes in E15.5 sorted fetal erythroid cells (Figure 1E; data not shown). Together, these data reveal a highly restricted spatial expression of E2f4 to the FL, the major site of erythropoiesis during embryonic development, and indicate that E2f4 contributes to the majority of E2F-dependent DNA-binding activity during fetal erythroid differentiation in vivo.
E2f4 is highly expressed in fetal liver erythroid cells. (A) E2f4 protein expression by immunohistochemistry in E12.5 E2f4+/+ (left panels) and E2f4–/– (right panels) embryo sections, counterstained with hematoxylin (blue). High-power images of the FL are shown in the bottom panels. Lack of detectable signal in the E2f4–/– littermate control demonstrates the specificity of the E2f4 antibody used in these studies. (B) Sorting protocol to isolate erythroid differentiation populations from E15.5 E2f4+/+ FL. Mature erythroblasts (Ter119hi/c-kitneg, M), proerythroblasts (Ter119dim/c-kitdim, P), and hematopoietic progenitors (Ter119neg/c-kithi, H; include the earliest erythroid progenitors, BFU-Es and CFU-Es). Morphology (benzidine and May-Grunwald-Giemsa stain) and cell cycle profiles by PI were used to characterize these populations. (C) Protein expression of E2F and pRB family members in sorted E15.5 E2f4+/+ FL subpopulations was assessed by Western blot; mSin3A indicates loading control. (D) Gel-shift analyses of E2F/pRb family complexes in E12.5 E2f4+/+ andE2f4–/– FL. (E) Gel-shift analysis of E2F/pRb family complexes in sorted E15.5 E2f4+/+ FL populations. Protein samples were derived from whole-cell extracts except where denoted. Nuc indicates nuclear extracts; WCE, whole-cell extracts. Solid arrowheads denote complexes supershifted by E2F4 polyclonal antibody, and clear arrowhead (Dii) denotes supershift with p107 mAb (SD6; a kind gift from N. Dyson), whereas an alternate p107 pAb (SC318X; Santa Cruz, Santa Cruz, CA) results in destruction of the complex (Diii,E).
E2f4 is highly expressed in fetal liver erythroid cells. (A) E2f4 protein expression by immunohistochemistry in E12.5 E2f4+/+ (left panels) and E2f4–/– (right panels) embryo sections, counterstained with hematoxylin (blue). High-power images of the FL are shown in the bottom panels. Lack of detectable signal in the E2f4–/– littermate control demonstrates the specificity of the E2f4 antibody used in these studies. (B) Sorting protocol to isolate erythroid differentiation populations from E15.5 E2f4+/+ FL. Mature erythroblasts (Ter119hi/c-kitneg, M), proerythroblasts (Ter119dim/c-kitdim, P), and hematopoietic progenitors (Ter119neg/c-kithi, H; include the earliest erythroid progenitors, BFU-Es and CFU-Es). Morphology (benzidine and May-Grunwald-Giemsa stain) and cell cycle profiles by PI were used to characterize these populations. (C) Protein expression of E2F and pRB family members in sorted E15.5 E2f4+/+ FL subpopulations was assessed by Western blot; mSin3A indicates loading control. (D) Gel-shift analyses of E2F/pRb family complexes in E12.5 E2f4+/+ andE2f4–/– FL. (E) Gel-shift analysis of E2F/pRb family complexes in sorted E15.5 E2f4+/+ FL populations. Protein samples were derived from whole-cell extracts except where denoted. Nuc indicates nuclear extracts; WCE, whole-cell extracts. Solid arrowheads denote complexes supershifted by E2F4 polyclonal antibody, and clear arrowhead (Dii) denotes supershift with p107 mAb (SD6; a kind gift from N. Dyson), whereas an alternate p107 pAb (SC318X; Santa Cruz, Santa Cruz, CA) results in destruction of the complex (Diii,E).
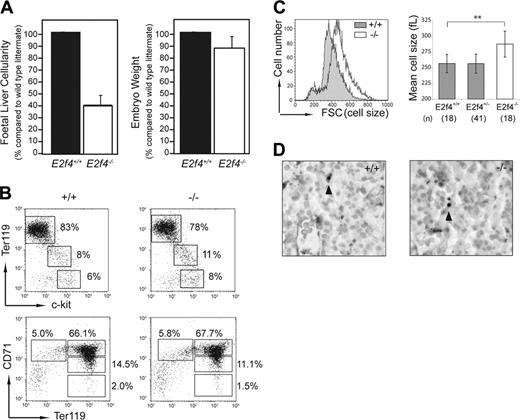
E2f4–/–embryos have a severely reduced fetal liver cellularity, which is not associated with a block in differentiation or increased apoptosis
The restricted expression of E2f4 pointed to the FL as a potential site where defects could account for the anemia of E2f4–/– animals. Consistent with the decreased number of peripheral erythrocytes observed in E2f4–/– embryos,13 we found that at E15.5, FL cellularity was reduced by approximately 60% in E2f4–/– embryos compared to E2f4+/+ littermate controls (Figure 2A). Of importance, this proportional decrease in FL cellularity far exceeded the reduction seen in overall body size for E2f4–/– embryos (approximately 12% reduction in body weight; Figure 2A). Similar results were obtained at E12.5 of gestation (data not shown).
To assess whether the decreased FL cellularity of E2f4–/– embryos was due to a significant block in erythroid differentiation, we used flow cytometry to determine the proportion of cells at various differentiation stages during erythropoiesis (Figure 2B). Whole FL single-cell suspensions from E2f4+/+ and E2f4–/– littermates (E15.5) were stained using a combination of Ter119, CD71, and c-kit cell surface markers.23,24 These data indicated no significant differences in the proportion of cells within each erythroid subpopulation, consistent with the normal erythroid differentiation observed histologically (Figure 1A). Of interest, as with the peripheral erythrocytes, the committed (Ter119+) erythroid cells of E2f4–/– FL were larger in size (macrocytic) than those of wild-type littermates (Figure 2C). No significant differences in levels of apoptosis were observed between E2f4+/+ and E2f4–/– FLs using both TUNEL on embryo sections in situ or trypan blue exclusion on FL suspensions ex vivo (Figure 2D; data not shown). Together, these data indicate that a block in differentiation or an increase in apoptosis cannot account for the reduction in FL cellularity in the E2f4–/– embryos.
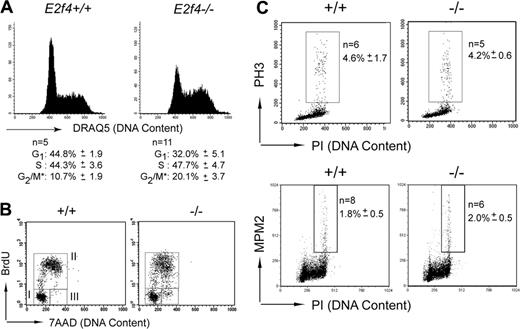
E2f4–/–fetal liver erythroid cells display cell cycle defects in vivo
In the absence of an obvious decrease in survival or a differentiation block, we investigated whether proliferation defects could account for the reduced numbers of E2f4–/– erythroid cells in vivo. Our previous studies, as well as those of other groups, failed to reveal proliferative defects in a number of cell types isolated from E2f4–/– mice, including mouse embryonic fibroblasts and lymphoid cells.13,14 However, in light of the high expression of E2f4 in FL and the reduced cellularity of E2f4–/– FL, we investigated the proliferative characteristics of E2f4–/– erythroid cells in vivo (Figure 3A). Using the DNA-interacting dye DRAQ5, we consistently observed an increase in the proportion of erythroid-committed cells (Ter119+) with 4N DNA content, indicative of an increased proportion of cells in late S and/or G2/M phase of the cell cycle.
Decreased cellularity of E2f4–/– fetal liver is not associated with a block in differentiation or decreased survival. (A) Whole FL cellularity (n = 8 litters) and whole embryo body weight (n = 4 litters) of E15.5 E2f4+/+ and E2f4–/– littermate embryos. Error bars indicate standard deviation from the mean. (B) Immunophenotyping of hematopoietic markers in E2f4+/+ and E2f4–/– E15.5 whole FLs by FACS. (C) Cell size of Ter119+/DRAQ5+ erythroid cells derived from E15.5 FL suspensions was determined by forward scatter measurement by FACS (left plot), and by mean cell volume (femtoliters) parameter on a Sysmex CD4-500 hematological cell counter (Sysmex America, Mundeleine, IL) (right plot); **P < .005, Student t test. Error bars indicate standard deviation from the mean. (D) Measurement of apoptosis by in situ TUNEL assay in E2f4+/+ and E2f4–/– E15.5 FLs. Arrowheads indicate TUNEL-positive cells.
Decreased cellularity of E2f4–/– fetal liver is not associated with a block in differentiation or decreased survival. (A) Whole FL cellularity (n = 8 litters) and whole embryo body weight (n = 4 litters) of E15.5 E2f4+/+ and E2f4–/– littermate embryos. Error bars indicate standard deviation from the mean. (B) Immunophenotyping of hematopoietic markers in E2f4+/+ and E2f4–/– E15.5 whole FLs by FACS. (C) Cell size of Ter119+/DRAQ5+ erythroid cells derived from E15.5 FL suspensions was determined by forward scatter measurement by FACS (left plot), and by mean cell volume (femtoliters) parameter on a Sysmex CD4-500 hematological cell counter (Sysmex America, Mundeleine, IL) (right plot); **P < .005, Student t test. Error bars indicate standard deviation from the mean. (D) Measurement of apoptosis by in situ TUNEL assay in E2f4+/+ and E2f4–/– E15.5 FLs. Arrowheads indicate TUNEL-positive cells.
To further investigate these differences, E15.5 fetuses were pulsed with BrdU in utero for 60 minutes, and FLs were harvested and analyzed by flow cytometry. Examination of E2f4–/– and wild-type littermate fetuses demonstrated that comparable numbers of Ter119+ cells enter or progress through S phase during the 1-hour labeling period (Figure 3B, gate II, and Table 1). However, erythroid cells from E2f4–/– fetuses consistently displayed an abnormal BrdU incorporation profile, with a spread in the level of BrdU-FITC fluorescence intensity, indicating that many cells have decreased efficiency of BrdU incorporation and therefore is suggestive of S phase progression defects (Figure 3B, compare left and right panels, gate II). Furthermore, although all E2f4+/+ cells that were in the G2/M phase of the cell cycle prior to BrdU exposure appear to undergo mitosis within an hour, there was a clear accumulation of BrdU-negative cells with 4N or less DNA content in E2f4-deficient FLs during the labeling period, suggesting delay in late S phase, G2, and/or mitosis (Figure 3B, compare left and right panels, gate III; E2f4+/+, 1.0 ± 0.5; E2f4–/–, 8.4 ± 2.1). To determine whether this delay was occurring at mitosis, the proportion of cells expressing phosphorylated histone H3 (PH3) or the phosphoepitope recognized by the MPM2 antibody, both markers of mitosis, was determined on whole FL by FACS. We observed no increase in the number of cells expressing phosphorylated histone H3 or recognized by the MPM2 antibody in the E2f4–/– FL, indicating that loss of E2f4 does not result in a delay or block in mitosis (Figure 3C). Together our analyses demonstrate that E2f4–/– erythroid cells show a delay through late S phase and/or G2 phase of the cell cycle in vivo.
FACS determination of BrdU incorporation by Ter119+ E15.5 FL cells following 1 hour in vivo BrdU pulse
Genotype . | n . | Gate I, BrdU-, %2n*† . | Gate II, BrdU+, %2n to 4n*† . | Gate III, BrdU-, %above 2n to 4n*† . |
|---|---|---|---|---|
| E2f4+/+ | 7 | 53.6 ± 11.2 | 45.0 ± 11.7 | 1.0 ± 0.5 |
| E2f4+/- | 12 | 49.9 ± 12.3 | 48.5 ± 12.1 | 1.3 ± 0.8 |
| E2f4-/- | 8 | 38.4 ± 3.7 | 52.5 ± 3.7 | 8.4 ± 2.1 |
Genotype . | n . | Gate I, BrdU-, %2n*† . | Gate II, BrdU+, %2n to 4n*† . | Gate III, BrdU-, %above 2n to 4n*† . |
|---|---|---|---|---|
| E2f4+/+ | 7 | 53.6 ± 11.2 | 45.0 ± 11.7 | 1.0 ± 0.5 |
| E2f4+/- | 12 | 49.9 ± 12.3 | 48.5 ± 12.1 | 1.3 ± 0.8 |
| E2f4-/- | 8 | 38.4 ± 3.7 | 52.5 ± 3.7 | 8.4 ± 2.1 |
Gates correspond to gates in Figure 3B.
DNA content where 2n is the DNA content of a diploid gene.
Cell cycle defects of E2f4–/– fetal liver erythroid cells in vivo. (A) Cell cycle profile of live Ter119+ gated E15.5 FL cells stained with DNA dye, DRAQ5. Percentage of cells in each cell cycle stage was determined by Modfit (*proportion of cells in G2/M, as determined by Modfit, may include cells in late S phase). (B) FACS determination of BrdU incorporation by Ter119+ E15.5 FL cells following 1 hour in vivo BrdU pulse. For quantitation, see Table 1. (C) Phosphohistone H3 and MPM2 staining was performed on whole E15.5 FL cells to determine proportion of cells undergoing mitosis. Numeric data are provided with the corresponding standard deviation.
Cell cycle defects of E2f4–/– fetal liver erythroid cells in vivo. (A) Cell cycle profile of live Ter119+ gated E15.5 FL cells stained with DNA dye, DRAQ5. Percentage of cells in each cell cycle stage was determined by Modfit (*proportion of cells in G2/M, as determined by Modfit, may include cells in late S phase). (B) FACS determination of BrdU incorporation by Ter119+ E15.5 FL cells following 1 hour in vivo BrdU pulse. For quantitation, see Table 1. (C) Phosphohistone H3 and MPM2 staining was performed on whole E15.5 FL cells to determine proportion of cells undergoing mitosis. Numeric data are provided with the corresponding standard deviation.
Defective proliferation of E2f4–/– erythroid progenitors in vitro
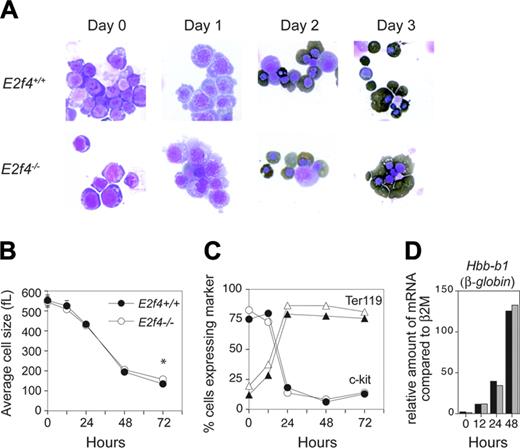
To remove any mitigating effects of nonerythroid tissue and to better define the function of E2f4 in erythropoiesis, we made use of a primary erythroid in vitro cell culture system that faithfully recapitulates the in vivo characteristics of terminal erythroid differentiation.16 We undertook our original studies in erythroid culture conditions favoring self-renewal (expansion conditions) and observed that although E2f4–/– FL erythroid cells displayed similar proliferation and differentiation characteristics in early stages of expansion cultures (up to day 7), by day 10 E2f4–/– erythroblasts had undergone premature abortive differentiation compared with days 15 to 18 for wild-type cultures (K.M.K and P.O.H., unpublished observation, May 2003). To better understand the requirement of E2f4 in erythroid proliferation and differentiation, we used culture conditions that allow terminal erythroid differentiation, thus providing a more accurate reflection of in vivo erythropoiesis.16,17 E12.5 E2f4+/+ and E2f4–/– FLs were harvested and expanded for 3 or 4 days and erythroid progenitors purified by depletion of Ter119+ cells using a MACS protocol. Equivalent purified populations were obtained from each sample as determined by FACS analysis using the cell surface markers c-kit, AA4.1, Ter119, and CD71 (data not shown). Purified populations were induced to terminally differentiate by switching to a differentiation inducing media. By multiple parameters, we found that the differentiation program in the E2f4–/– cultures was virtually identical to the E2f4+/+ controls (Figure 4). Morphologically, both E2f4+/+ and E2f4–/– cultures decreased in size, became hemoglobinized, progressively decreased their nuclear to cytoplasmic ratio, and compacted their chromatin with a high proportion becoming enucleated (Figure 4A). In both E2f4+/+ and E2f4–/– cultures, we observed a reduction in cell size from approximately 450 to 150 fL (Figure 4B). Of interest, consistent with the macrocytosis observed in vivo, the average cell size of E2f4–/– cells on the final day was significantly larger than E2f4+/+ cells (E2f4+/+: 138.9 ± 5.7 fL; E2f4–/–: 158.5 ± 3.8 fL, n = 4; P < .002, Student t test). FACS analysis of the cultures shows that the majority of cells rapidly up-regulated the erythroid marker Ter119 and down-regulated c-kit within the first day of culture (Figure 4C). In addition, both E2f4+/+ and E2f4–/– cultures showed the same kinetics of induction of Hbb-b1 (β-globin) gene regulation (Figure 4D). In summary, by the criterion measured here, we observed that E2f4–/– erythroid cultures show comparable kinetics of differentiation to that of wild-type cultures.
In contrast, the proliferation parameters showed significant differences between E2f4+/+ and E2f4–/– cultures (Figure 5). In the first 48 hours, E2f4+/+ cultures undergo 3 to 4 rounds of differentiation-associated cell division that results in an 8- to 12-fold increase in cell numbers by day 3 (Figure 5A). This proliferative burst is markedly reduced in the E2f4–/– culture (Figure 5A). Of importance, the cultures had similar viability, demonstrating that the decrease in cell number was not due to decreased cell survival (Figure 5B). Cell cycle analysis of E2f4+/+ and E2f4–/– cultures show that there was a significant decrease in cells in S phase at 24 hours, in addition to an increase in cells with 4N DNA content from 24 to 72 hours (Figure 5C; individual cell cycle profiles shown in Figure S1). BrdU labeling of cells at 24 hours confirmed these cell cycle defects. We observed a significant decrease in the proportion of cells incorporating BrdU in E2f4–/– cultures compared with E2f4+/+ cultures and similar to that observed in vivo, an approximate 2.5-fold increase in nonlabeled cells with 4N DNA content in E2f4–/– cultures compared with E2f4+/+ cultures (Figure 5D). The underlying reason for the differences in the proportion of E2f4–/– erythroid cells incorporating BrdU observed in vivo and in culture is not currently clear but may reflect the high enrichment of specific differentiation stages achieved in culture compared with the heterogenous populations analyzed in vivo. Alternatively, these differences could reflect dissimilarities between culture conditions and the FL microenvironment, highlighting a differential requirement for E2f4 in S phase progression in response to specific factors present or absent in the culture conditions used.
Normal differentiation kinetics of E2f4–/– fetal liver erythroid cells in vitro. FL erythroid cells pooled from E2f4+/+ and E2f4–/– E12.5 embryos were expanded and induced to differentiate in vitro. Features of differentiation were measured at 0, 12, 24, 48, and 72 hours following differentiation induction. (A) Morphologic analysis of cytospins following erythroid differentiation by May-Grunwald-Giemsa and benzidine (dark brown) staining. (B) Average cell size (fL) determined by Coulter counter analysis. (C) Proportion of cells expressing hematopoietic markers c-kit (circles) and Ter119 (triangles) by FACS. (D) Relative abundance of mRNA transcript of erythroid differentiation marker β-globin compared with β 2M, assessed by real-time PCR; E2f4+/+, ▪; E2f4–/–, ##. For all other graphs, E2f4+/+, closed symbols; E2f4–/–, open symbols. Error bars indicate standard deviation.
Normal differentiation kinetics of E2f4–/– fetal liver erythroid cells in vitro. FL erythroid cells pooled from E2f4+/+ and E2f4–/– E12.5 embryos were expanded and induced to differentiate in vitro. Features of differentiation were measured at 0, 12, 24, 48, and 72 hours following differentiation induction. (A) Morphologic analysis of cytospins following erythroid differentiation by May-Grunwald-Giemsa and benzidine (dark brown) staining. (B) Average cell size (fL) determined by Coulter counter analysis. (C) Proportion of cells expressing hematopoietic markers c-kit (circles) and Ter119 (triangles) by FACS. (D) Relative abundance of mRNA transcript of erythroid differentiation marker β-globin compared with β 2M, assessed by real-time PCR; E2f4+/+, ▪; E2f4–/–, ##. For all other graphs, E2f4+/+, closed symbols; E2f4–/–, open symbols. Error bars indicate standard deviation.
Impaired proliferation of differentiating E2f4–/– fetal erythroid cells in vitro. FL erythroid cells pooled from E2f4+/+ and E2f4–/– E12.5 embryos were expanded and induced to differentiate in vitro. Proliferation, viability, and cell cycle parameters were measured at 0, 12, 24, 48, and 72 hours following differentiation induction. (A) Cumulative proliferation was assessed by measurement of live cell numbers in each culture by trypan blue exclusion assay. (B) Level of viability was determined by trypan blue exclusion. (C) Cell cycle profiles were measured by PI analysis, and the percentage of cells in each cell cycle stage was determined by Modfit. (D) FACS determination of BrdU incorporation by Ter119+ E15.5 FL cells following 1 hour in vitro BrdU pulse at 24 hours after differentiation induction. For all graphs, E2f4+/+, closed symbols; E2f4–/–, open symbols. Error bars indicate standard deviation.
Impaired proliferation of differentiating E2f4–/– fetal erythroid cells in vitro. FL erythroid cells pooled from E2f4+/+ and E2f4–/– E12.5 embryos were expanded and induced to differentiate in vitro. Proliferation, viability, and cell cycle parameters were measured at 0, 12, 24, 48, and 72 hours following differentiation induction. (A) Cumulative proliferation was assessed by measurement of live cell numbers in each culture by trypan blue exclusion assay. (B) Level of viability was determined by trypan blue exclusion. (C) Cell cycle profiles were measured by PI analysis, and the percentage of cells in each cell cycle stage was determined by Modfit. (D) FACS determination of BrdU incorporation by Ter119+ E15.5 FL cells following 1 hour in vitro BrdU pulse at 24 hours after differentiation induction. For all graphs, E2f4+/+, closed symbols; E2f4–/–, open symbols. Error bars indicate standard deviation.
These in vitro analyses demonstrate that E2f4–/– erythroid cells showed impaired S phase progression as well as a delay through late S phase and/or G2 phase of the cell cycle. Together these data indicate that loss of E2f4 results in impaired cell cycle progression and reduced proliferation during terminal erythroid differentiation.
E2f4 loss results in expression changes in genes associated with cell cycle control, gene regulation, and erythroid differentiation
To begin investigating the underlying molecular mechanism by which E2f4 regulates FL erythropoiesis, we compared the “global” gene expression patterns in E2f4+/+ and E2f4–/– FLs using cDNA microarray analysis. Our data identified 84 genes reproducibly deregulated in E2f4-deficient FLs that could be categorized in 3 clearly delineated functional groups, with remaining genes corresponding to genes of miscellaneous or unknown function and ESTs (Table 2). The deregulation of gene expression observed by microarray closely matched that observed by real-time PCR. Comparative quantitation of gene expression for a selection of genes is shown in Figure 6A.
Expression changes in E2f4-/- FL determined by cDNA microarray
Expression change in E2f4-/- FL/unigene . | Gene name . | Gene symbol . | Gene deregulated, no. arrays . | E2F site* . | Description . |
|---|---|---|---|---|---|
| Cell cycle | |||||
| Down-regulation | |||||
| Mm.034554 | E2f4 | E2f4 | 7 | -1 | Cell cycle |
| Mm.206841 | SMC4-like 1 | Smc4/1 | 5 | ND | Chromosome segregation |
| Mm.26412 | SMC1-like 1 | Smc1/1 | 4 | Yes | Chromosome segregation |
| Mm.16711†‡ | Mini chromosome maintenance deficient 2 | Mcm2 | 4 | Yes | DNA replication |
| Mm.4189†§∥ | Cyclin A2 | Ccna2 | 4 | -1 | Cell cycle |
| Mm.4538 | Lamin B receptor | Lbr | 4 | Yes | Chromatin/nuclear matrix |
| Mm.124†‡ | Thymopoietin/LAP2 | Tmpo | 3 | Yes | Chromatin/nuclear matrix |
| Mm.42196 | Ubiquitin-like, containing ring finger domain 1 | Uhrf1/ICBP90 | 3 | Yes | Replication/DNA damage |
| Mm.37562 | Polymerase (DNA directed), delta 3 | Pold3 | 3 | Yes | DNA replication |
| Mm.197520‡ | F-box only protein 5/Emi1 | Fbxo5 | 3 | Yes | Mitosis |
| Up-regulation | |||||
| Mm.3141 | Cyclin D2 | Ccnd2 | 4 | -1 | Cell cycle |
| Mm.6856‡ | Pituitary tumor-transforming 1/securin | Pttg1 | 3 | No | Chromosome segregation |
| Gene regulation (transcription and translation) | |||||
| Down-regulation | |||||
| Mm.261664 | Histone 1, H4m | Hist1h4m | 5 | ND | Chromatin assembly |
| Mm.6775 | Ornithine decarboxylase antizyme inhibitor | Oazin | 5 | -1 | Cell growth/metabolism |
| Mm.16519 | Nuclear receptor 2F2/COUP TFII | Nr2f2 | 4 | -1 | Transcriptional factor |
| Mm.643 | Ribosomal protein S15 | Rps15 | 4 | -1 | Ribosome biogenesis |
| Mm.2291 | Cytotoxic granule-associated RNA BP 1 | Tia1 | 4 | No | |
| Mm.28003†∥¶ | Chromobox homolog 5 (Drosophila Hp1α) | Cbx5 | 3 | Yes | Chromatin assembly |
| Mm.641 | Activating transcription factor 4/CREB2 | Atf4 | 3 | -1 | Cell cycle/proliferation |
| Mm.142822 | Ewing sarcoma homolog | Ewsh | 3 | Yes | Transcription |
| Mm.25877 | Putative homeodomain transcription factor | Phtf | 3 | -1 | Transcription |
| Mm.1693 | High mobility group box 2 | Hmgb2 | 3 | -1 | Chromosome organization |
| Mm.2238 | Eukaryotic translation initiation factor 3 | Eif3 | 3 | -1 | Protein biosynthesis |
| Mm.260384 | Eukaryotic translation initiation factor 5 | Eif5 | 3 | ND | Protein biosynthesis |
| Mm.46401 | Son cell proliferation protein | Son | 3 | -1 | |
| Mm.196782 | Zinc finger protein 212 | Znf212 | 3 | ND | |
| Up-regulation | |||||
| Mm.4419 | Ribosomal protein L5 | Rpl5 | 3 | No | Ribosome biogenesis |
| Mm.42767 | Ribosomal protein S17 | Rps17 | 3 | No | Ribosome biogenesis |
| Erythropoiesis | |||||
| Down-regulation | |||||
| Mm.7248 | Band 3 | Slc4a1 | 7 | -1 | Anion transport |
| Mm.28683 | Transferrin receptor | Trfr | 6 | No | Iron ion transport |
| Mm.143731 | ABC-me | Abcb10 | 5 | Yes | Heme transport |
| Mm.196110 | Hemoglobin alpha, adult chain 1 | Hba-a1 | 5 | No | Oxygen transport |
| Mm.233825 | Hemoglobin, beta adult major chain | Hbb-b1 | 4 | No | Oxygen transport |
| Mm.249983 | Hemoglobin, beta adult minor chain | Hbb-b2 | 3 | No | Oxygen transport |
| Mm.1090 | Glutathione peroxidase 1 | Gpx1 | 3 | -1 | Oxidative stress response |
| Up-regulation | |||||
| Mm.35830 | Hemoglobin Y, beta-like embryonic chain | Hbb-y | 5 | No | Oxygen transport |
| Miscellaneous | |||||
| Down-regulation | |||||
| Mm.6417 | CD24a antigen | Cd24a | 5 | -1 | Defense response |
| Mm.14802 | H19 fetal liver mRNA | H19 | 5 | ND | |
| Mm.9086 | Solute carrier family 16, member 1 | Slc16a1 | 4 | Yes | Lactate metabolism |
| Mm.8649 | Ag identified by mAb MRC OX-2 | Mox2 | 4 | No | |
| Mm.45436 | Lysozyme | Lyzs | 4 | No | Cell wall catabolism |
| Mm.103038 | Axonal-associated cell adhesion molecule | Axcam | 4 | No | Cell adhesion |
| Mm.297 | Actin, beta, cytoplasmic | Actb | 4 | -1 | Cytoplasm organization |
| Mm.6354 | Phosphatidylinositol glycan, class Q | Pigq | 3 | -1 | Transferase activity |
| Mm.36710 | Mitochondrial solute carrier protein | Mscp | 3 | ND | |
| Mm.3350 | Microtubule-actin crosslinking factor 1 | Macf1 | 3 | ND | |
| Mm.153481 | Calnexin | Canx | 3 | -1 | Ca++ ion storage and binding |
| Mm.2128 | S100 calcium binding protein A9 | S100a9 | 3 | No | Ca++ ion binding |
| Mm.133872 | Stefin A1 | Stfa1 | 3 | ND | Cysteine protease inhibitor |
| Mm.201778 | Ubiquitin specific protease 7 | Usp7 | 3 | ND | Protein catabolism |
| Mm.103199 | Pleiomorphic adenoma gene-like 2 24 ESTs | Plagl2 | 3 | -1 | |
| Up-regulation | |||||
| Mm.43831 | Galectin-1 | Lgals1 | 6 | -1 | Myoblast differentiation |
| Mm.667 | Glutathione S-transferase, mu 5 | Gstm5 | 5 | -1 | Reactive oxygen metabolism |
| Mm.5567 | PDZ and LIM domain 1 | Pdlim1 | 3 | -1 | Intracellular signaling |
| Mm.256034 | Chaperonin subunit 3 (gamma) | Cct3 | 3 | ND | Chaperone activity |
| Mm.1779 | Sterol carrier protein 2, liver 4 ESTs | Scp2 | 3 | ND | Peroxisome organization |
Expression change in E2f4-/- FL/unigene . | Gene name . | Gene symbol . | Gene deregulated, no. arrays . | E2F site* . | Description . |
|---|---|---|---|---|---|
| Cell cycle | |||||
| Down-regulation | |||||
| Mm.034554 | E2f4 | E2f4 | 7 | -1 | Cell cycle |
| Mm.206841 | SMC4-like 1 | Smc4/1 | 5 | ND | Chromosome segregation |
| Mm.26412 | SMC1-like 1 | Smc1/1 | 4 | Yes | Chromosome segregation |
| Mm.16711†‡ | Mini chromosome maintenance deficient 2 | Mcm2 | 4 | Yes | DNA replication |
| Mm.4189†§∥ | Cyclin A2 | Ccna2 | 4 | -1 | Cell cycle |
| Mm.4538 | Lamin B receptor | Lbr | 4 | Yes | Chromatin/nuclear matrix |
| Mm.124†‡ | Thymopoietin/LAP2 | Tmpo | 3 | Yes | Chromatin/nuclear matrix |
| Mm.42196 | Ubiquitin-like, containing ring finger domain 1 | Uhrf1/ICBP90 | 3 | Yes | Replication/DNA damage |
| Mm.37562 | Polymerase (DNA directed), delta 3 | Pold3 | 3 | Yes | DNA replication |
| Mm.197520‡ | F-box only protein 5/Emi1 | Fbxo5 | 3 | Yes | Mitosis |
| Up-regulation | |||||
| Mm.3141 | Cyclin D2 | Ccnd2 | 4 | -1 | Cell cycle |
| Mm.6856‡ | Pituitary tumor-transforming 1/securin | Pttg1 | 3 | No | Chromosome segregation |
| Gene regulation (transcription and translation) | |||||
| Down-regulation | |||||
| Mm.261664 | Histone 1, H4m | Hist1h4m | 5 | ND | Chromatin assembly |
| Mm.6775 | Ornithine decarboxylase antizyme inhibitor | Oazin | 5 | -1 | Cell growth/metabolism |
| Mm.16519 | Nuclear receptor 2F2/COUP TFII | Nr2f2 | 4 | -1 | Transcriptional factor |
| Mm.643 | Ribosomal protein S15 | Rps15 | 4 | -1 | Ribosome biogenesis |
| Mm.2291 | Cytotoxic granule-associated RNA BP 1 | Tia1 | 4 | No | |
| Mm.28003†∥¶ | Chromobox homolog 5 (Drosophila Hp1α) | Cbx5 | 3 | Yes | Chromatin assembly |
| Mm.641 | Activating transcription factor 4/CREB2 | Atf4 | 3 | -1 | Cell cycle/proliferation |
| Mm.142822 | Ewing sarcoma homolog | Ewsh | 3 | Yes | Transcription |
| Mm.25877 | Putative homeodomain transcription factor | Phtf | 3 | -1 | Transcription |
| Mm.1693 | High mobility group box 2 | Hmgb2 | 3 | -1 | Chromosome organization |
| Mm.2238 | Eukaryotic translation initiation factor 3 | Eif3 | 3 | -1 | Protein biosynthesis |
| Mm.260384 | Eukaryotic translation initiation factor 5 | Eif5 | 3 | ND | Protein biosynthesis |
| Mm.46401 | Son cell proliferation protein | Son | 3 | -1 | |
| Mm.196782 | Zinc finger protein 212 | Znf212 | 3 | ND | |
| Up-regulation | |||||
| Mm.4419 | Ribosomal protein L5 | Rpl5 | 3 | No | Ribosome biogenesis |
| Mm.42767 | Ribosomal protein S17 | Rps17 | 3 | No | Ribosome biogenesis |
| Erythropoiesis | |||||
| Down-regulation | |||||
| Mm.7248 | Band 3 | Slc4a1 | 7 | -1 | Anion transport |
| Mm.28683 | Transferrin receptor | Trfr | 6 | No | Iron ion transport |
| Mm.143731 | ABC-me | Abcb10 | 5 | Yes | Heme transport |
| Mm.196110 | Hemoglobin alpha, adult chain 1 | Hba-a1 | 5 | No | Oxygen transport |
| Mm.233825 | Hemoglobin, beta adult major chain | Hbb-b1 | 4 | No | Oxygen transport |
| Mm.249983 | Hemoglobin, beta adult minor chain | Hbb-b2 | 3 | No | Oxygen transport |
| Mm.1090 | Glutathione peroxidase 1 | Gpx1 | 3 | -1 | Oxidative stress response |
| Up-regulation | |||||
| Mm.35830 | Hemoglobin Y, beta-like embryonic chain | Hbb-y | 5 | No | Oxygen transport |
| Miscellaneous | |||||
| Down-regulation | |||||
| Mm.6417 | CD24a antigen | Cd24a | 5 | -1 | Defense response |
| Mm.14802 | H19 fetal liver mRNA | H19 | 5 | ND | |
| Mm.9086 | Solute carrier family 16, member 1 | Slc16a1 | 4 | Yes | Lactate metabolism |
| Mm.8649 | Ag identified by mAb MRC OX-2 | Mox2 | 4 | No | |
| Mm.45436 | Lysozyme | Lyzs | 4 | No | Cell wall catabolism |
| Mm.103038 | Axonal-associated cell adhesion molecule | Axcam | 4 | No | Cell adhesion |
| Mm.297 | Actin, beta, cytoplasmic | Actb | 4 | -1 | Cytoplasm organization |
| Mm.6354 | Phosphatidylinositol glycan, class Q | Pigq | 3 | -1 | Transferase activity |
| Mm.36710 | Mitochondrial solute carrier protein | Mscp | 3 | ND | |
| Mm.3350 | Microtubule-actin crosslinking factor 1 | Macf1 | 3 | ND | |
| Mm.153481 | Calnexin | Canx | 3 | -1 | Ca++ ion storage and binding |
| Mm.2128 | S100 calcium binding protein A9 | S100a9 | 3 | No | Ca++ ion binding |
| Mm.133872 | Stefin A1 | Stfa1 | 3 | ND | Cysteine protease inhibitor |
| Mm.201778 | Ubiquitin specific protease 7 | Usp7 | 3 | ND | Protein catabolism |
| Mm.103199 | Pleiomorphic adenoma gene-like 2 24 ESTs | Plagl2 | 3 | -1 | |
| Up-regulation | |||||
| Mm.43831 | Galectin-1 | Lgals1 | 6 | -1 | Myoblast differentiation |
| Mm.667 | Glutathione S-transferase, mu 5 | Gstm5 | 5 | -1 | Reactive oxygen metabolism |
| Mm.5567 | PDZ and LIM domain 1 | Pdlim1 | 3 | -1 | Intracellular signaling |
| Mm.256034 | Chaperonin subunit 3 (gamma) | Cct3 | 3 | ND | Chaperone activity |
| Mm.1779 | Sterol carrier protein 2, liver 4 ESTs | Scp2 | 3 | ND | Peroxisome organization |
ND indicates not determined.
The promoter regions were analyzed using the Ensemble Mouse Genome Browser (www.ensembl.org/Mus_musculus/). Sequences were scanned for consensus E2F binding sites (TTTC/GC/GCGC) allowing for 1 mismatch (-1) in the TTT triplet sequence and spanned 1000 base pairs upstream of the transcription start site to the end of intron 1 of each gene.
Ren et al.26
Balciunaite et al.29
Takahashi et al.25
Rayman et al.32
Weinman et al.27
The first group of genes consistently deregulated in E2f4–/– FL includes genes required for normal cell cycle progression (Table 2; for the complete microarray data set, refer to Table S1). These include down-regulation of genes previously identified as direct E2f4 targets, such as Ccna2 (encoding cyclin A2), Smc4l1, and Emi1,25-29 as well as other E2F-regulated genes, such as Mcm2,30 and up-regulation of Ccnd2 and Pttg1(securin) mRNA. The second group consists of genes required for gene regulation, including genes with functions in chromatin assembly and modification, transcription, and translation. The chromatin remodeling genes include Cbx5/Hp1α (another described direct E2f4 target)26,27 and Hist1h4m. Protein synthetic machinery, such as elongation factors Eif3 and Eif5, was also found up-regulated, while some ribosomal protein mRNAs were down-regulated.
The third group represents genes associated with terminal erythroid differentiation. Of the erythroid-specific genes deregulated, many are associated with hemoglobin production. E15.5 E2f4–/– FL showed increased embryonic globin gene expression (Hbb-y) and concurrent decrease in adult globin expression (Hbb-b1, Hbb-b2, and Hba-a1). This may reflect the anemia present in E2f4–/– fetuses and the proportional difference in “primitive” erythrocytes (residual yolk sac–derived erythroid cells that express predominantly embryonic globins) versus “definitive” erythrocytes (FL-derived erythroid cells that express predominantly adult globins that will constitute the adult erythroid compartment) observed in E2f4–/– FL. In addition, a selection of additional erythroid-specific genes is down-regulated, including Src4a1 (encoding the band 3 protein), Trfr (transferrin receptor), Abc-me, and Gpx1 (glutathione peroxidase). Other markers expressed solely on mature erythrocytes, such as AlasII and Nfe2, are each present on the array and remain unchanged in E2f4–/– FL. That only a selection of genes associated with mature erythroid differentiation is deregulated is consistent with the absence of a severe block in erythroid differentiation in E2f4–/– FL erythropoiesis. Furthermore, we found that the majority of the erythroid-specific genes deregulated in E2f4–/– FL do not contain an E2F binding site consistent with the notion that at least some of these genes are not directly regulated by E2f4.
Of note, of the 84 genes deregulated in E2f4–/– FL, the vast majority (70) were found to be down-regulated in E2f4–/– FL, consistent with a possible role for E2f4 as a transcriptional activator in FL erythroid cells. Of importance, the down-regulation of a large group of genes involved in cell cycle progression in the E2f4–/– FL supports the notion of a role for E2f4 in promoting cellular proliferation.
E2f4 directly regulates the transcriptional expression of a subset of cell cycle genes in erythroid cells
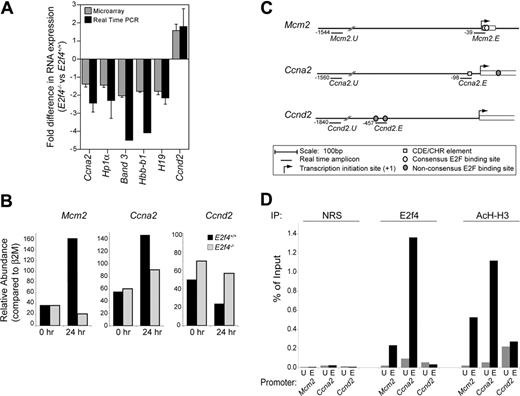
The down-regulation of genes previously reported to be direct E2F4 target genes suggested that E2f4 could be directly regulating the expression of genes involved in cell cycle progression in erythroid cells. To examine this, we undertook chromatin immunoprecipitation (ChIP) studies on in vitro differentiating erythroid progenitors. The CCNA2 gene has previously been shown to be bound and directly regulated by E2F4 in human fibroblasts; however, it must be noted that, in this context, E2F4 is thought to regulate the transcriptional repression rather than activation of these genes.25,31,32 We also examined Mcm2, previously described as being E2F regulated,30 and as a control, the Ccnd2 gene, which contains 2 nonconsensus E2F sites (Figure 6C) but is thought to be primarily regulated through myc rather than E2F.33
Consistent with data from whole FL analysis, E2f4–/– erythroid cultures showed decreased expression of Ccna2 (∼1.7-fold) and Mcm2 (∼ 8-fold) and increased expression of Ccnd2 (∼ 2.5-fold) at day 1 of in vitro differentiation, the time at which the cell cycle defects first become apparent in E2f4–/– erythroid cells (Figure 6B). To determine whether E2f4 was directly bound to the promoters of these genes at this time, wild-type erythroid cultures were harvested at day 1 of differentiation and analyzed by ChIP for E2f4 binding. As can be seen in Figure 6D, E2f4 was consistently enriched at Ccna2 and Mcm2 promoters but not at upstream control sites. The signal was E2f4 specific as samples immunoprecipitated with nonimmune rabbit serum or a control rabbit antibody directed to yeast Gal4 did not produce any significant enrichment (Figure 6D and data not shown). Consistent with the lack of E2F consensus sites, no enrichment for E2f4 was detected for the Ccnd2 promoter. In addition, in the same experiment, we examined the acetylation status of the gene promoters as a surrogate marker for gene activation and transcription. All 3 genes were found enriched for acetylated histone H3 (AcH-H3), indicating an open chromatin structure and consistent with transcriptional expression of these genes in erythroid cells at the time point examined.
E2F4 directly binds to promoter regions of Ccna2 and Mcm2 but notCcnd2. (A) Fold difference in RNA expression between E2f4+/+ and E2f4–/– E15.5 FLs determined by microarray analysis (##; n = 7) and real-time PCR (▪; n = 3), the latter normalized to GAPDH expression. Data represent 2 to 3 individual FLs pooled per experiment. (B) Relative abundance of Mcm2, Ccna2, and Ccnd2 mRNA transcript in E2f4+/+ and E2f4–/– erythroid cells at 0 and 24 hours after induction of differentiation, normalized to β 2M expression. Data shown are representative of 3 independent experiments. (C) For ChIP experiments, real-time PCR primers were designed to detect genomic DNA spanning putative E2F binding sites present in the Mcm2, Ccna2, and Ccnd2 promoters (Mcm2.E, Ccna2.E, and Ccnd2.E), in addition to nonspecific upstream sites (Mcm2.U, Ccna2.U, and Ccnd2.U), as marked. (D) ChIP experiments were performed on wild-type erythroid cells 24 hours after induction of differentiation in vitro. Cross-linked DNA/protein was immunoprecipitated using nonimmune rabbit serum (NRS), or rabbit polyclonal antibodies recognizing E2F4 or acetylated histone H3 (AcH-H3). Precipitated DNA samples were amplified with primers recognizing upstream promoter nonspecific sites (U) and the E2F consensus sites (E) of candidate genes. Quantification of precipitated DNA was measured by real-time PCR, where the amount of immunoprecipitated promoter DNA is depicted as a percentage of the amount present in the total input chromatin sample. Data shown are representative of 5 independent experiments. Error bars indicate standard deviation.
E2F4 directly binds to promoter regions of Ccna2 and Mcm2 but notCcnd2. (A) Fold difference in RNA expression between E2f4+/+ and E2f4–/– E15.5 FLs determined by microarray analysis (##; n = 7) and real-time PCR (▪; n = 3), the latter normalized to GAPDH expression. Data represent 2 to 3 individual FLs pooled per experiment. (B) Relative abundance of Mcm2, Ccna2, and Ccnd2 mRNA transcript in E2f4+/+ and E2f4–/– erythroid cells at 0 and 24 hours after induction of differentiation, normalized to β 2M expression. Data shown are representative of 3 independent experiments. (C) For ChIP experiments, real-time PCR primers were designed to detect genomic DNA spanning putative E2F binding sites present in the Mcm2, Ccna2, and Ccnd2 promoters (Mcm2.E, Ccna2.E, and Ccnd2.E), in addition to nonspecific upstream sites (Mcm2.U, Ccna2.U, and Ccnd2.U), as marked. (D) ChIP experiments were performed on wild-type erythroid cells 24 hours after induction of differentiation in vitro. Cross-linked DNA/protein was immunoprecipitated using nonimmune rabbit serum (NRS), or rabbit polyclonal antibodies recognizing E2F4 or acetylated histone H3 (AcH-H3). Precipitated DNA samples were amplified with primers recognizing upstream promoter nonspecific sites (U) and the E2F consensus sites (E) of candidate genes. Quantification of precipitated DNA was measured by real-time PCR, where the amount of immunoprecipitated promoter DNA is depicted as a percentage of the amount present in the total input chromatin sample. Data shown are representative of 5 independent experiments. Error bars indicate standard deviation.
These data therefore indicate that E2f4 binds directly to the promoter of Ccna2 and Mcm2 genes at a time that they are actively transcribed. As loss of E2f4 results in decreased expression of Ccna2 and Mcm2 genes, we propose that E2f4 contributes to the transcriptional activation of Ccna2 and Mcm2 in primary erythroid cells. These data support the notion that the contribution of E2f4 to the transcriptional activation of genes required for cell cycle progression may be responsible for the proliferation defects of E2f4–/– erythroblasts.
Discussion
Prior to the current study, the vast majority of reports on E2f4 had assigned its function as an inhibitor of cellular proliferation through promotion of cell cycle exit and differentiation. We have examined the basis of the defective FL erythropoiesis observed in E2f4-deficient embryos. Our studies demonstrate a new role for E2f4 in promoting cell cycle progression and proliferation of erythroid cells in vivo and provide a functional explanation for the macrocytic anemia observed in E2f4–/– mice.
A novel role for E2f4 in promoting erythroid proliferation
Data presented here demonstrate that the major role of E2f4 in erythropoiesis is in the regulation of proliferation. As cell division and the reduction in cell size (mass) are tightly linked in erythroid differentiation, our data suggest that this reduction in division results in the macrocytosis observed in E2f4–/– erythroid cells in vitro and in vivo. We therefore propose that the fetal macrocytic anemia of E2f4–/– mice is a direct consequence of reduced proliferative expansion of the erythroid-committed compartment of the FL. Supporting this conclusion, mice deficient for positive regulators of the cell cycle, cyclin D2 and D3, or Cdk4 and Cdk6, show a fetal anemia remarkably similar to that of E2f4–/– mice.34,35 This suggests that E2f4 may be a downstream effector of cyclin D–Cdk complexes in fetal erythropoiesis. Furthermore, combined loss of the transcriptionally “activating” E2F family members, E2f1 and E2f2, results in a marked macrocytic anemia with megaloblastic features in adult mice.36 In view of the data presented here, we propose an analogous role for E2f4 in activating transcription and promoting cell cycle progression in fetal erythropoiesis. Our findings demonstrate that the function of individual E2F family members in promoting or inhibiting cell cycle progression is highly dependent on the cellular and biochemical context. Therefore, in the particular case of E2f4 in the erythroid lineage, E2f4's major function is to promote cellular proliferation rather than cell cycle exit as has been proposed in fibroblasts.5
What may be giving rise to the cell cycle defects observed in E2f4–/– erythroid cells? The fetal macrocytic anemia of E2f4–/– mice has a number of striking similarities to human megaloblastic anemia. These include erythroid macrocytosis, anemia, and the increased presence of Howell-Jolly bodies within mature erythrocytes.13 However, these megaloblastic features appear restricted to erythroid lineage as the neutropenia or thrombocytopenia (decreased myeloid and platelet numbers, respectively) usually seen in a megaloblastic anemia patient is not observed in E2f4–/– mice.13 The cellular defects characteristic of megaloblastic anemia are thought to result from slow replication due to decreased dietary or metabolic availability of folate or vitamin B12 and the ensuing limited pool of nucleotides.37 Although folate metabolism enzymes have been described as E2F targets,38-40 we suspect defects in folate metabolism are not the underlying cause of the E2f4–/– erythroid phenotype. First, we have observed no differences in the expression of Dhfr, Tk, and Ts between E2f4+/+ and E2f4–/– FL cells in our cDNA microarrays or Northern analysis (see Table S1 and data not shown). In addition, we do not observe the large increase in apoptosis that has been previously reported for mouse models of megaloblastic anemia arising from dietary folate deficiency.41 Finally, in preliminary experiments, leucovorin, a folate metabolite that bypasses the requirement for much of the enzymatic processing of dietary folate, did not rescue the E2f4–/– erythroid phenotype in vitro at physiologic and supraphysiologic doses (K.M.K. and P.O.H., unpublished observation, October 2003).
Another possibility is that the delayed cell cycle progression of E2f4–/– erythroid cells results from the activation of a DNA damage response. The appearance of increased Howell-Jolly bodies in the E2f4 peripheral blood erythrocytes13 and the increased number of cells present in late S and/or G2 phase in E2f4–/– erythroid cells could be indicative of such an increase in DNA damage, or an inability to repair DNA damage sufficiently. E2Fs have previously been implicated to have direct roles in checkpoint maintenance and DNA repair.1,42 Further experiments will be required to examine whether an aberrant activation of checkpoint response can contribute to the E2f4–/– erythroid phenotype.
Notwithstanding this last possibility, the data presented in this study strongly support a model by which E2f4 promotes cell cycle progression through the direct transcriptional regulation of cell cycle genes. A number of genes deregulated in the E2f4–/– FL play essential roles in replication and progression through mitosis including Mcm2, Ccna2, Smc1 and 4, Emi-1, and Pttg1/securin. Of importance, our ChIP studies indicate that at least for a subset of these genes deregulation of their expression does not merely reflect the E2f4–/– erythroid cell cycle defects but instead most likely results from loss of E2f4 at their promoters. Therefore, deregulated protein levels of one or more of the cell cycle control genes identified in the array experiments could result in slow progression through the cell cycle and would lead to the phenotype observed in E2f4–/– cells.
A potential role for E2f4 in transcriptional activation in erythroid cells
The presence of E2F4, pRB family members, and associated chromatin modifiers at E2F promoters of cells held in G0, and the subsequent loss of E2F4 and repressive complexes at these promoters at the G1/S transition, have been key findings supporting the transcriptional repressive function of E2F4.25,32 However, overexpression experiments have previously shown that expression of nuclear E2F4 can effectively induce the transcription of E2F-dependent promoters (and promote cell cycle progression),43 indicating an intrinsic ability for E2f4 to be a transcriptional activator. Our studies provide the first evidence in vivo that E2f4 may behave primarily as a transcriptional activator in the context of differentiating erythroid cells.
First, our data indicate that E2f4 is relatively free of regulation by pRB family members in differentiating erythroid cells. Despite high expression of p107 and pRb in erythroid cells, our gel-shift experiments indicate that the vast majority of DNA-bound E2f4 is in “free” form with only a small proportion of DNA-bound p107/E2f4 complexes present and no DNA-bound pRb/E2f4 complexes detectable (Figure 1). In addition, the minimal overlap in genes deregulated in E2f4–/–, Rb–/–, or p130–/–p107–/– FL (K.M.K. and P.O.H., unpublished observation, December 2002) is also consistent with the minor contribution of E2f4-pRB family–repressive complexes in differentiating erythroid cells. This property would contribute to E2f4's ability to promote proliferation in this tissue and would also facilitate E2f4 to function as a transcriptional activator. Of importance, loss of E2f4 resulted in reduced levels of several well-characterized E2F-responsive genes, and our ChIP analyses confirmed that E2f4 was bound to the promoter of a subset of these genes, as they were actively transcribed. E2f4 is therefore likely to contribute directly to their transcriptional activation. Detailed characterization of promoter occupancy and transcriptional regulation of direct E2f4 targets in erythroid cell populations synchronized in cell cycle and differentiation stages will be required to determine exactly how the relative contribution of the activator and repressor functions of E2f4 contribute to gene regulation in erythroid cells.
Our original analysis of E2f4–/– mice revealed a critical requirement for E2f4 in the control of erythropoiesis; however, how E2f4 was regulating this process remained unknown.13 We have now identified the cellular basis for the requirement for E2f4 in fetal erythropoiesis. Our studies have revealed a novel and unexpected function for E2f4 in promoting cell proliferation in a tissue compartment that has been genetically determined to be dependent on E2f4 function. These studies not only provide new insights into the regulation of fetal erythropoiesis but also have revealed new modes of cell cycle regulation by the pRb-E2F pathway in development.
Prepublished online as Blood First Edition Paper, April 4, 2006; DOI 10.1182/blood-2005-09-008656.
K.M.K. was supported by a Melbourne University Postgraduate Scholarship and P.O.H. was supported by a Special Fellowship of the Leukemia and Lymphoma Society of America and a Career Development Award from the National Health and Medical Research Council of Australia. This work was funded by a project grant from the National Health and Medical Research Council of Australia.
K.M.K, A.J.C., R.M.I., and P.O.H contributed to the design, performance, and analysis of research; K.M.K. and P.O.H. wrote the paper; and all authors checked the final version of the paper.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Hartmut Beug for advice with setting up the erythroid culture system. Many thanks to Andrew Holloway, Ryan van Laar, Richard Tothill, and the Peter MacCallum Microarray Facility for technical assistance with microarray analysis; Olivia Cakebread for technical assistance; Jodie Hayes and Mary Thorpe for animal husbandry; Andrew Fryga and Ralph Rossi for assistance with FACS; Gretchen Poortinga and Jo White for advice with ChIP analysis; and Jian-Guo Zhang and Helene Martin for the kind gift of reagents. We would also like to thank Grant McArthur, Helena Richardson, Louise Purton, and Sarah Russell for comments and critical reading of paper.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal