Abstract
Aberrant somatic hypermutation (SHM) has been identified as a mechanism for genomewide instability in diffuse large B-cell lymphoma (DLBCL). To assess whether aberrant SHM plays a role in the molecular pathogenesis of Hodgkin lymphoma (HL), we investigated microdissected neoplastic cells of nodular lymphocyte-predominant HL (NLPHL; n = 10) and classic HL (cHL; n = 9) for the presence of mutations in the 5′ sequences of 4 previously identified aberrant SHM targets (PIM1, PAX5, RhoH/TTF, c-MYC). Mutations in one or more genes were detected in 80% of NLPHLs and 55% of cHLs, with 50% and 30% of patients carrying mutations in 2 or more genes, respectively. The most frequently involved protooncogene was PAX5, mutated in 7 of 9 patients with NLPHL and 2 of 9 patients with cHL. In total, 34 mutations were detected in NLPHL (frequency, 1.04/1000 bp) and 35 were detected in patients with cHL (frequency, 1.92/1000 bp). Mutations were of somatic origin because they were absent in control T cells and shared most of the features of the immunoglobulin variable (IGV) gene–associated SHM mechanism—ie, single nucleotide substitutions (n = 63) with rare deletions/insertions (n = 6) and a predominance of transitions over transversions with preferential targeting motifs. Our finding that NLPHL and cHL are targeted by aberrant SHM, as is DLBCL, suggests that these lymphomas may share common molecular pathogenetic events.
Introduction
Hodgkin lymphoma is a neoplasm that encompasses 2 distinct clinicopathologic entities, lymphocyte-predominant (NLPHL) and classic Hodgkin lymphoma (cHL).1 It is now widely recognized that cHL (more than 95% of cases) and NLPHL are derived from germinal center B cells, probably at different stages of differentiation.2-6 A number of clinical and histologic features suggest that HL should be distinguished from other germinal center–derived B-cell lymphomas.1 However, an overlap is occasionally observed between HL and B-cell non-Hodgkin lymphoma (NHL), and this remains an unsolved issue.
During T-cell–dependent immune responses, antigen-activated B cells undergo clonal expansion in germinal centers, where the immunoglobulin variable (IGV) region genes are subject to a physiologic process of somatic hypermutation (SHM).7 This process introduces mainly single nucleotide substitutions to create variants with higher affinity for the antigen, which will be selected to become memory B cells or plasma cells.8
The SHM process is not restricted to the immunoglobulin loci because other genes expressed in the germinal centers, such as BCL6,9-11 CD95/FAS,12 CD79a, and CD79b,13 have been found mutated in normal germinal center B cells, though the significance of these mutations is unknown.
A few years ago, the SHM process was found to malfunction in more than 50% of diffuse large B-cell lymphomas (DLBCLs) in immunocompetent hosts and immunocompromised hosts.14-17 As a consequence, a number of genes, including the known protooncogenes PIM1, c-MYC, RhoH/TTF (ARHH), and PAX5, are aberrantly targeted by mutations in their 5′ regions, including coding sequences. The 4 hypermutable genes are also susceptible to chromosomal translocations in the same region, consistent with a role for hypermutation in generating translocations through DNA double-strand breaks.14,18,19
Although NLPHL and most cHLs are now known to originate from germinal center B cells,1,5,6,20 no studies have been performed to investigate the presence of aberrant SHM in this disease type, primarily because of technical difficulties in analyzing tumor cells that usually represent less than 5% of cells in the infiltrated tissue.1
To assess whether the SHM process functions aberrantly in HL, we investigated tumor cells microdissected from frozen lymph node samples of NLPHL and cHL for the presence of mutations in the PIM1, PAX5, RhoH/TTF, and c-MYC genes. We report that, as in DLBCL, aberrant SHM is a frequent event in NLPHL and in cHL, suggesting that these lymphoma entities may share common molecular pathogenetic events.
Materials and methods
Tissue samples
The study was carried out on frozen lymph node specimens from patients with typical NLPHL (n = 10) and cHL (n = 9) who met the criteria of WHO classification.1 Frozen samples were provided by the Institutes of Pathology in Perugia, Pavia, Aviano, and Bologna, Italy; Toulouse, France; and Barcelona, Spain. Each case had been diagnosed by an expert hemopathologist and was reviewed at the University of Perugia by 2 of the authors (B.F., A.L.). The protocols of this study were approved by the Department of Clinical and Experimental Medicine at the University of Perugia.
Immunostaining procedure
Frozen tissue sections were cut at 9-μm thickness, dried overnight, and fixed at room temperature in acetone for 10 minutes. Lymphocytic/histiocytic (L/H) cells in patients with NLPHL were immunostained with a specific anti-CD20 monoclonal antibody (DakoCytomation, Glostrup, Denmark). Immunolabeling of Hodgkin and Reed-Sternberg cells in patients with cHL was performed with an anti-CD30 (Ber-H2) monoclonal antibody kindly provided by Prof. Harald Stein (Berlin, Germany). Control T cells were labeled with a specific anti-CD3 polyclonal antibody purchased from DakoCytomation. All immunostainings were performed using the immunoalkaline phosphatase (alkaline phosphatase anti–alkaline phosphatase [APAAP]) technique.21 Images were collected by using an Olympus BX51 microscope equipped with a UPlanF1 100×/1.3 oil-immersion objective lens; an Olympus Camedia camera was used to capture images, and Camedia Master Pro 4 software was used to process them (all equipment from Olympus, Melville, NY).
EBV analysis
Immunostaining for latent membrane protein-1 (LMP1) and in situ hybridization for Epstein-Barr virus (EBV)–encoded small RNA (EBER) was carried out according to standard procedures.
Cell microdissection and DNA extraction
Single tumor cells from 2 patients with HL were dissected with a hydraulic micromanipulator and were collected in a final volume of 10 μLof 10 mM Tris-HCl, pH 7.4, as previously described.6 In 11 patients with HL, single tumor cells were microdissected with the use of a pulsed ultraviolet laser (microbeam with Palm “inside” technology; Olympus, Milan, Italy). Cells were laser-pressure catapulted into a cap positioned and moved with a joystick. The cap surface had been previously coated with 20 to 40 μL of 10 mM Tris-HCl, pH 7.4. Catapulted cells were centrifuged into 500-μL DNAse-free tubes previously coated with 1% BSA/10 mM Tris-HCl, pH 7.4. In 6 patients, cells were dissected with the use of a hydraulic micromanipulator and laser technology.
For each patient, 1 to 4 pools, each containing 10 to 50 tumor cells, were collected, for a total of 50 to 200 cells per patient. Pools of 30 to 50 T cells were also microdissected from each patient as a negative control for the somatic origin of the mutations. Isolated cells were frozen and cryopreserved at –80°C. Before polymerase chain reaction (PCR) analysis, DNA was extracted from the laser-microdissected cells by incubation with 400 μg/mL proteinase K/0.2% Tween-20 at 55°C for 4 hours and subsequent inactivation of the enzyme at 95°C for 10 minutes.
Mutational analysis of PIM1, PAX5, RhoH/TTF, c-MYC, and IGV genes
Mutational analysis of the 4 proto-oncogenes was performed on selected regions that contained greater than 90% of the mutations found in DLBCL of immunocompetent and immunocompromised hosts.14,16
Amplification of all genes was performed by a nested PCR protocol; this was followed by direct sequencing. The first round of amplification consisted of 2 multiplex reactions amplifying c-MYC exon 1, RhoH/TTF, and PAX5 in one reaction and c-MYC exon 2 and PIM1 in the other reaction. PCR was performed with the use of the Qiagen Multiple PCR mastermix (Qiagen, Milan, Italy) in a final volume of 50 μL on 5 to 10 μL digested cells containing 20 to 40 tumor cells. In the second round of amplification, each gene was amplified in separate reactions with 2 μL of the first round of reaction and internal primers. Oligonucleotide sequences used as primers for the first and second rounds of amplification are provided in Tables S1 and S2 (available on the Blood website; see the Supplemental Tables link at the top of the online article). Genes that could not be amplified by the multiplex approach were amplified individually in separate reactions by nested PCR.
Rearranged IGHV genes were amplified with a seminested PCR approach. The first amplification round was carried out with an IGHV family-specific primer mix complementary to IGHV framework region 1 and combined to a JH outer primer mix. A 3-μL aliquot of the first round was reamplified in a second round in separate reactions for each of the 6 IGHV families, with IGHV family-specific primers combined to a JH inner primer mix.
PCR products were separated by agarose gel electrophoresis, purified with the Perfectprep Gel Cleanup kit (Eppendorf, Hamburg, Germany), and directly sequenced with the ABI Prism Big Dye Terminator version 1.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Milan, Italy). Automated sequencing of samples was performed on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The presence of mutations was always confirmed on independent PCR products. Nucleotide changes caused by previously reported polymorphisms or present in normal DNA from the same patient were disregarded in the assessment of the mutation frequency.
Clonal analysis of PIM1, PAX5, RhoH/TTF, and c-MYC exon 1
The presence of intraclonal heterogeneity, indicative of ongoing hypermutation, was assessed in 3 patients with NLPHL and in 2 patients with cHL previously shown to harbor mutations in PIM1, PAX5, RhoH/TTF, or c-MYC exon 1 by DNA direct sequencing. PIM1, PAX5, and RhoH/TTF were analyzed in the same regions explored by direct sequencing, whereas the subcloning of c-MYC exon 1 was restricted to sequences included from nucleotide +2298 to nucleotide +3088. Each gene was amplified from pools containing approximately 30 cells with the use of Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). After 45 cycles, PCR products were gel purified, incubated with 0.1 mM dATP and Taq polymerase (Qiagen) for 15 minutes at 72°C, and cloned into the pCR4-TOPO plasmid vector (Invitrogen, Paisley, United Kingdom) as recommended by the manufacturer. For each proto-oncogene, at least 20 clones were analyzed. To determine the Pfu Turbo DNA polymerase error rate in our experimental strategy, 20 clones of c-MYC exon 1, 20 clones of PAX5, 20 clones of RhoH/TTF, and 15 clones of PIM1 were generated from a pool of 40 fibroblast cells (derived from the MRC-5 cell line) according to the same PCR and cloning procedures used for HL cells. The average Pfu Turbo error frequency varied from 1.3 × 10–4 for c-MYC exon 1 to 2.4 × 10–4 for PAX5. For the evaluation of ongoing somatic hypermutation, the frequency of mutation in the tumor clones was compared with the frequency of mutation of the MRC-5 clones, and statistical significance was assessed by the Student t test. Other definitions used were unconfirmed mutations (mutations observed in only one clone) and confirmed mutations (mutations observed in more than one clone).
Statistical analysis
Mutation data were managed with Excel software (Microsoft, Redmond, WA). SPSS software (6.0 for Windows; SPSS, Chicago, IL) was used for statistical analysis. Differences in the prevalence of mutated cases and mutation frequency were defined as statistically significant for P values less than or equal to .05. The prevalence of aberrant SHM in NLPHL was compared with the prevalence of aberrant SHM in cHL by χ2 test with Bonferroni adjustment for multiple comparison. For each of the 4 protooncogenes, differences in mutation frequency between NLPHL and cHL were analyzed by the t test. Mutation frequencies were calculated on the nucleotide lengths of the sequences analyzed. Mutation frequencies of individual nucleotides were compared with the expected mutation frequency by the goodness-of-fit χ2 test. Mutation frequencies of nucleotides occurring in the context of an RGYW/WRCY motif was compared with the expected mutation frequency by the goodness-of fit χ2 test.
Results
Frequency of aberrant SHM in NLPHL and cHL
We studied 19 patients with HL (10 NLPHL, 9 cHL) whose clinicopathologic characteristics are shown in Table 1. LMP1 and EBER were negative in 7 of 7 and 5 of 5 NLPHL patients tested, respectively. Results of EBER and LMP1 staining were available in 8 of 9 cHL patients. EBER was positive in 3 of 8 cHL patients (2 of whom were LMP1 positive; the rest of the patients were LMP1 negative).
Clinicopathologic characteristics of the 19 patients with HL
Patient no. . | Histology . | Age, y/sex . | Disease stage . | . | Therapy . | PR/relapse* . | . | Outcome† . | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NLPHL | 52/M | CS-IIA | RT | Rel.;n = 2 | Alive | |||
| 2 | NLPHL | 42/F | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 3 | NLPHL | 14/M | CS-IIB | 4COPP/ABV+RT;36Gy | Rel.;n = 1; | ABMT | Alive | ||
| 4 | NLPHL | 22/M | CS-IA | NA | No | NA | |||
| 5 | NLPHL‡ | 15/M | CS-IIA | NA | Rel.;n = 1 | Alive | |||
| 6 | NLPHL | 72/F | CS-IIIA | 8 | CVPP | No | Alive | ||
| 7 | NLPHL | 27/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 8 | NLPHL | 63/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 9 | NLPHL | 42/M | CS-IIA | RT | No | Alive | |||
| 10 | NLPHL | 11/M | CS-IA | NA | NA | NA | |||
| 11 | HL-NS | 22/F | CS-IIA | 6 | ABVD | No | Alive | ||
| 12 | HL-NS | 40/F | CS-IVA | 6 | ABVD | No | Alive | ||
| 13 | HL-NS | 17/F | CS-IIIB | 4 | ABVD | PR; IEV + ABMT | Alive | ||
| 14 | HL-NS | 33/F | CS-IVA | 8 | ABVD+ RT | No | Alive | ||
| 15 | HL-MC | 67/F | CS-IIIB | 6 | ABVD+ RT | No | Alive | ||
| 16 | HL-NS | 24/F | CS-IVB | 8 | BEACOPP+ RT | No | Alive | ||
| 17 | HL-MC | 28/M | CS-IA | RT | No | Alive | |||
| 18 | HL-MC | 35/M | CS-IIB | 6 | ABVD | No | Alive | ||
| 19 | HL-NS | 67/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
Patient no. . | Histology . | Age, y/sex . | Disease stage . | . | Therapy . | PR/relapse* . | . | Outcome† . | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NLPHL | 52/M | CS-IIA | RT | Rel.;n = 2 | Alive | |||
| 2 | NLPHL | 42/F | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 3 | NLPHL | 14/M | CS-IIB | 4COPP/ABV+RT;36Gy | Rel.;n = 1; | ABMT | Alive | ||
| 4 | NLPHL | 22/M | CS-IA | NA | No | NA | |||
| 5 | NLPHL‡ | 15/M | CS-IIA | NA | Rel.;n = 1 | Alive | |||
| 6 | NLPHL | 72/F | CS-IIIA | 8 | CVPP | No | Alive | ||
| 7 | NLPHL | 27/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 8 | NLPHL | 63/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
| 9 | NLPHL | 42/M | CS-IIA | RT | No | Alive | |||
| 10 | NLPHL | 11/M | CS-IA | NA | NA | NA | |||
| 11 | HL-NS | 22/F | CS-IIA | 6 | ABVD | No | Alive | ||
| 12 | HL-NS | 40/F | CS-IVA | 6 | ABVD | No | Alive | ||
| 13 | HL-NS | 17/F | CS-IIIB | 4 | ABVD | PR; IEV + ABMT | Alive | ||
| 14 | HL-NS | 33/F | CS-IVA | 8 | ABVD+ RT | No | Alive | ||
| 15 | HL-MC | 67/F | CS-IIIB | 6 | ABVD+ RT | No | Alive | ||
| 16 | HL-NS | 24/F | CS-IVB | 8 | BEACOPP+ RT | No | Alive | ||
| 17 | HL-MC | 28/M | CS-IA | RT | No | Alive | |||
| 18 | HL-MC | 35/M | CS-IIB | 6 | ABVD | No | Alive | ||
| 19 | HL-NS | 67/M | CS-IA | 4 | ABVD+ RT | No | Alive | ||
RT indicates radiation therapy; ABVD, Adriamycin, bleomycin, vinblastine, dacarbazine; COPP, cyclophosphamide, Oncovin, procarbazine, prednisone; ABV, Adriamycin, bleomycin, vinblastine; CVPP, cyclophosphamide, vincristine, prednisone, procarbazine; BEACOPP, bleomycin, etoposide, doxorubicin, cyclophosphamide, vincristine, procarbazine, prednisone; PR, partial remission; CR, complete remission; Rel., relapse; NA, not available; NLPHL, nodular lymphocyte predominant Hodgkin lymphoma; HL-NS, Hodgkin lymphoma nodular sclerosis; HL-MC, Hodgkin lymphoma, mixed cellularity.
If relapse occurred, number of instances and subsequent treatment are also listed. If PR occurred, treatment is also listed.
All indications of “Alive” entailed CR.
Nodular and diffuse.
To evaluate the presence of aberrant SHM, tumor cells were isolated from lymph node frozen sections of the 19 HL patients and were subjected to PCR amplification and direct sequencing of the PIM1, RhoH/TTF, PAX5, and c-MYC genes, as described in “Materials and methods.” In 4 patients, mutations were confirmed on independent PCR products from the same pool of cells. In the remaining patients, mutations were confirmed on independent PCR products from 2 different pools of cells. Results obtained from the analysis of different pools of cells were identical. For each patient, multiplex PCR amplification was performed on a minimum of 20 tumor cells and was successful in 7 patients. In the remaining patients, 1 to 3 single genes were amplified from different pools containing at least 15 cells.
Analysis revealed the presence of mutations targeting at least 1 of the 4 proto-oncogenes in 8 (80.0%) of 10 NLPHL and 5 (55.5%) of 9 cHL patients, with 5 (62.5%) of 8 NLPHL and 3 (60.0%) of 5 cHL patients carrying mutations in more than one gene (Tables 2, 3). The most frequently involved proto-oncogene in NLPHL was PAX5, found mutated in 7 (77.8%) of 9 patients, followed by PIM1 (3 [37.5%] of 8 patients), c-MYC exon 2 (3 [33.3%] of 9 patients), and c-MYC exon 1 and RhoH/TTF, each mutated in 1 patient (10.0% and 11.0%, respectively) (Table 4). The most frequently involved proto-oncogene in cHL was c-MYC exon 1, found mutated in 4 (44.4%) of 9 patients, whereas no mutation was detected in RhoH/TTF (Table 4). In all patients, mutations were somatic in origin because they were not found in T cells amplified from the same patients (data not shown). No correlation between EBV status and aberrant SHM process was observed.
Mutational analysis of IGHV, c-MYC, RhoH/TTF, PAX5, and PIM1 in NLPHL
. | IGH gene analysis . | . | . | . | . | Mutation position . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGHV . | IGHD . | IGHJ . | Mutation, % . | Functional status . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||||||
| NLPHL 1 | 3-30 | 6-06 | 5b | 25.2 | + | - | - | - | - | - | ||||||||
| NLPHL 2 | 3-33 | 5-24 + 5-18 | 6b | 7.14 | + | - | - | G261A; G395A | G1323A | G729A; C1026G; G1425C; C1436G | ||||||||
| NLPHL 3 | 4-34 | 2-15 | 4b | 4.54 | Crippled | - | G4580T | - | - | C674T; C848T; del18bp (1317-1334); C1357G | ||||||||
| NLPHL 4 | 4-34 | 3-03 | 6b | 12.3 | + | - | T4312A | - | - | NA | ||||||||
| NLPHL 5 | 3-07 | 4-17 + 3-03 + 7-27 | 6b | 6.73 | + | - | NA | - | NA | G910A; C953T; C962T; G1070C; G1342A | ||||||||
| NLPHL 6 | 4-59 | 4-17 | 4b | 9.18 | + | - | - | - | - | C731T; T805G | ||||||||
| NLPHL 7 | NA | ND | ND | ND | ND | G3019A; G3056A | - | - | NA | A762G | ||||||||
| NLPHL 8 | 3-33 | D5 | 6c | 5.36 | + | delG3028 | - | NA | T1365C, C1682T | G847A, C958T | ||||||||
| NLPHL 9 | 4-39 | D4 | 4b | 5.94 | + | - | G4650A | - | del1137-1140 | C674G, T771G, C853G, A1033G | ||||||||
| NLPHL 10 | 3-74 | 2-02 | 4b | 9.33 | + | - | - | - | - | - | ||||||||
. | IGH gene analysis . | . | . | . | . | Mutation position . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGHV . | IGHD . | IGHJ . | Mutation, % . | Functional status . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||||||
| NLPHL 1 | 3-30 | 6-06 | 5b | 25.2 | + | - | - | - | - | - | ||||||||
| NLPHL 2 | 3-33 | 5-24 + 5-18 | 6b | 7.14 | + | - | - | G261A; G395A | G1323A | G729A; C1026G; G1425C; C1436G | ||||||||
| NLPHL 3 | 4-34 | 2-15 | 4b | 4.54 | Crippled | - | G4580T | - | - | C674T; C848T; del18bp (1317-1334); C1357G | ||||||||
| NLPHL 4 | 4-34 | 3-03 | 6b | 12.3 | + | - | T4312A | - | - | NA | ||||||||
| NLPHL 5 | 3-07 | 4-17 + 3-03 + 7-27 | 6b | 6.73 | + | - | NA | - | NA | G910A; C953T; C962T; G1070C; G1342A | ||||||||
| NLPHL 6 | 4-59 | 4-17 | 4b | 9.18 | + | - | - | - | - | C731T; T805G | ||||||||
| NLPHL 7 | NA | ND | ND | ND | ND | G3019A; G3056A | - | - | NA | A762G | ||||||||
| NLPHL 8 | 3-33 | D5 | 6c | 5.36 | + | delG3028 | - | NA | T1365C, C1682T | G847A, C958T | ||||||||
| NLPHL 9 | 4-39 | D4 | 4b | 5.94 | + | - | G4650A | - | del1137-1140 | C674G, T771G, C853G, A1033G | ||||||||
| NLPHL 10 | 3-74 | 2-02 | 4b | 9.33 | + | - | - | - | - | - | ||||||||
Mutational analysis of IGHV, c-MYC, RhoH/TTF, PAX5, and PIM1 in classic HL
. | IGH gene analysis . | . | . | . | . | Mutation position . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGHV . | IGHD . | IGHJ . | Mutation, % . | Functional status . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||||||
| cHL 1 | 1-69 | 3-09 + 1-26 | 4 | 24.1 | + | - | - | - | - | - | ||||||||
| cHL 2 | 4-39 | NA | 4 | 7.69 | + | - | - | - | G1255A, G1554A | - | ||||||||
| cHL 3 | 3-33 | 3-03 | 6b | 6.77 | + | - | - | - | - | - | ||||||||
| cHL 4 | 4-39 | NA | 5b | 13.6 | + | - | - | - | - | - | ||||||||
| cHL 5 | 3-11 | NA | 6c | 1.24 | Out-of-frame | G2471A, G2851A | - | - | - | - | ||||||||
| cHL 6 | 1-69 | NA | 6b | 14.4 | + | - | - | - | - | - | ||||||||
| cHL 7 | 4-39 | 3-22 | 4b | 0 | + | T2486C, A2528G, T2771A, G2805A, T2868A, T3152C, T3268C, A3305T, insT3426 | - | NA | - | C718T, C871T | ||||||||
| cHL 8 | NA | ND | ND | ND | ND | T2456G, G2709T, A2730G, C2732A, C2770T, C2778G, T2868C, G2886A, del38bp 2891-2928, G2933A | A4692G | - | - | - | ||||||||
| cHL 9 | 5-a | 6-13 | 6c | 0 | + | A2490G, C3319T | - | - | - | T788C, G847A, C848G, C853A, G971A, del3bp 1045-1047, G1425A | ||||||||
. | IGH gene analysis . | . | . | . | . | Mutation position . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | IGHV . | IGHD . | IGHJ . | Mutation, % . | Functional status . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||||||
| cHL 1 | 1-69 | 3-09 + 1-26 | 4 | 24.1 | + | - | - | - | - | - | ||||||||
| cHL 2 | 4-39 | NA | 4 | 7.69 | + | - | - | - | G1255A, G1554A | - | ||||||||
| cHL 3 | 3-33 | 3-03 | 6b | 6.77 | + | - | - | - | - | - | ||||||||
| cHL 4 | 4-39 | NA | 5b | 13.6 | + | - | - | - | - | - | ||||||||
| cHL 5 | 3-11 | NA | 6c | 1.24 | Out-of-frame | G2471A, G2851A | - | - | - | - | ||||||||
| cHL 6 | 1-69 | NA | 6b | 14.4 | + | - | - | - | - | - | ||||||||
| cHL 7 | 4-39 | 3-22 | 4b | 0 | + | T2486C, A2528G, T2771A, G2805A, T2868A, T3152C, T3268C, A3305T, insT3426 | - | NA | - | C718T, C871T | ||||||||
| cHL 8 | NA | ND | ND | ND | ND | T2456G, G2709T, A2730G, C2732A, C2770T, C2778G, T2868C, G2886A, del38bp 2891-2928, G2933A | A4692G | - | - | - | ||||||||
| cHL 9 | 5-a | 6-13 | 6c | 0 | + | A2490G, C3319T | - | - | - | T788C, G847A, C848G, C853A, G971A, del3bp 1045-1047, G1425A | ||||||||
Frequency of aberrant somatic hypermutation in NLPHL and cHL
. | No. mutated/tested (%) . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Histology . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||
| NLPHL; n = 10 | 2/10 (10.0) | 3/9 (33.3) | 1/9 (11.1) | 3/8 (37.5) | 7/9 (77.8)* | ||||
| cHL; n = 9 | 4/9 (44.4) | 1/9 (11.1) | 0/8 (0) | 1/9 (11.1) | 2/9 (22.2) | ||||
. | No. mutated/tested (%) . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Histology . | c-MYC exon 1 . | c-MYC exon 2 . | RhoH/TTF . | PIM1 . | PAX5 . | ||||
| NLPHL; n = 10 | 2/10 (10.0) | 3/9 (33.3) | 1/9 (11.1) | 3/8 (37.5) | 7/9 (77.8)* | ||||
| cHL; n = 9 | 4/9 (44.4) | 1/9 (11.1) | 0/8 (0) | 1/9 (11.1) | 2/9 (22.2) | ||||
Corrected P = .096 (PAX5 mutations in NLPHL compared with cHL). All (4 of 4) hypermutated cases were mixed cellularity. Only 1 of 5 hypermutated cases was nodular sclerosis (P = .025).
Comparison between NLPHL and cHL revealed no significant differences in the prevalence of mutations affecting the 4 genes, though a trend toward higher prevalence of PAX5 mutations was observed in NLPHL compared with cHL (uncorrected, P = .024; Bonferroni-corrected, P = .096).
Distribution of the mutations in the 4 genes analyzed is graphically represented in Figure 1, and detailed descriptions of the mutations found in NLPHL and cHL are given in Tables 2 and 3, respectively.
In total, 34 events were observed in 8 NLPHL patients, corresponding to an average mutation frequency (calculated in mutated cases only) of 1.04/1000 bp (range, 0.24/1000 bp in c-MYC exon 1 to 1.82/1000 bp in PAX5) (Table 5). In cHL patients, 35 mutational events were observed that were distributed in 5 patients (average mutation frequencies, 1.92/1000 bp; range, 0.428/1000 bp in c-MYC exon 2 to 2.61/1000 bp in PAX5) (Table 5). Overall, the average mutation frequency of c-MYC was significantly higher in cHL than in NLPHL patients (1.22/1000 bp vs 0.24/1000 bp; P = .03). A representative example of NLPHL-bearing mutations affecting the RhoH/TTF and PAX5 genes (Table 2; patient 2) is shown in Figure 2. Overall, these findings demonstrate that PIM1, PAX5, RhoH/TTF, and c-MYC are frequent targets of aberrant SHM in the neoplastic cells of NLPHL and cHL, analogous to what was originally observed in DLBCL.14
Features of c-MYC, RhoH/TTF, PIM1, and PAX5 mutations in NLPHL and cHL
. | Mutation frequency/1000 bp (range) . | . | Deletions/insertions . | . | Single-bp substitutions . | . | Transitions/transversions (ratio; P)* . | . | G>C/A>T (ratio) . | . | RGYW/WRCY (P)* . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | ||||||
| c-MYC | 0.24 (0.20-0.40) | 1.22 (0.40-2.23) | 1/0 | 1/1 | 5 | 22 | 3/2 (1.5) | 15/7 (2.1; .02) | 4/1 (4.0) | 10/12 (0.83) | 2 | 13 (.03) | ||||||
| Exon 1 | 0.58 (0.38-0.77) | 2.21 (0.77-3.84) | 1/0 | 1/1 | 2 | 21 | 2/0 | 14/7 (2.0; .04) | 2/0 | 10/11 (0.91) | 1 | 13 (.04) | ||||||
| Exon 2 | 0.428 | 0.428 | 0/0 | 0/0 | 3 | 1 | 1/2 (0.5) | 1/0 | 2/1 (2.0) | 0/1 | 1 | 0 | ||||||
| RhoH/TTF | 0.902 | 0 | 0/0 | 0 | 2 | 0 | 2/0 | 0/0 | 2/0 | 0/0 | 1 | 0 | ||||||
| PIM1 | 0.661 (0.49-0.99) | 0.991 | 1/0 | 0 | 3 | 2 | 3/0 | 2/0 | 2/1 (2.0) | 2/0 | 2 | 0 | ||||||
| PAX5 | 1.82 (0.58-2.90) | 2.61 (1.16-4.07) | 1/0 | 1/0 | 21 | 8 | 12/9 (1.33) | 6/2 (3.0) | 17/4 (3.2) | 7/1 (1.67) | 9 | 3 | ||||||
| All genes | 1.04 | 1.92 | 3/0 | 2/1 | 31 | 32 | 20/11 (1.82; .017) | 23/9 (2.55; .01) | 25/6 (4.17) | 19/13 (1.46) | 14 | 16 | ||||||
. | Mutation frequency/1000 bp (range) . | . | Deletions/insertions . | . | Single-bp substitutions . | . | Transitions/transversions (ratio; P)* . | . | G>C/A>T (ratio) . | . | RGYW/WRCY (P)* . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | NLPHL . | cHL . | ||||||
| c-MYC | 0.24 (0.20-0.40) | 1.22 (0.40-2.23) | 1/0 | 1/1 | 5 | 22 | 3/2 (1.5) | 15/7 (2.1; .02) | 4/1 (4.0) | 10/12 (0.83) | 2 | 13 (.03) | ||||||
| Exon 1 | 0.58 (0.38-0.77) | 2.21 (0.77-3.84) | 1/0 | 1/1 | 2 | 21 | 2/0 | 14/7 (2.0; .04) | 2/0 | 10/11 (0.91) | 1 | 13 (.04) | ||||||
| Exon 2 | 0.428 | 0.428 | 0/0 | 0/0 | 3 | 1 | 1/2 (0.5) | 1/0 | 2/1 (2.0) | 0/1 | 1 | 0 | ||||||
| RhoH/TTF | 0.902 | 0 | 0/0 | 0 | 2 | 0 | 2/0 | 0/0 | 2/0 | 0/0 | 1 | 0 | ||||||
| PIM1 | 0.661 (0.49-0.99) | 0.991 | 1/0 | 0 | 3 | 2 | 3/0 | 2/0 | 2/1 (2.0) | 2/0 | 2 | 0 | ||||||
| PAX5 | 1.82 (0.58-2.90) | 2.61 (1.16-4.07) | 1/0 | 1/0 | 21 | 8 | 12/9 (1.33) | 6/2 (3.0) | 17/4 (3.2) | 7/1 (1.67) | 9 | 3 | ||||||
| All genes | 1.04 | 1.92 | 3/0 | 2/1 | 31 | 32 | 20/11 (1.82; .017) | 23/9 (2.55; .01) | 25/6 (4.17) | 19/13 (1.46) | 14 | 16 | ||||||
The frequency of mutations within RGYW/WRCY motifs was compared with the frequency of mutations outside RGYW/WRCY motifs by the χ2 goodness-of-fit test.
P values are reported only when significant.
Mutational profiles of c-MYC, PIM1, RhoH/TTF, and PAX5 in NLPHL and cHL
General features of the mutations observed in NLPHL and cHL are summarized in Table 5. In both HL subtypes, the overwhelming majority of mutations were represented by single base-pair substitutions (31 in NLPHL and 32 in cHL), whereas deletions or insertions of a short DNA stretch were observed only in 4 patients (c-MYC in cHL8, PIM1 in NLPHL9, and PAX5 in NLPHL3 and cHL9), and 2 patients displayed a single base-pair insertion or deletion in the c-MYC exon 1 sequence (cHL7, NLPHL8) (Tables 2, 3). Of the 31 single base-pair substitutions observed in NLPHL, 20 were transitions and 11 were transversions, with a transition/transversion ratio of 1.82 (expected ratio, 0.5; P < .01) (Table 5). Of the 32 single base-pair substitutions observed in cHL, 23 were transitions and 9 were transversions, with a transition/transversion ratio of 2.55 (expected ratio, 0.5; P < .01) (Table 5). Fourteen of 31 single base-pair substitutions detected in NLPHL were located within RGYW/WRCY motifs (mutation frequency of 0.52% within RGYW/WRCY motifs compared with 0.21% outside RGYW/WRCY motifs; P = .08) (Table 5). In cHL, 16 of 32 (50%) single base-pair substitutions affected RGYW/WRCY motifs. The frequency of mutations targeting RGYW/WRCY motifs was significantly higher than the frequency of mutations occurring outside RGYW/WRCY motifs (0.60% vs 0.19%; P = .042) (Table 5).
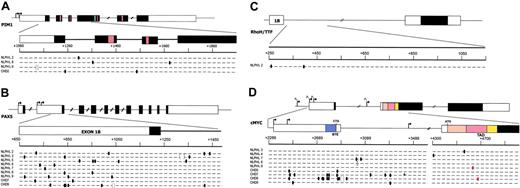
Mutational analyses.PIM1 (A), PAX5 (B), RhoH/TTF (C), and c-MYC (D) in NLPHL and cHL. Genomic loci are shown, with untranslated (open boxes) and translated (filled boxes) sequences; hatched boxes are relevant protein functional domains. Arrows indicate transcriptional start sites. Regions amplified for analysis are expanded for each gene and aligned with sequences of mutated NLPHL and cHL cases, shown below the loci. One line represents 2 alleles, where each small segment represents a 20-bp (PIM1, PAX5, RhoH/TTF) or a 40-bp (c-MYC) interval (numbering according to the relative GenBank accession numbers). Ovals indicate 1-bp substitution (red indicates resulted in missense mutation); brackets indicate deletions; plus indicates insertions. BTE indicates segment controlling block of transcriptional elongation; TAD, transactivation domain.
Mutational analyses.PIM1 (A), PAX5 (B), RhoH/TTF (C), and c-MYC (D) in NLPHL and cHL. Genomic loci are shown, with untranslated (open boxes) and translated (filled boxes) sequences; hatched boxes are relevant protein functional domains. Arrows indicate transcriptional start sites. Regions amplified for analysis are expanded for each gene and aligned with sequences of mutated NLPHL and cHL cases, shown below the loci. One line represents 2 alleles, where each small segment represents a 20-bp (PIM1, PAX5, RhoH/TTF) or a 40-bp (c-MYC) interval (numbering according to the relative GenBank accession numbers). Ovals indicate 1-bp substitution (red indicates resulted in missense mutation); brackets indicate deletions; plus indicates insertions. BTE indicates segment controlling block of transcriptional elongation; TAD, transactivation domain.
Aberrant SHM introduces missense mutations in c-MYC
Although most of the mutations were distributed in the 5′ noncoding sequences of the affected genes, 2 distinct nucleotide substitutions in c-MYC were found to affect the exon 2 sequences, encoding for the transcriptional activation domain. Specifically, NLPHL9 carried a single base-pair substitution leading to a change of Ala for Thr at position 44 of the amino acid sequence (Figure 1; Table 2), and in cHL8 one missense mutation led to the substitution of Thr for Ala at position 58 (Figure 1; Table 3). The latter is a known phosphorylation site that is critical for the control of c-MYC protein stability and whose loss is associated with increased transforming ability.22-24
Relationship between aberrant SHM and IGHV gene status
The presence of mutations in the c-MYC, RhoH/TTF, PIM1, and PAX5 proto-oncogenes was related to the mutation status of IGHV genes in NLPHL and cHL. Although all these mutations are thought to depend on the SHM mechanism active in germinal center B cells, IGHV gene mutations are acquired physiologically,7,8 whereas mutations of c-MYC, RhoH/TTF, PIM1, and PAX5 appear to be restricted to lymphomas.14
All patients were analyzed for IGHV gene rearrangements, and a PCR product could be amplified in 9 of 10 NLPHL patients and in 8 of 9 cHL patients. A functional IGHV rearrangement was obtained from 8 of 9 NLPHL and 7 of 8 cHL patients (Tables 2, 3). In NLPHL3, a stop-codon was detected within the originally productive IGHV rearrangement, leading to a crippled IGHV sequence (Table 2), whereas in cHL5, only an out-of-frame IGHV rearrangement was amplified (Table 3). SHM was observed in all but 2 productive IGHV rearrangements and ranged from 4.54% to 25.2% in NLPHL patients (Table 2) and from 7.69% to 24.1% in cHL patients (Table 3). In 2 cHL patients, only an unmutated, potentially functional IGHV rearrangement was identified. Both patients showed aberrant SHM. Overall, the average mutation frequency of c-MYC, RhoH/TTF, PIM1, and PAX5 was 50-fold to 500-fold lower than that of IGHV genes but still significantly higher than the background mutation frequency in mammalian cells (Tables 2, 3, 5). Moreover, among patients with IGHV mutations, comparison of the mutation frequencies revealed no correlations between IGHV mutations and the occurrence of c-MYC, RhoH/TTF, PIM1, and PAX5 mutations, supporting the hypothesis that aberrant SHM may be caused by a qualitative rather than a quantitative defect of the SHM mechanism.
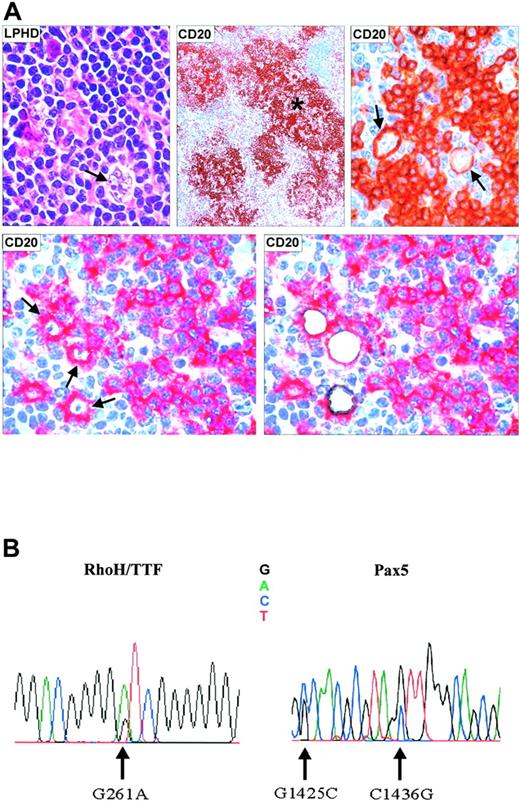
Microdissection and mutational analysis of a representative patient with NLPHL. (A) Lymph node biopsy showing a neoplastic nodule containing characteristic L/H cells with popcorn appearance of the nucleus (top left, hematoxylineosin stain; magnification, 800 ×). (Top middle, immunoperoxidase technique; magnification, ×150. L/H tumor cells are CD20+ (arrows) and are surrounded by a rim of CD20– rosette T cells (top right, immunoperoxidase technique; magnification, 800 ×). CD20+ L/H cells before and after laser microdissection (bottom, right and left) (APAAP technique). *CD20+ tumor nodule. (B) Electropherogram showing mutations of the RhoH/TTF and PAX5 genes in microdissected L/H cells (Table 2, patient 2).
Microdissection and mutational analysis of a representative patient with NLPHL. (A) Lymph node biopsy showing a neoplastic nodule containing characteristic L/H cells with popcorn appearance of the nucleus (top left, hematoxylineosin stain; magnification, 800 ×). (Top middle, immunoperoxidase technique; magnification, ×150. L/H tumor cells are CD20+ (arrows) and are surrounded by a rim of CD20– rosette T cells (top right, immunoperoxidase technique; magnification, 800 ×). CD20+ L/H cells before and after laser microdissection (bottom, right and left) (APAAP technique). *CD20+ tumor nodule. (B) Electropherogram showing mutations of the RhoH/TTF and PAX5 genes in microdissected L/H cells (Table 2, patient 2).
Clonal analysis of PIM1, PAX5, RhoH/TTF, and c-MYC
To investigate whether aberrant somatic hypermutation is ongoing in HL, we selected 3 NLPHL patients and 2 cHL patients shown by direct sequencing to carry mutations in PIM1, PAX5, RhoH/TTF, or c-MYC exon 1. These patients were analyzed for the presence of intraclonal heterogeneity by the sequencing of cloned PCR products generated using the proofreading Pfu polymerase. As a control for background error rate, the fibroblast MRC-5 cell line was used. Statistical significance was assessed by the Student t test.
Results showed that, in all tumors analyzed, 1 or 2 predominant alleles recapitulated the mutations observed by direct sequencing of the PCR product, confirming their presence in the tumor clone and revealing their frequent biallelic distribution (not shown). A significant number of intraclonal variants were found in the RhoH/TTF and PAX5 sequences from 1 of 3 patients with NLPHL (NLPHL2) and in the c-MYC exon 1 sequence from 2 patients with cHL (cHL7 and cHL8). In the remaining 2 patients, the frequency of mutations did not differ significantly from the polymerase error rate (Table S2). These data indicate that aberrant somatic hypermutation activity may be ongoing in at least a fraction of NLPHLs and cHLs.
Discussion
In this study, we provide evidence that the aberrant SHM process, originally reported to affect the proto-oncogenes PIM1, c-MYC, RhoH/TTF (ARHH), and PAX5 in DLBCL,14 is also operating in the neoplastic cells of NLPHL and cHL. Specifically, we found that 80% of NLPHLs and 55% of cHLs are targeted by aberrant SHM in at least 1 of these 4 proto-oncogenes and that, analogous to DLBCL,14 the molecular profile of aberrant SHM is reminiscent of the mutational spectrum of IGV genes.25 In fact, mutations in the c-MYC, RhoH/TTF, PIM1, and PAX5 proto-oncogenes are predominantly represented by single nucleotide substitutions and have occasional deletions and insertions, display a preference for transitions over transversions, and display a preferential distribution within the RGYW/WRCY motifs, with elevated ratios of G>C over A>T substitutions.
Our study also shows the presence of intraclonal diversity in a few samples, representative of both NLPHL and cHL, indicating that aberrant hypermutation activity may be ongoing in at least a fraction of patients, consistent with previous analyses performed on IGV genes.3,4,6 The presence of ongoing hypermutation in NLPHL is also in agreement with the observation that this disease type consistently expresses activation-induced cytidine deaminase (AID),26 the enzyme indispensable for SHM.27,28 Conversely, AID expression has been reported only infrequently in cHL (3 of 47 patients26 ). Because immunohistochemical staining for AID could not be performed in cHL7 and cHL8, we do not know whether AID was present in at least a fraction of neoplastic cells. Alternatively, it is possible that AID had been active in earlier stages of tumor development, causing intraclonal diversification of the protooncogenes investigated, but that the expression/hypermutation activity was turned off in later phases of the disease.
In this study, aberrant SHM was also detected in 2 cHL patients carrying unmutated IGHV genes. The presence of unmutated, potentially functional IGHV rearrangements in cHL has been previously reported29 and may be explained by the physiologic presence of normal, unmutated GC founder cells (from which the lymphoma can arise) or by alterations in the IGHV locus that may impair the somatic hypermutation in IGV genes, but not in other genes. This finding is also consistent with the hypothesis that a qualitative deficit of the SHM machinery (eg, loss of target specificity) rather than an overall increase in its activity may be responsible for these mutations in B-cell lymphomas.
Activation of c-MYC, RhoH/TTF, PIM1, and PAX5 may be relevant for B-cell lymphomagenesis14 because these genes encode for signal transducers (PIM1 and RhoH/TTF) and transcription factors (PAX5 and c-MYC) involved in B-cell development/differentiation or in the regulation of proliferation and apoptosis, and they have also been implicated in lymphoma-associated chromosomal translocations.30-33
In NLPHL and cHL, aberrant SHM may alter the function of these genes by at least 2 modalities. Because mutations cluster around the 5′ untranslated region of the genes, it is conceivable that mutations may deregulate gene transcription by affecting specific regulatory regions, with a mechanism analogous to that observed for BCL6 and c-MYC in DLBCL and Burkitt lymphoma, respectively.34,35 In our study, a high number of mutations identified in NLPHL cluster in a restricted region of PAX5 (nucleotides +650 to +1000), localized immediately upstream of the translated region of exon 1B. Sequence analysis of this region showed evidence of putative binding sites for various transcriptional factors, including SP1, ETS, and AP2 (Transcription Element Search Software; http://www.cbil.upenn.edu/tess).
In some genes, such as PIM1 and c-MYC, some changes introduced by aberrant SHM may also lead to amino acid substitutions and, consequently, may alter the biochemical or structural properties of the protein.16 In particular, in vitro and in vivo studies have shown that mutations in the c-MYC transactivation domain can deregulate c-MYC function by interfering with its phosphorylation or protein stability or with repression of its transactivation activity by the Rb-related protein p107.23,24,36 More recently, c-MYC alleles carrying mutations at threonine 58 or at proline 57 have been documented in vivo to show decreased proapoptotic function because of their inability to induce Bim1 gene expression.22 In our series, 2 samples (NLPHL9 and cHL8) displayed missense mutations within the sequences of c-MYC exon 2 that encode the transactivation domain. Notably, 1 of these 2 mutations affects threonine 58, indicating that aberrant somatic hypermutation may contribute to deregulated c-MYC activity.
The finding that NLPHL and cHL tumor cells are targeted by the aberrant SHM, a distinctive feature of DLCBL, is intriguing. In fact, despite some overlapping features,37-41 NLPHL and cHL represent distinct diseases that clearly differ from DLBCL in terms of biologic properties, morphologic appearance, clinical course, and response to therapy.1,42 The most probable explanation is that genetic alterations in addition to aberrant SHM may eventually concur in the pathogenesis of HL. Interestingly, BCL6 gene rearrangements have been detected in approximately 40% of NLPHL and DLBCL but not in classic HL.39,43 Moreover, the prevalence of genes targeted by the aberrant SHM process appears to differ in the diverse lymphomas. In particular, PAX5 mutations occur at a higher frequency in DLBCL and NLPHL than in cHL. Conversely, the RhoH/TTF gene, which is mutated at high frequencies in DLBCL,14-16 is rarely affected in either NLPHL or cHL. Thus, it is conceivable that targeting of other yet unrecognized germinal center–associated genes by the aberrant SHM process, possibly in combination with other unknown genetic lesions or variations in the host immune response, may contribute to the remarkable differences in the biologic and clinical features of NLPHL, cHL, and DLBCL.
Prepublished online as Blood First Edition Paper, April 13, 2006; DOI 10.1182/blood-2005-10-3949.
Supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) and by the Ricerca Sanitaria Finalizzata Regione Piemonte, Torino, Italy; Progetti di Ricerca di Interesse Nazionale (PRIN) 2004, Ministero dell'Università e della Ricerca (MIUR), Rome, Italy; and Novara–Associazione Italiana contro le Leucemie (AIL); Agenzia per le Organizzazioni Non Lucrative di Utilità Sociale (OnLus), Novara, Italy. E.T. was supported by a fellowship from Livia Bendetti (Perugia, Italy).
A.L., D.C., and T.M. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Annunziata Gloghini (Centro di Riferimento Oncologico, Aviano, Italy) for providing frozen tissue samples for the study. Laura Pasqualucci is a Special Fellow of the Leukemia and Lymphoma Society.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal