To determine whether gene expression profiling could improve risk classification and outcome prediction in older acute myeloid leukemia (AML) patients, expression profiles were obtained in pretreatment leukemic samples from 170 patients whose median age was 65 years. Unsupervised clustering methods were used to classify patients into 6 cluster groups (designated A to F) that varied significantly in rates of resistant disease (RD; P < .001), complete response (CR; P = .023), and disease-free survival (DFS; P = .023). Cluster A (n = 24), dominated by NPM1 mutations (78%), normal karyotypes (75%), and genes associated with signaling and apoptosis, had the best DFS (27%) and overall survival (OS; 25% at 5 years). Patients in clusters B (n = 22) and C (n = 31) had the worst OS (5% and 6%, respectively); cluster B was distinguished by the highest rate of RD (77%) and multidrug resistant gene expression (ABCG2, MDR1). Cluster D was characterized by a “proliferative” gene signature with the highest proportion of detectable cytogenetic abnormalities (76%; including 83% of all favorable and 34% of unfavorable karyotypes). Cluster F (n = 33) was dominated by monocytic leukemias (97% of cases), also showing increased NPM1 mutations (61%). These gene expression signatures provide insights into novel groups of AML not predicted by traditional studies that impact prognosis and potential therapy.
Introduction
In most patients, particularly those over 55 years of age, acute myeloid leukemia (AML) is a highly resistant disease (RD) and overall outcomes remain extremely poor.1-5 While improved survival has been achieved in younger AML patients or in selected cytogenetic subsets, older patients are either unable to receive intensive chemotherapy or such therapy results in remission rates of only 25% to 55% and overall survival (OS) rates of 10% or less.1,6-10 In addition to age and white blood cell (WBC) count, the presence of recurring cytogenetic abnormalities provides the most important prognostic information in AML. Unfortunately, cytogenetic abnormalities associated with favorable outcomes account for only 5% to 12% (t(8;21)), 5% to 8% (inv(16)), and 10% to 12% (t(15;17)) of all AML cases and are disproportionately seen in younger patients.11,12 In contrast, approximately 50% to 70% of all AMLs have normal or risk-indeterminate karyotypes.11,13,14
Gene mutations confer additional prognostic information that may be useful in refining cytogenetic risk classification.15-19 The most frequently acquired mutation in AML is a mutation at exon 12 of the nucleophosmin (NPM1) gene. This multifunctional, nucleocytoplasmic shuttling protein primarily resides in the nucleolus, playing a role in maintenance of genomic integrity, ARF-p53 pathway regulation, and centrosome duplication.20,21 Mutated NPM1 relocates to the cytoplasm and disrupts normal NPM1 function. Approximately 25% to 35% of AML patients have NPM1 mutations,22-24 with a higher percentage (47%-60%) seen among those with a normal karyotype.22,25-26 The impact on survival is variable, but likely favorable, with secondary influences such as concurrent FLT3 mutations having potentially significant roles.23,24,26,27 The FLT3 mutations occur as internal tandem duplications (ITDs), observed in 15% to 35% of AML, or point mutations of the intracellular tyrosine-kinase domain (TKD), seen in an additional 5% to 10% of patients.19 The prognostic impact of FLT3 mutations trends toward decreased survivals or increased relapse rates primarily for patients with FLT3-ITDs.28-30
In contrast to traditional cytogenetic analysis or the detection of mutations in individual genes, global gene expression profiling provides a powerful method to probe the marked biologic heterogeneity of AML. Comprehensive expression profiles have the power to provide new insights into mechanisms of leukemogenesis and to enhance risk classification and therapeutic targeting in AML. A number of laboratories using supervised learning algorithms have identified unique gene expression signatures associated with karyotypic abnormalities, normal karyotypes, and NPM1 mutation status.31-39 In contrast, we wished to determine whether gene expression profiling using an entirely unsupervised approach could reveal intrinsic biologic groups of AML among a set of well-characterized older AML patients, with a high frequency of normal and unfavorable cytogenetic abnormalities. We further wished to determine whether the gene expression signatures we derived were useful in risk classification and therapeutic targeting in this poor-risk disease.
Patients, materials, and methods
Patients
This study used pretreatment samples from patients with previously untreated de novo or secondary AML by French-American-British (FAB) criteria who were registered to Southwest Oncology Group (SWOG) clinical trials for patients over the age of 55 years (studies S9031, S9333), patients aged 15 to 55 years (S9034, S9500), and patients with secondary AML (S9126). Trial details have been previously reported.2,9,40-42 All trials except S9031 excluded patients with acute promyelocytic leukemia (FAB-M3); S9031 evaluation was limited to non-M3 AML patients who received induction chemotherapy with Ara-C and an anthracycline. Case selection was restricted to patients with cryopreserved blood or bone marrow containing more than 80% leukemic blasts, stored in the SWOG Myeloid Leukemia Repository (University of New Mexico) after appropriate informed consent. Microarrays were performed for 185 eligible patients between February 2003 and September 2003, and 170 had high-quality gene expression data that fulfilled technical criteria for study inclusion (outlined in “Gene expression profiling”). Clinical, morphologic, cytogenetic, and outcome data on the 170 patients, along with all gene expression profiles, are provided at the National Cancer Institute Gene Expression Data Portal website. Conventional cytogenetic banding was performed in SWOG-approved laboratories with review and risk classification assessment performed by members of the SWOG Cytogenetic Committee per published criteria.11 For studies S9031, S9126, S9333, and S9500, response to induction chemotherapy was assessed according to SWOG criteria.43 Study S9034, an intergroup trial coordinated by the Eastern Cooperative Oncology Group (E3489), used slightly different response criteria.
Gene expression profiling
RNA was prepared from thawed cryopreserved samples with the Qiagen RNeasy mini kit (Qiagen, Valencia, CA). All specimens had more than 80% blasts as confirmed by microscopic review of Wright-stained cytospin preparations of the thawed cell suspensions. Total RNA concentration was quantified with the RiboGreen assay (Molecular Probes, Eugene, OR); RNA integrity and DNA contamination were evaluated as described at the University of New Mexico Cancer Research and Treatment Center website.44 The isolated RNA was reverse transcribed into cDNA and retranscribed into cRNA after double amplification using a modification reported by Ivanova et al to enhance detection of low-abundance genes.44,45 Biotinylated cRNA was fragmented and hybridized to HG_U95Av2 oligonucleotide microarrays (Affymetrix, Santa Clara, CA).44 After analysis with Affymetrix Microarray Suite (MAS 5.0), the data were scaled to minimize experimental variation.44 Technical criteria for case inclusion of the 185 initial specimens evaluated included adequate total RNA more than 2.5 μg, good quality cRNA, good quality scanned images, and good experimental quality. Experimental quality was assessed by GAPDH at least 1800, at least 10% expressed genes, and GAPDH 3′/5′ amplification ratios of 4 or below. High-quality expression data were obtained on 170 of the 185 specimens, 133 from marrow and 37 from peripheral blood. Of the original 12 625 probe sets in the Affymetrix HG_U95Av2 probe sets, 9463 genes were “present” in at least 1 case; these genes were further analyzed after transformation to Savage rank scores (VxInsight).44
NPM1 and FLT3 mutational status
Samples were evaluated for NPM1 mutations using cDNA amplified to generate a 249 bp fragment spanning portions of exons 11 and 12 (Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).44 The polymerase chain reaction (PCR) products were subjected to dissociation analysis (65°C to 80°C) with appropriate controls. Samples with characteristic melting profiles underwent agarose gel electrophoresis and hybridization with NPM1 variant A probe or a pool of 13 probes for variants B to Q.26 Cases were also evaluated for FLT3-ITDs in exons 14 and 15 as previously described and screened for FLT3-TKD in exon 20 using 2 methods (see Document S1).44,46 Suspected FLT3-ITDs and TKD mutations were confirmed by sequencing.46
Statistical analysis
VxInsight,47 developed at Sandia National Laboratories for extremely large datasets, was the primary unsupervised data mining tool used in this study.48-51 Using a force-directed placement algorithm, clusters were formed 100 times using different starting conditions for the random number generator. The most representative single ordination (the most central member of the whole set) was then determined by measurement of the total overlap of local neighborhoods around the individual genes. Analysis of variance (ANOVA) was used to identify rank-ordered gene lists characterizing each cluster; bootstrap resampling was applied to estimate the stability of these lists.51 Receiver operator characteristic (ROC) curves and genetic algorithm K-nearest neighbor method (GA/KNN) were additionally employed to identify top characterizing genes for the VxInsight-derived clusters, as further explained in Tables S1-S14.44 The full rank-ordered gene lists derived from ANOVA with bootstrapping, ROC, and GA/KNN are provided in Tables S1-S14.44 Principal component analysis (PCA) and hierarchical clustering were performed using MATLAB (MathWorks, Natick, MA).52,53 Concordance between VxInsight and hierarchical clusters was measured by the adjusted Rand index, with Monte Carlo estimation of statistical significance (n = 10 000 replications).54
Comparisons between clusters were based on the Kruskal-Wallis test for continuous variables (age, laboratory values) and on the χ2 approximation of the Fisher exact test and Pearson χ2 test for independence for dichotomous and categoric variables (complete response [CR], resistant disease [RD], FAB classification, cytogenetic characteristics, FLT3 mutations). Overall survival (OS) was measured from registration on treatment study until death from any cause, with observation censored for patients last known alive. Disease-free survival (DFS) was measured from the date the CR was established until the relapse of leukemia or death from any cause, with observation censored for patients last known to be alive without report of relapse. Distributions of OS and DFS were estimated by the method of Kaplan and Meier55 and compared between clusters using the log-rank test.56 Multivariate analyses of cluster differences and prognostic factors were based on logistic regression models for CR and RD and on proportional hazards regression models for OS and DFS.57 In logistic regression models, differences in proportions between clusters are represented as odds ratios relative to a defined cluster. This permits the cluster differences to be compared on a consistent scale regardless of other terms in the model. The hazard ratio plays an analogous role for proportional hazards regression models. All P values were 2-tailed and, in view of the exploratory nature of these analyses, were calculated without adjustment for multiple testing.
Results
AML cohort
Gene expression profiles were obtained from a retrospective cohort of 170 patients with previously untreated AML. Clinical, morphologic, cytogenetic, and mutation status of the cohort, outlined in Table 1, showed no sex predominance and most patients (80%) over the age of 55 years with a median age of 65 years (range, 20-84 years). Thirty-two cases (19%) were judged by clinical history to have secondary AML, while 104 (61%) had clinically de novo AML (clinical onset was not recorded in the 2 trials for patients of age 15 to 55, and in none of the other trials was secondary AML further classified as myelodysplastic syndrome [MDS]–versus treatment-related). All FAB subtypes were included except AML-M3, with a preponderance of acute myeloblastic leukemia with maturation (FAB-M2, 35%). Adequate cytogenetic analyses were obtained on 141 (83%) of the patients, and 139 of these could be assigned to cytogenetic risk categories. Most cytogenetically evaluable cases fell into the intermediate cytogenetic risk group (59%) due to the high percentage of patients with normal karyotypes (46%).
Clinical, morphologic, cytogenetic, and mutation characteristics of adult AML cohort (N = 170 patients)
Characteristic . | Data . |
|---|---|
| Age, no. patients (%) | |
| Younger than 56 y | 34 (20) |
| 56 y or older | 136 (80) |
| Sex, no. patients (%) | |
| Female | 85 (50) |
| Male | 85 (50) |
| FAB classification, no. patients (%) | |
| M1 | 40 (24) |
| M2 | 60 (35) |
| M4 | 42 (25) |
| M5 | 13 (8) |
| M6 | 1 (1) |
| M7 | 2 (1) |
| M0 | 10 (6) |
| Other | 2 (1) |
| Evaluable cytogenics, no. patients (%) | |
| No | 29 (17) |
| Yes | 141 (83) |
| Cytogenetic risk category, no. patients (%)* | |
| Favorable | 12 (9) |
| Intermediate | 83 (59) |
| Unfavorable | 44 (31) |
| Not assigned | 2 (1) |
| Specific cytogenetic features, no. patients (%)† | |
| Normal | 65 (46) |
| t(8;21) | 8 (6) |
| inv(16) | 4 (3) |
| NPM1 mutation status, no. patients (%)‡; | |
| Type A | 45 (27) |
| Non-type A§ | 5 (3) |
| Type A or non-A | 50 (30) |
| FLT3 mutation status, no. patients (%)∥ | |
| ITD | 46 (27) |
| TKD | 13 (12) |
| Median age, y (range) | 65 (20-84) |
| Median WBC count, × 109/L (range) | 22.9 (0.7-272.5) |
| Median peripheral blasts, % (range) | 43 (0-99) |
| Median marrow blasts, % (range) | 70 (5-99) |
| Median platelet count, × 109/L (range) | 53 (2-1052) |
| Median hemoglobin level, g/dL (range) | 9.1 (4.3-14.4) |
Characteristic . | Data . |
|---|---|
| Age, no. patients (%) | |
| Younger than 56 y | 34 (20) |
| 56 y or older | 136 (80) |
| Sex, no. patients (%) | |
| Female | 85 (50) |
| Male | 85 (50) |
| FAB classification, no. patients (%) | |
| M1 | 40 (24) |
| M2 | 60 (35) |
| M4 | 42 (25) |
| M5 | 13 (8) |
| M6 | 1 (1) |
| M7 | 2 (1) |
| M0 | 10 (6) |
| Other | 2 (1) |
| Evaluable cytogenics, no. patients (%) | |
| No | 29 (17) |
| Yes | 141 (83) |
| Cytogenetic risk category, no. patients (%)* | |
| Favorable | 12 (9) |
| Intermediate | 83 (59) |
| Unfavorable | 44 (31) |
| Not assigned | 2 (1) |
| Specific cytogenetic features, no. patients (%)† | |
| Normal | 65 (46) |
| t(8;21) | 8 (6) |
| inv(16) | 4 (3) |
| NPM1 mutation status, no. patients (%)‡; | |
| Type A | 45 (27) |
| Non-type A§ | 5 (3) |
| Type A or non-A | 50 (30) |
| FLT3 mutation status, no. patients (%)∥ | |
| ITD | 46 (27) |
| TKD | 13 (12) |
| Median age, y (range) | 65 (20-84) |
| Median WBC count, × 109/L (range) | 22.9 (0.7-272.5) |
| Median peripheral blasts, % (range) | 43 (0-99) |
| Median marrow blasts, % (range) | 70 (5-99) |
| Median platelet count, × 109/L (range) | 53 (2-1052) |
| Median hemoglobin level, g/dL (range) | 9.1 (4.3-14.4) |
ITD indicates internal tandem duplication; TKD, point mutations of intracellular tyrosine-kinase domain.
To convert hemoglobin level from grams per deciliter to grams per liter, multiply grams per deciliter by 10.
n = 141. Cytogenetic risk categories are defined by the following cytogenetic abnormalities (abn): favorable: inv(16)/t(16;16)/del(16q), t(8;21), or t(15;17) with any additional abn; intermediate: +8, -Y, +6, del(12p), or normal karyotype; unfavorable; -5/del(5q), -7/del(7q), inv(3q), abn 11q, t(6;9), t(9;22), abn 17p, or complex karyotype defined as more than 3 abn. Other findings are listed as not assigned or nonevaluable.12
n = 141.
n = 165.
Non—type A includes 4 cases that hybridized to probes for 13 known variants (B to Q)26,44 and 1 case that was sequenced.
n = 105.
Unsupervised clustering algorithms
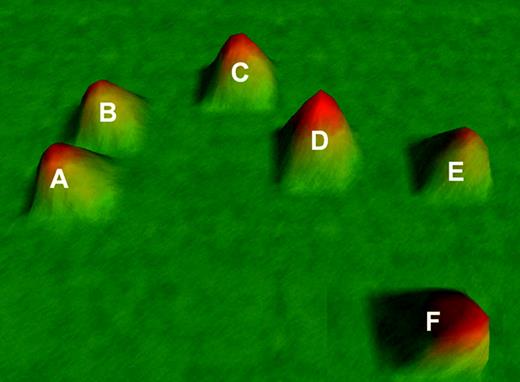
VxInsight analysis partitioned the AML patients into 6 distinct and stable groups based on strong similarities in gene expression among the 9463 genes, visualized in Figure 1.
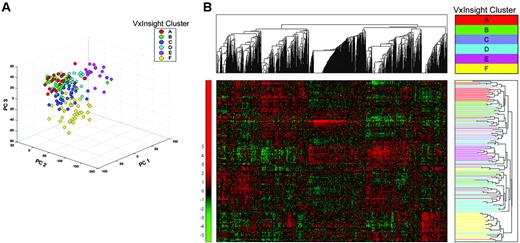
Membership among clusters ranged from a low of 18 patients (cluster E, 11%) to a high of 42 patients (cluster D, 25%). Clusters derived from PCA and unsupervised hierarchical clustering showed significant levels of concordance with the VxInsight-derived clusters (P < .001) (Figure 2).
VxInsight cluster membership, treatment outcomes, and clinical correlates
DFS varied significantly between VxInsight clusters (Figure 3; P = .023). Clusters A and C had the lowest and highest hazard ratios, respectively, for relapse or death in remission (Table 2), and all 3 remitting patients in cluster B relapsed within 16 months. Of the 170 patients, 145 have died and the remaining 25 were last known to be alive at 13 months to 10.9 years after starting treatment (median, 6.1 years). OS did not vary significantly among clusters (P = .40) but generally paralleled the DFS results, with cluster A having the best OS and clusters B and C generally the worst (Table 2; Figure 3).
Treatment outcomes of 170 adult AML patients by VxInsight cluster
. | A . | B . | C . | D . | E . | F . | P* . |
|---|---|---|---|---|---|---|---|
| No. patients | 24 | 22 | 31 | 42 | 18 | 33 | |
| Disease-free survival | |||||||
| Events | 8 | 3 | 17 | 12 | 5 | 13 | .023 |
| 5 y, %† | 27 | 0 | 6 | 23 | 19 | 32 | |
| 95% CI, % | 6-61 | 0-71 | 0-29 | 2-45 | 0-52 | 10-54 | |
| HR‡ | 1.00 | 2.56 | 3.58 | 1.73 | 1.50 | 1.25 | |
| 95% CI | — | 0.67-9.81 | 1.52-8.44 | 0.71-4.25 | 0.49-4.59 | 0.52-3.03 | |
| Overall survival | |||||||
| Deaths | 18 | 21 | 29 | 34 | 15 | 28 | .40 |
| 5 y, %† | 25 | 5 | 6 | 18 | 15 | 17 | |
| 95% CI, % | 8-42 | 0-23 | 0-15 | 6-30 | 0-32 | 4-30 | |
| HR‡ | 1.00 | 1.75 | 1.62 | 1.21 | 1.73 | 1.39 | |
| 95% CI | — | 0.93-3.29 | 0.90-2.94 | 0.68-2.15 | 0.87-3.44 | 0.77-2.51 | |
| Resistant disease | |||||||
| No. (%) | 8 (33) | 17 (77) | 5 (16) | 19 (45) | 6 (33) | 5 (15) | < .001 |
| 95% CI, % | 16-55 | 55-92 | 5-34 | 30-61 | 13-59 | 5-32 | |
| CR§ | 1.00 | 6.80 | 0.39 | 1.65 | 1.00 | 0.36 | |
| 95% CI | — | 1.94-27.4 | 0.10-1.35 | 0.59-4.86 | 0.27-3.66 | 0.09-1.25 | |
| Complete response | |||||||
| No. (%) | 11 (46) | 3 (14) | 18 (58) | 16 (38) | 7 (39) | 18 (55) | .023 |
| 95% CI, % | 26-67 | 3-35 | 39-75 | 24-54 | 17-64 | 36-72 | |
| CR§ | 1.00 | 0.19 | 1.64 | 0.73 | 0.75 | 1.42 | |
| 95% CI | — | 0.04-0.80 | 0.56-4.79 | 0.26-2.01 | 0.22-2.60 | 0.49-4.08 |
. | A . | B . | C . | D . | E . | F . | P* . |
|---|---|---|---|---|---|---|---|
| No. patients | 24 | 22 | 31 | 42 | 18 | 33 | |
| Disease-free survival | |||||||
| Events | 8 | 3 | 17 | 12 | 5 | 13 | .023 |
| 5 y, %† | 27 | 0 | 6 | 23 | 19 | 32 | |
| 95% CI, % | 6-61 | 0-71 | 0-29 | 2-45 | 0-52 | 10-54 | |
| HR‡ | 1.00 | 2.56 | 3.58 | 1.73 | 1.50 | 1.25 | |
| 95% CI | — | 0.67-9.81 | 1.52-8.44 | 0.71-4.25 | 0.49-4.59 | 0.52-3.03 | |
| Overall survival | |||||||
| Deaths | 18 | 21 | 29 | 34 | 15 | 28 | .40 |
| 5 y, %† | 25 | 5 | 6 | 18 | 15 | 17 | |
| 95% CI, % | 8-42 | 0-23 | 0-15 | 6-30 | 0-32 | 4-30 | |
| HR‡ | 1.00 | 1.75 | 1.62 | 1.21 | 1.73 | 1.39 | |
| 95% CI | — | 0.93-3.29 | 0.90-2.94 | 0.68-2.15 | 0.87-3.44 | 0.77-2.51 | |
| Resistant disease | |||||||
| No. (%) | 8 (33) | 17 (77) | 5 (16) | 19 (45) | 6 (33) | 5 (15) | < .001 |
| 95% CI, % | 16-55 | 55-92 | 5-34 | 30-61 | 13-59 | 5-32 | |
| CR§ | 1.00 | 6.80 | 0.39 | 1.65 | 1.00 | 0.36 | |
| 95% CI | — | 1.94-27.4 | 0.10-1.35 | 0.59-4.86 | 0.27-3.66 | 0.09-1.25 | |
| Complete response | |||||||
| No. (%) | 11 (46) | 3 (14) | 18 (58) | 16 (38) | 7 (39) | 18 (55) | .023 |
| 95% CI, % | 26-67 | 3-35 | 39-75 | 24-54 | 17-64 | 36-72 | |
| CR§ | 1.00 | 0.19 | 1.64 | 0.73 | 0.75 | 1.42 | |
| 95% CI | — | 0.04-0.80 | 0.56-4.79 | 0.26-2.01 | 0.22-2.60 | 0.49-4.08 |
— indicates not applicable.
P value for heterogeneity among 6 clusters based on Pearson χ2 test for independence (CR, RD) or log-rank test (OS, DFS).
Kaplan-Meier estimate of probability of OS or DFS at 5 years.
Hazard ratio, relative to cluster A.
Odds ratio, relative to cluster A.
Response to induction chemotherapy varied significantly among the 6 clusters (Table 2). Sixty (35%) of the 170 patients were resistant to their protocol induction chemotherapy, with a significantly different RD rate seen between clusters (P < .001). This was largely due to an exceptionally high RD rate in cluster B (77%) compared with all other clusters combined (43 of 148; 29%), although heterogeneity among the remaining 5 clusters was also significant (P = .021). Roughly complementary results were observed for CR. Seventy-three patients (43%) achieved CR, and the CR rate varied significantly among clusters (P = .023), being lowest in cluster B (14%). Forty-seven of the remitting patients have relapsed, and 11 others have died without report of relapse.
VxInsight cluster membership was not significantly correlated with patient age or de novo versus secondary onset of disease (Figure 4; Table 3). Despite the absence of a significant association with age, it was noteworthy that only 4% of patients in the 2 clusters with worse outcomes (B, C) were under age 56, compared with 27% (32 of 117) of patients in the remaining clusters.
Clinical and laboratory correlates of VxInsight clusters derived from 170 adult AMLs
. | A . | B . | C . | D . | E . | F . | P* . |
|---|---|---|---|---|---|---|---|
| No. patients | 24 | 22 | 31 | 42 | 18 | 33 | — |
| Median age, y (range) | 67 (22-76) | 68 (58-76) | 65 (44-84) | 62 (20-83) | 60 (21-81) | 64 (34-83) | .27 |
| No. with secondary AML (%) | 5 (25) | 7 (32) | 5 (17) | 6 (20) | 2 (22) | 7 (27) | .87 |
| Median pretreatment laboratory values | |||||||
| WBC count, × 109/L | 29 | 6 | 14 | 20 | 33 | 57 | < .001 |
| % PB blasts | 76 | 28 | 38 | 48 | 85 | 11 | < .001 |
| % BM blasts | 82 | 59 | 55 | 71 | 80 | 70 | .004 |
| Platelet count, × 109/L | 36 | 91 | 62 | 42 | 52 | 62 | .002 |
| Hemoglobin level, g/dL | 9.8 | 8.7 | 9.4 | 8.7 | 9.5 | 9.3 | .20 |
| Cytogenetic risk group, no. (%)* | |||||||
| Favorable | 0 (0) | 0 (0) | 1 (3) | 10 (29) | 1 (8) | 0 (0) | < .001 |
| t(8;21) | 0 (0) | 0 (0) | 0 (0) | 8 (24) | 0 (0) | 0 (0) | < .001 |
| Intermediate | 16 (80) | 15 (79) | 19 (66) | 9 (27) | 7 (54) | 17 (71) | < .001 |
| Abnormal | 1 (5) | 7 (37) | 3 (10) | 1 (3) | 1 (8) | 5 (21) | — |
| Normal | 15 (75) | 8 (42) | 6 (55) | 8 (24) | 6 (46) | 12 (50) | .011 |
| Unfavorable, no. (%)† | 4 (20) | 4 (21) | 9 (31) | 15 (44) | 5 (38) | 7 (29) | .41 |
| NPM1 mutation status, no. (%)‡ | |||||||
| NPM1 + (%) | 18/23 (78) | 1/22 (5) | 4/30 (13) | 2/41 (5) | 6/18 (33) | 19/31 (61) | < .001 |
| FLT3 mutation status, no. (%)§ | |||||||
| ITD+ | 8/24 (33) | 3/22 (14) | 6/31 (19) | 13/42 (31) | 5/18 (28) | 11/32 (34) | .467 |
| TKD+ | 5/19 (26) | 0/9 (0) | 2/18 (11) | 3/27 (11) | 1/8 (13) | 2/24 (8) | .404 |
| Both NPM1+ and FLT3-ITD | 7/23 (30) | 1/22 (5) | 2/30 (7) | 2/41 (5) | 3/18 (17) | 10/31 (32) | .003 |
. | A . | B . | C . | D . | E . | F . | P* . |
|---|---|---|---|---|---|---|---|
| No. patients | 24 | 22 | 31 | 42 | 18 | 33 | — |
| Median age, y (range) | 67 (22-76) | 68 (58-76) | 65 (44-84) | 62 (20-83) | 60 (21-81) | 64 (34-83) | .27 |
| No. with secondary AML (%) | 5 (25) | 7 (32) | 5 (17) | 6 (20) | 2 (22) | 7 (27) | .87 |
| Median pretreatment laboratory values | |||||||
| WBC count, × 109/L | 29 | 6 | 14 | 20 | 33 | 57 | < .001 |
| % PB blasts | 76 | 28 | 38 | 48 | 85 | 11 | < .001 |
| % BM blasts | 82 | 59 | 55 | 71 | 80 | 70 | .004 |
| Platelet count, × 109/L | 36 | 91 | 62 | 42 | 52 | 62 | .002 |
| Hemoglobin level, g/dL | 9.8 | 8.7 | 9.4 | 8.7 | 9.5 | 9.3 | .20 |
| Cytogenetic risk group, no. (%)* | |||||||
| Favorable | 0 (0) | 0 (0) | 1 (3) | 10 (29) | 1 (8) | 0 (0) | < .001 |
| t(8;21) | 0 (0) | 0 (0) | 0 (0) | 8 (24) | 0 (0) | 0 (0) | < .001 |
| Intermediate | 16 (80) | 15 (79) | 19 (66) | 9 (27) | 7 (54) | 17 (71) | < .001 |
| Abnormal | 1 (5) | 7 (37) | 3 (10) | 1 (3) | 1 (8) | 5 (21) | — |
| Normal | 15 (75) | 8 (42) | 6 (55) | 8 (24) | 6 (46) | 12 (50) | .011 |
| Unfavorable, no. (%)† | 4 (20) | 4 (21) | 9 (31) | 15 (44) | 5 (38) | 7 (29) | .41 |
| NPM1 mutation status, no. (%)‡ | |||||||
| NPM1 + (%) | 18/23 (78) | 1/22 (5) | 4/30 (13) | 2/41 (5) | 6/18 (33) | 19/31 (61) | < .001 |
| FLT3 mutation status, no. (%)§ | |||||||
| ITD+ | 8/24 (33) | 3/22 (14) | 6/31 (19) | 13/42 (31) | 5/18 (28) | 11/32 (34) | .467 |
| TKD+ | 5/19 (26) | 0/9 (0) | 2/18 (11) | 3/27 (11) | 1/8 (13) | 2/24 (8) | .404 |
| Both NPM1+ and FLT3-ITD | 7/23 (30) | 1/22 (5) | 2/30 (7) | 2/41 (5) | 3/18 (17) | 10/31 (32) | .003 |
P values were determined using χ2 test.
To convert hemoglobin level from grams per deciliter to grams per liter, multiply grams per deciliter by 10.
PB indicates peripheral blood; BM, bone marrow.
n = 139.
11 q23 abnormalities were seen in 9 cases and distributed in clusters A to F as 0, 1, 2, 1, 1, 3, respectively.
n = 165; values given indicate number of patients with the mutation out of number of patients tested.
For ITD, n = 169; for TKD, n = 105. Values given indicate number of patients with the mutation out of number of patients tested.
VxInsight clusters in adult AML. Six distinct clusters of AML patients are identified based on gene expression profiles and designated A to F. The data are visualized as a 3-dimensional terrain map with 2-dimensional distances reflecting gene expression profile correlates and the third dimension representing cluster membership density. Additional information on VxInsight is available at the University of New Mexico Health Sciences Center website and Sandia National Laboratories website.44,47
VxInsight clusters in adult AML. Six distinct clusters of AML patients are identified based on gene expression profiles and designated A to F. The data are visualized as a 3-dimensional terrain map with 2-dimensional distances reflecting gene expression profile correlates and the third dimension representing cluster membership density. Additional information on VxInsight is available at the University of New Mexico Health Sciences Center website and Sandia National Laboratories website.44,47
Alternative clustering algorithms of adult AML cohort. (A) A multidimensional scatterplot generated using principal component analysis (PCA) reduces the dimensionality of the data by projecting the expression data matrix into 3 dimensions. The largest sources of gene expression signal variance are represented as principal components (labeled PC1, PC2, PC3).52 (B) Two-dimensional unsupervised clustering dendrograms and “heat map” of gene expression data from the 170 AML cases using 9463 genes. Pearson correlation coefficient was used to compute gene and patient similarity. The cluster-to-cluster distance was computed using the average linkage. The relative gene expression scale is depicted on the left with the normalized scores ranging from –5 to more than 5. Gene cluster and patient cluster dendrograms are plotted to the top and right sides of the heat map, respectively. After PCA and hierarchical clustering was performed, the individual patients were color coded for comparison with their VxInsight cluster membership. These methods showed a significant degree of concordance with VxInsight cluster membership (adjusted Rand index = 0.3457, P < .001) (Table S15).44
Alternative clustering algorithms of adult AML cohort. (A) A multidimensional scatterplot generated using principal component analysis (PCA) reduces the dimensionality of the data by projecting the expression data matrix into 3 dimensions. The largest sources of gene expression signal variance are represented as principal components (labeled PC1, PC2, PC3).52 (B) Two-dimensional unsupervised clustering dendrograms and “heat map” of gene expression data from the 170 AML cases using 9463 genes. Pearson correlation coefficient was used to compute gene and patient similarity. The cluster-to-cluster distance was computed using the average linkage. The relative gene expression scale is depicted on the left with the normalized scores ranging from –5 to more than 5. Gene cluster and patient cluster dendrograms are plotted to the top and right sides of the heat map, respectively. After PCA and hierarchical clustering was performed, the individual patients were color coded for comparison with their VxInsight cluster membership. These methods showed a significant degree of concordance with VxInsight cluster membership (adjusted Rand index = 0.3457, P < .001) (Table S15).44
Clinical and laboratory parameters that showed significant correlation with VxInsight cluster membership were pretreatment WBC counts, blast percentages, platelet counts, FAB classification, normal or t(8;21) karyotypes, and NPM1 mutation status (Tables 1 and 3). The lower WBC and blast counts in the poor-risk clusters (B, C) suggest underlying marrow damage. Clusters were segregated by their degree of blast maturation and more specifically by myeloid versus monocytic derivation (FAB classification) (Figure 4). Cluster F consisted almost entirely of monocytic leukemias, with 97% of members having FAB-M4 or FAB-M5, although monocytic leukemias were present in lower proportions in the 5 other clusters. Cytogenetic risk groups varied with cluster membership (Table 3; Figure 4). Cluster A, with the best OS, had the highest percentage of normal karyotypes (75%). In contrast, cluster D had the highest percentage of karyotypic abnormalities (76%), including those associated with both favorable (8 of 8 patients with t(8;21) and 2 of 4 with inv(16)) and unfavorable risk.
Estimated distributions for disease-free survival and overall survival of AML patients by VxInsight cluster membership. (A) DFS varied significantly among the 6 clusters (log-rank, P = .023). (B) OS showed a trend that paralleled the DFS findings among the 6 clusters (log-rank, P = .40). Tick marks indicate censored observations for patients last known to be alive without report of relapse.
Estimated distributions for disease-free survival and overall survival of AML patients by VxInsight cluster membership. (A) DFS varied significantly among the 6 clusters (log-rank, P = .023). (B) OS showed a trend that paralleled the DFS findings among the 6 clusters (log-rank, P = .40). Tick marks indicate censored observations for patients last known to be alive without report of relapse.
NPM1 mutations were present in 30% (50 of 165) of cases with significant differences observed between VxInsight clusters (Tables 1 and 3; Figure 5). The highest prevalences were seen in cluster A (78%), which also had the highest percentage of females and normal karyotypes, and in cluster F (61%) with the predominance of monocytic leukemias. FLT3-ITD mutations were identified in 27% of cases (Table 1) with no significant differences among VxInsight groups (Table 3). A significant number of patients with FLT3-ITDs also had NPM1 mutations (Table 3). FLT3-TKDs were found in 12% of the AMLs investigated; cluster A had the highest percentage of point mutations (FLT3-TKDs).
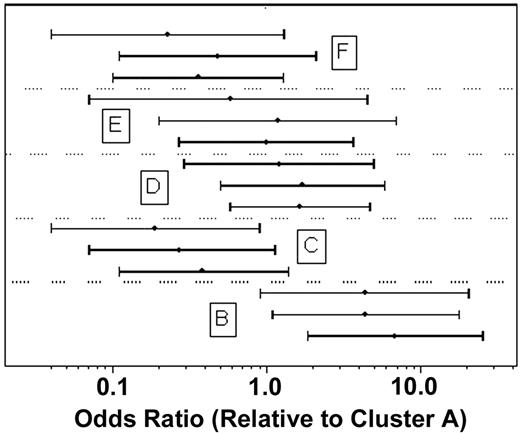
Further analyses were performed to investigate whether comparisons of outcomes between the clusters might be biased by confounding effects of the other factors considered. In multivariate logistic regression analysis, increasing age (P = .024), secondary AML onset (P = .010), and unfavorable cytogenetic risk category (P = .030) had independent detrimental prognostic effects on RD. AML onset and/or cytogenetic risk group were unknown for 57 of the 170 patients. Therefore, to allow for the possibility that results might be biased by the exclusion of these patients, the association between RD rate and cluster was estimated with and without adjustment for age, AML onset, and cytogenetic category for the 113 patients with complete data. The results, shown in Figure 6, confirm that heterogeneity of RD rates among the 6 clusters remained statistically significant (P < .001) after adjusting for possible confounding. The variation of CR rates among the 6 clusters remained marginally significant after adjusting for age, AML onset, and unfavorable cytogenetics (P = .051). In proportional hazards regression analyses adjusting for age, AML onset, and cytogenetic risk category, the variation of OS among clusters remained nonsignificant (P = .56). DFS also did not vary significantly among clusters after accounting for similar effects (P = .22); however, this analysis was inconclusive because only 49 remitting patients had both AML onset and cytogenetic risk group data.
Genes distinguishing VxInsight clusters
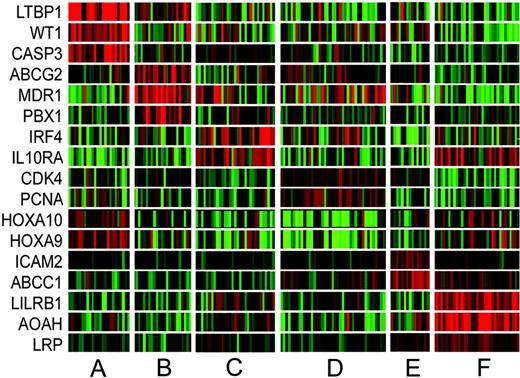
Using ANOVA with bootstrapping, gene lists were derived that define the VxInsight clusters. The 50 most significant discriminating genes for each cluster are provided in Tables S1-S6,44 with a summary of these lists, including the most significantly up-regulated and down-regulated genes, given in Table 4. Gene expression patterns for a subset of these genes are highlighted in Figure 7. The top 50 ranked genes for clusters B, D, and F are primarily up-regulated (90%, 92%, 98% of genes, respectively) in comparison with the down-regulation of several significant differentiating genes for clusters A, C, and E (36%, 54%, 14% of genes, respectively). Cluster D, containing virtually all of the cases with favorable cytogenetic abnormalities and a large percentage of intermediate and unfavorable karyotypes, is defined by high expression (top 46 characterizing genes are overexpressed) of a number of genes involved in DNA replication (GART, MCM3, PCNA), control of cell proliferation (CDK4, ODC1, STMN1), transcription (POLR2H, EIR2S1, HTATSF1), and DNA repair (UNG, CHEK2, APEX1, ADPRT). This gene expression signature may be reflective of high “proliferative” activity. An interesting finding is the decreased expression of homeobox A9 (HOXA9) and A10 (HOXA10) in cluster D compared with the other AMLs. This may relate in part to the low incidence of NPM1 mutations.39
Rank-ordered gene lists characterizing each cluster; genes are up-regulated or down-regulated in relation to the other clusters
Cluster and order . | P . | Gene . | Probe set . | Description . |
|---|---|---|---|---|
| Up-regulated genes | ||||
| A | ||||
| 1 | .002 | LTBP1 | 1495_at | Latent transforming growth factor beta binding protein 1 |
| 2 | .003 | CASP3 | 36143_at | Caspase-3, apoptosis-related cysteine protease |
| 4 | .011 | FTO | 37242_at | Fatso |
| 5 | .015 | FOXC1 | 41027_at | Forkhead box C1 |
| 6 | .003 | COL4A5 | 32667_at | Collagen, type IV, alpha 5 (Alport syndrome) |
| 11 | .015 | RASGRP3 | 34748_at | RAS guanyl-releasing protein 3 (calcium and DAG-regulated) |
| 19 | .025 | MYCN | 35158_at | v-myc myelocytomatosis viral-related oncogene, neuroblastoma derived |
| B | ||||
| 1 | .005 | BIA2 | 36713_at | BIA2 |
| 2 | .003 | CXorf6 | 38916_at | Chromosome X open reading frame 6 |
| 3 | .014 | PLOD2 | 34795_at | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase (lysine hydroxylase) 2 |
| 4 | .011 | OPTN | 41744_at | Optineurin |
| 5 | .016 | CLIC2 | 40013_at | Chloride intracellular channel 2 |
| 6 | .017 | RHD | 37164_at | Rhesus blood group, D antigen |
| 7 | .021 | CDC42BPA | 39962_at | CDC42 binding protein kinase alpha (DMPK-like) |
| 8 | .038 | ANK3 | 36965_at | Ankyrin 3, node of Ranvier (ankyrin G) |
| C | ||||
| 1 | .003 | SDR1 | 40782_at | Short-chain dehydrogenase/reductase 1 |
| 2 | .01 | SDS | 40390_at | Serine dehydratase |
| 6 | .058 | SERPINF1 | 40856_at | Serine (or cysteine) proteinase inhibitor, clade F |
| 8 | .092 | MALT1 | 38575_at | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 |
| 9 | .051 | HERPUD1 | 39733_at | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| 10 | .056 | IRF4 | 37625_at | Interferon regulatory factor 4 |
| 11 | .034 | IL10RA | 1062_g_at | Interleukin-10 receptor, alpha |
| 12 | .035 | BCS1L | 31842_at | BCS1-like (yeast) |
| 13 | .054 | RAB9A | 39628_at | RAB9A, member RAS oncogene family |
| D | ||||
| 1 | .001 | RNASEP1 | 37471_at | Ribonuclease P1 |
| 2 | .006 | PGDS | 35523_at | Prostaglandin D2 synthase, hematopoietic |
| 3 | .006 | NHP2L1 | 41746_at | NHP2 nonhistone chromosome protein 2-like 1 (S cerevisiae) |
| 4 | .010 | UNG | 37686_s_at | Uracil-DNA glycosylase |
| 5 | .011 | POP1 | 38513_at | Processing of precursors 1 |
| 6 | .008 | HSU79274 | 31838_at | Protein predicted by clone 23733 |
| 7 | .005 | CGI-51 | 34845_at | CGI-51 protein |
| 8 | .010 | NASP | 33255_at | Nuclear autoantigenic sperm protein (histone-binding) |
| 9 | .010 | CDK4 | 1942_s_at | Cyclin-dependent kinase 4 |
| E | ||||
| 1 | .002 | CAPN1 | 33908_at | Calpain 1, (mu/l) large subunit |
| 2 | .010 | HSF1 | 40200_at | Heat shock transcription factor 1 |
| 3 | .005 | ACTN4 | 41753_at | Actinin, alpha 4 |
| 5 | .007 | TNRC11 | 40998_at | Trinucleotide repeat containing 11 |
| 6 | .007 | G2AN | 37040_at | Alpha glucosidase II alpha subunit |
| 7 | .005 | NFIC | 440_at | Nuclear factor I/C (CCAAT-binding transcription factor) |
| F | ||||
| 1 | .001 | EPB41L3 | 41385_at | Erythrocyte membrane protein band 4.1-like 3 |
| 2 | .001 | FCGR2A | 37688_f_at | Fc fragment of IgG, low affinity IIa, receptor for (CD32) |
| 3 | .001 | HK3 | 36372_at | Hexokinase 3 (white cell) |
| 4 | .002 | CSPG2 | 31682_s_at | Chondroitin sulfate proteoglycan 2 (versican) |
| 5 | .005 | PGAM1 | 41221_at | Phosphoglycerate mutase 1 (brain) |
| 6 | .003 | LILRB1 | 32475_at | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| 7 | .006 | CYBB | 37975_at | Cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| 9 | .004 | CD86 | 36270_at | CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) |
| Down-regulated genes | ||||
| A | ||||
| 3 | .004 | HLA-DPB1 | 38095_i_at | Major histocompatibility complex, class II, DP beta 1 |
| 7 | .010 | HLA-DMA | 37344_at | Major histocompatibility complex, class II, DM alpha |
| 8 | .012 | HLA-DPB1 | 38096_f_at | Major histocompatibility complex, class II, DP beta 1 |
| 9 | .007 | CD74 | 35016_at | CD74 antigen (invariant polypeptide of MHC, class II antigen-associated) |
| 12 | .001 | HLA-DRB3 | 41723_s_at | Major histocompatibility complex, class II, DR beta 3 |
| 16 | .009 | HLA-DRA | 37039_at | Major histocompatibility complex, class II, DR alpha |
| 20 | .007 | RAB31 | 33371_s_at | RAB31, member RAS oncogene family |
| B | ||||
| 12 | .01 | LGALS1 | 33412_at | Lectin, galactoside-binding, soluble, 1 (galectin 1) |
| 34 | .046 | STX4A | 37911_at | Syntaxin 4A (placental) |
| 37 | .032 | PTPRC | 40520_g_at | Protein tyrosine phosphatase, receptor type, C |
| 40 | .036 | EMP3 | 39182_at | Epithelial membrane protein 3 |
| 44 | .026 | CORO1A | 38976_at | Coronin, actin binding protein, 1A |
| 51 | .048 | INPPL1 | 36598_s_at | Inositol polyphosphate phosphatase-like 1 |
| C | ||||
| 3 | .015 | DNCL1 | 34891_at | Dynein, cytoplasmic, light polypeptide 1 |
| 4 | .022 | RAB9P40 | 109_at | Rab9 effector p40 |
| 5 | .035 | BDH | 37211_at | 3-hydroxybutyrate dehydrogenase (heart, mitochondrial) |
| 7 | .018 | CGI-87 | 41590_at | CGI-87 protein |
| 12 | .035 | BCS1L | 31842_at | BCS1-like (yeast) |
| 14 | .027 | POP5 | 39516_at | RNase MRP/RNase P protein-like |
| D | ||||
| 24 | .047 | HOXA10 | 41448_at | Homeobox A10 |
| 34 | .028 | — | 32021_at | H sapiens transcribed sequence with weak similarity to protein ref: NP_060265.1(H sapiens) hypothetical protein FLJ20378 |
| 40 | .046 | KIAA0669 | 41788_i_at | KIAA0669 gene product |
| 42 | .056 | HOXA9 | 37809_at | Homeobox A9 |
| E | ||||
| 4 | .002 | DPM1 | 34879_at | Dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| 16 | .010 | MCP | 38441_s_at | Membrane cofactor protein (CD46, trophoblast-lymphocyte cross-reactive antigen) |
| 19 | .019 | STXBP3 | 37962_r_at | Syntaxin binding protein 3 |
| 25 | .007 | PSMC6 | 949_s_at | Proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| 35 | .033 | CHUK | 33770_at | Conserved helix-loop-helix ubiquitous kinase |
| 36 | .019 | COPB | 34326_at | Coatomer protein complex, subunit beta |
| F | ||||
| 50 | .019 | CCND2 | 36650_at | Cyclin D2 |
| 57 | .020 | ERG | 914_g_at | v-ets erythroblastosis virus E26 oncogene like (avian) |
| 80 | .035 | IMPDH2 | 36624_at | IMP (inosine monophosphate) dehydrogenase 2 |
| 82 | .022 | 6-SEP | 38826_at | Septin 6 |
| 84 | .025 | RPL17 | 32440_at | Ribosomal protein L17 |
Cluster and order . | P . | Gene . | Probe set . | Description . |
|---|---|---|---|---|
| Up-regulated genes | ||||
| A | ||||
| 1 | .002 | LTBP1 | 1495_at | Latent transforming growth factor beta binding protein 1 |
| 2 | .003 | CASP3 | 36143_at | Caspase-3, apoptosis-related cysteine protease |
| 4 | .011 | FTO | 37242_at | Fatso |
| 5 | .015 | FOXC1 | 41027_at | Forkhead box C1 |
| 6 | .003 | COL4A5 | 32667_at | Collagen, type IV, alpha 5 (Alport syndrome) |
| 11 | .015 | RASGRP3 | 34748_at | RAS guanyl-releasing protein 3 (calcium and DAG-regulated) |
| 19 | .025 | MYCN | 35158_at | v-myc myelocytomatosis viral-related oncogene, neuroblastoma derived |
| B | ||||
| 1 | .005 | BIA2 | 36713_at | BIA2 |
| 2 | .003 | CXorf6 | 38916_at | Chromosome X open reading frame 6 |
| 3 | .014 | PLOD2 | 34795_at | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase (lysine hydroxylase) 2 |
| 4 | .011 | OPTN | 41744_at | Optineurin |
| 5 | .016 | CLIC2 | 40013_at | Chloride intracellular channel 2 |
| 6 | .017 | RHD | 37164_at | Rhesus blood group, D antigen |
| 7 | .021 | CDC42BPA | 39962_at | CDC42 binding protein kinase alpha (DMPK-like) |
| 8 | .038 | ANK3 | 36965_at | Ankyrin 3, node of Ranvier (ankyrin G) |
| C | ||||
| 1 | .003 | SDR1 | 40782_at | Short-chain dehydrogenase/reductase 1 |
| 2 | .01 | SDS | 40390_at | Serine dehydratase |
| 6 | .058 | SERPINF1 | 40856_at | Serine (or cysteine) proteinase inhibitor, clade F |
| 8 | .092 | MALT1 | 38575_at | Mucosa-associated lymphoid tissue lymphoma translocation gene 1 |
| 9 | .051 | HERPUD1 | 39733_at | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| 10 | .056 | IRF4 | 37625_at | Interferon regulatory factor 4 |
| 11 | .034 | IL10RA | 1062_g_at | Interleukin-10 receptor, alpha |
| 12 | .035 | BCS1L | 31842_at | BCS1-like (yeast) |
| 13 | .054 | RAB9A | 39628_at | RAB9A, member RAS oncogene family |
| D | ||||
| 1 | .001 | RNASEP1 | 37471_at | Ribonuclease P1 |
| 2 | .006 | PGDS | 35523_at | Prostaglandin D2 synthase, hematopoietic |
| 3 | .006 | NHP2L1 | 41746_at | NHP2 nonhistone chromosome protein 2-like 1 (S cerevisiae) |
| 4 | .010 | UNG | 37686_s_at | Uracil-DNA glycosylase |
| 5 | .011 | POP1 | 38513_at | Processing of precursors 1 |
| 6 | .008 | HSU79274 | 31838_at | Protein predicted by clone 23733 |
| 7 | .005 | CGI-51 | 34845_at | CGI-51 protein |
| 8 | .010 | NASP | 33255_at | Nuclear autoantigenic sperm protein (histone-binding) |
| 9 | .010 | CDK4 | 1942_s_at | Cyclin-dependent kinase 4 |
| E | ||||
| 1 | .002 | CAPN1 | 33908_at | Calpain 1, (mu/l) large subunit |
| 2 | .010 | HSF1 | 40200_at | Heat shock transcription factor 1 |
| 3 | .005 | ACTN4 | 41753_at | Actinin, alpha 4 |
| 5 | .007 | TNRC11 | 40998_at | Trinucleotide repeat containing 11 |
| 6 | .007 | G2AN | 37040_at | Alpha glucosidase II alpha subunit |
| 7 | .005 | NFIC | 440_at | Nuclear factor I/C (CCAAT-binding transcription factor) |
| F | ||||
| 1 | .001 | EPB41L3 | 41385_at | Erythrocyte membrane protein band 4.1-like 3 |
| 2 | .001 | FCGR2A | 37688_f_at | Fc fragment of IgG, low affinity IIa, receptor for (CD32) |
| 3 | .001 | HK3 | 36372_at | Hexokinase 3 (white cell) |
| 4 | .002 | CSPG2 | 31682_s_at | Chondroitin sulfate proteoglycan 2 (versican) |
| 5 | .005 | PGAM1 | 41221_at | Phosphoglycerate mutase 1 (brain) |
| 6 | .003 | LILRB1 | 32475_at | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| 7 | .006 | CYBB | 37975_at | Cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| 9 | .004 | CD86 | 36270_at | CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) |
| Down-regulated genes | ||||
| A | ||||
| 3 | .004 | HLA-DPB1 | 38095_i_at | Major histocompatibility complex, class II, DP beta 1 |
| 7 | .010 | HLA-DMA | 37344_at | Major histocompatibility complex, class II, DM alpha |
| 8 | .012 | HLA-DPB1 | 38096_f_at | Major histocompatibility complex, class II, DP beta 1 |
| 9 | .007 | CD74 | 35016_at | CD74 antigen (invariant polypeptide of MHC, class II antigen-associated) |
| 12 | .001 | HLA-DRB3 | 41723_s_at | Major histocompatibility complex, class II, DR beta 3 |
| 16 | .009 | HLA-DRA | 37039_at | Major histocompatibility complex, class II, DR alpha |
| 20 | .007 | RAB31 | 33371_s_at | RAB31, member RAS oncogene family |
| B | ||||
| 12 | .01 | LGALS1 | 33412_at | Lectin, galactoside-binding, soluble, 1 (galectin 1) |
| 34 | .046 | STX4A | 37911_at | Syntaxin 4A (placental) |
| 37 | .032 | PTPRC | 40520_g_at | Protein tyrosine phosphatase, receptor type, C |
| 40 | .036 | EMP3 | 39182_at | Epithelial membrane protein 3 |
| 44 | .026 | CORO1A | 38976_at | Coronin, actin binding protein, 1A |
| 51 | .048 | INPPL1 | 36598_s_at | Inositol polyphosphate phosphatase-like 1 |
| C | ||||
| 3 | .015 | DNCL1 | 34891_at | Dynein, cytoplasmic, light polypeptide 1 |
| 4 | .022 | RAB9P40 | 109_at | Rab9 effector p40 |
| 5 | .035 | BDH | 37211_at | 3-hydroxybutyrate dehydrogenase (heart, mitochondrial) |
| 7 | .018 | CGI-87 | 41590_at | CGI-87 protein |
| 12 | .035 | BCS1L | 31842_at | BCS1-like (yeast) |
| 14 | .027 | POP5 | 39516_at | RNase MRP/RNase P protein-like |
| D | ||||
| 24 | .047 | HOXA10 | 41448_at | Homeobox A10 |
| 34 | .028 | — | 32021_at | H sapiens transcribed sequence with weak similarity to protein ref: NP_060265.1(H sapiens) hypothetical protein FLJ20378 |
| 40 | .046 | KIAA0669 | 41788_i_at | KIAA0669 gene product |
| 42 | .056 | HOXA9 | 37809_at | Homeobox A9 |
| E | ||||
| 4 | .002 | DPM1 | 34879_at | Dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| 16 | .010 | MCP | 38441_s_at | Membrane cofactor protein (CD46, trophoblast-lymphocyte cross-reactive antigen) |
| 19 | .019 | STXBP3 | 37962_r_at | Syntaxin binding protein 3 |
| 25 | .007 | PSMC6 | 949_s_at | Proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| 35 | .033 | CHUK | 33770_at | Conserved helix-loop-helix ubiquitous kinase |
| 36 | .019 | COPB | 34326_at | Coatomer protein complex, subunit beta |
| F | ||||
| 50 | .019 | CCND2 | 36650_at | Cyclin D2 |
| 57 | .020 | ERG | 914_g_at | v-ets erythroblastosis virus E26 oncogene like (avian) |
| 80 | .035 | IMPDH2 | 36624_at | IMP (inosine monophosphate) dehydrogenase 2 |
| 82 | .022 | 6-SEP | 38826_at | Septin 6 |
| 84 | .025 | RPL17 | 32440_at | Ribosomal protein L17 |
ANOVA was used to identify rank-ordered gene lists with bootstrap resampling to estimate the stability of these lists. P represents the estimated fraction of time that a gene was ranked at or above its observed position after tabulation of rankings from 1000 bootstrap resamplings (Document S1).44
— indicates gene symbol not available.
Genes associated with cell signaling (IL12, ranked 29), apoptosis (LTBP1, caspase-3), leukemic transformation (MEIS, ranked 30; WT1, ranked 22; FOXC1), and multidrug resistance (MRP2, ranked 40) are overexpressed by cluster A. The top-ranking gene, latent transforming growth factor (TGF) beta binding protein (LTBP1), activates latent TGF-β, a modulator of apoptosis that is independent of caspase-3–mediated mechanisms.58-60 FOXC1 is a TGF-β1 responsive gene that possibly functions as a tumor suppressor gene.61 Notably absent in cluster A is expression of the major histocompatibility complex (MHC) II alleles.
Cluster B, with the poorest clinical outcomes, shows increased expression of the multidrug resistance gene ABCG2 (ranked 18). The multidrug resistance membrane transporter (MDR1) is concurrently overexpressed (Figure 7). Additional genes of interest are PBX1 and serine/threonine protein kinase 17A (STK17A, ranked 23). STK17A plays a role in the regulation of apoptosis; PBX1 is a cofactor in genetic mechanisms that prevent myeloid differentiation but appears to lack inherent transformation ability in isolation.62,63 Cluster C shows expression of genes involved in immunoregulation (IRF4, IL10R, MALT), including several probe sets for gamma interferon and interferon-inducible genes.
Inhibitors of apoptotic function (ICAM2, ranked 34; DFFA/DFF45, ranked 33) are overexpressed among cluster E members. This cluster showed variable expression of genes related to immune function with up-regulation of some genes (SPN, IRF3, IFITM2) and down-regulation of others (MCP, CHUK). Finally, cluster F has the most distinguishing genetic profile due to the significant number of genes associated with monocyte differentiation and function (LILRB1, AOAH, TIL3, CASP1, LGALS3). The multidrug resistant gene for vault-transporter lung resistance protein (LRP, ranked 54) is also found in this group.
Clinical characteristics of 170 AML patients separated by VxInsight cluster membership. Each horizontal row represents an individual AML patient, and each column is the clinical variable for that individual. Age is presented as a continuum with the lightest color (white) representing the youngest patients and the darkest color (dark red) representing the oldest patients. Discode relates to AML onset; it is color coded and categorized similar to the remaining clinical variables, as described below the associated columns. Distribution of FAB classification varied significantly among clusters (P < .001).
Clinical characteristics of 170 AML patients separated by VxInsight cluster membership. Each horizontal row represents an individual AML patient, and each column is the clinical variable for that individual. Age is presented as a continuum with the lightest color (white) representing the youngest patients and the darkest color (dark red) representing the oldest patients. Discode relates to AML onset; it is color coded and categorized similar to the remaining clinical variables, as described below the associated columns. Distribution of FAB classification varied significantly among clusters (P < .001).
Alternative gene lists using different statistical and normalization methods are provided in Tables S7-S14.44 These show extensive overlap with the ANOVA-derived gene lists.
Discussion
We used a novel unsupervised clustering algorithm (VxInsight) to analyze gene expression profiles from older AML patients with a high proportion of intermediate- and poor-risk outcome factors. This type of analysis, without knowledge of prior class definitions, allows for identification of fundamental subsets of patients sharing similar expression signatures. Unanticipated similarities between cytogenetically diverse patient groups, as discovered in this study and reported by others,35 would have been harder to detect with a more restrictive supervised approach. The result is an interesting separation of the AML cases into 6 distinct clusters with outcome differences.
In contrast to previous studies using unsupervised computational methods alone,32,34,35 we found significant outcome differences between the clusters defined by gene expression for RD after induction therapy (P < .001), CR rate after induction therapy (P = .023), and DFS (P = .023). The heterogeneity of RD and CR rates among clusters was not explained by confounding effects of age, AML onset, and unfavorable cytogenetics, indicating that the clustering conveyed prognostic information independent of the other factors. For some patients, data were absent regarding prognostic factors, particularly de novo versus secondary onset of AML and cytogenetics. However, in the multivariate regression analyses of treatment outcomes, it was evident that excluding the patients with incomplete data did not markedly influence the magnitude or statistical significance of differences between clusters. This was most clearly evident for RD, for which both the statistical significance of cluster differences and the ORs representing the magnitudes of those differences were essentially unchanged by the adjustment for covariates. Evaluation of DFS was limited by the small number of remitting patients with complete data. For CR, adjusting for the covariates decreased the statistical significance from P = .023 to .051, which is not a profound change, especially given the necessity of excluding patients with incomplete data from the multivariate analysis.
Members of cluster A had the best DFS and OS: 27% and 25%, respectively, at 5 years. The striking finding for this group was the high percentage of NPM1 mutations (78%). This group has many of the characteristics emerging for cases of AML with NPM1 mutations, including the disproportionate number of women (67%), increase in normal karyotypes (75%), older age (but not significantly different than other cluster groups), and higher WBC counts.22,24-27 Genes responsible for the better outcome were not clearly identified, but significant overlap was discovered between top genes predicting for cluster A and for those previously reported for NPM1 mutations based on the data by Alcalay et al (Tables S18-S19).39,44 For example, 8 of the top 15 ranked genes (53%) for cluster A were also genes found to be predictive of NPM1 mutations.39 This finding is particularly striking given the use of different Affymetrix platforms with different probe sets (see Supplement). Genes predictive of cluster A were also examined in the AML dataset of Valk et al34 ; their cluster group 6 showed a similar gene expression pattern to cluster A as well as a high incidence of NPM1 mutations (100%) (Table S17 and Figure S2).33,44
Nucleophosmin (NPM1) gel analysis/hybridization. (A) Hybridization to wild-type NPM1 probe confirms the presence of wild-type NPM1 in 7 patient specimens and 3 cell lines (specimens 8 to 10). All 165 AML samples contained wild-type NPM1. (B) Patient specimen (1, 3-5, 7) hybridized to NPM1 variant A probe consistent with an NPM1 variant type A mutation (see Supplement).44
Nucleophosmin (NPM1) gel analysis/hybridization. (A) Hybridization to wild-type NPM1 probe confirms the presence of wild-type NPM1 in 7 patient specimens and 3 cell lines (specimens 8 to 10). All 165 AML samples contained wild-type NPM1. (B) Patient specimen (1, 3-5, 7) hybridized to NPM1 variant A probe consistent with an NPM1 variant type A mutation (see Supplement).44
Estimated odds ratios for resistant disease in each of clusters B through F, relative to cluster A. For each cluster 3 estimates are shown: based on all 170 patients without adjustment for other factors (bottom); based on 113 patients with known AML onset (de novo versus secondary) and cytogenetic risk category without adjustment for other factors (middle); and based on the same 113 patients but with adjustment for age, AML onset, and cytogenetic risk category (unfavorable versus favorable/intermediate) (top). Bars indicate 95% confidence intervals. Results of the 3 analyses are generally consistent and, in particular, the heterogeneity of RD rates among the 6 clusters remained statistically significant after adjusting for possible confounding (P < .001).
Estimated odds ratios for resistant disease in each of clusters B through F, relative to cluster A. For each cluster 3 estimates are shown: based on all 170 patients without adjustment for other factors (bottom); based on 113 patients with known AML onset (de novo versus secondary) and cytogenetic risk category without adjustment for other factors (middle); and based on the same 113 patients but with adjustment for age, AML onset, and cytogenetic risk category (unfavorable versus favorable/intermediate) (top). Bars indicate 95% confidence intervals. Results of the 3 analyses are generally consistent and, in particular, the heterogeneity of RD rates among the 6 clusters remained statistically significant after adjusting for possible confounding (P < .001).
Gene expression profiles associated with NPM1 mutations are dominated by a stem-cell molecular signature.39 Activation of HOX genes and TALE partner genes (ie, MEIS) is found in NPM1 gene signatures.39 The reportedly favorable impact of NPM1 mutations on survival has included higher CR rates23,26 and a trend to longer OS and EFS.26 However, other studies have observed either no significant effect22,25 or an impact only when NPM1-mutated cases are also FLT3-ITD negative.24,26,27 While AML with NPM1 mutations is associated with increased FLT3 mutations,22-24 this relationship was not observed for cluster A members. Cluster A had a disproportionate number of FLT3 mutations involving TKDs rather than ITDs, but the overall FLT3 mutation incidence was similar to the other VxInsight groups. FLT3-TKDs have been linked to increased release of IL-12 by leukemic blasts; IL-12A was overexpressed by members of cluster A.64 IL-12 has antiangiogenic and antitumor effects and, unless offset by an increased level of proangiogenic regulators, may have a role in improving outcomes.64,65
Differential expression of select genes among VxInsight clusters. The columns represent the 170 AML samples ordered by VxInsight membership. Rows represent select genes that are differentially expressed among the VxInsight clusters. Red indicates high expression relative to the mean; green, low expression relative to the mean.
Differential expression of select genes among VxInsight clusters. The columns represent the 170 AML samples ordered by VxInsight membership. Rows represent select genes that are differentially expressed among the VxInsight clusters. Red indicates high expression relative to the mean; green, low expression relative to the mean.
Cluster A members had overexpression of Wilms tumor (WT1) gene; this gene is overexpressed at variable levels in 75% to 100% of AMLs at diagnosis.18,66 A lower level of expression of WT1 has been seen among the more differentiated AMLs in most but not all series.18,67 Because of the increased WT1 expression, patients in cluster A may be more likely to benefit from WT1-specific immunotherapy than other AML patients, either in the form of a T-cell approach or a vaccine.14,67 One potential problem is that a number of the MHC class II genes were down-regulated in cluster A. Because tumor cells are poorly immunogenic when deficient in MHC class II molecule expression, the leukemic cells may escape host immunity. Cluster A also had overexpression of genes that promote apoptosis (LTBP1, CASP3), with LTBP1 being a particularly important gene for predicting cluster A membership (ranked 1) and for predicting NPM1 mutations.24,39
Patients in clusters B and C had the worst DFS, with estimated probabilities of 0% and 6%, respectively, at 5 years. They also had the poorest OS, although OS did not vary significantly among clusters. Cluster B, in particular, is an interesting group of 22 patients: 77% were unresponsive to induction chemotherapy, and its 3 remitting patients all relapsed within 16 months. This group of patients might be considered prone to disease resistance, because the patients had the highest median age (68 years) and highest incidence of secondary disease (32%), yet these factors did not vary significantly in the 6 clusters. Despite 42% of cases having a normal karyotype, only one individual (5%) had an NPM1 mutation. One gene overexpressed by these patients was ABCG2. ABCG2, also termed breast cancer resistance protein (BCRP) and mitoxantrone resistance protein (MRX), is a member of the ATP-binding cassette (ABC) superfamily of membrane transporters that function as drug efflux pumps to remove chemotherapeutic agents from cells.68 ABCG2 is expressed by approximately one third of adult AMLs when measured using semiquantitative reverse transcriptase (RT)–PCR or flow cytometric analysis.69-72 Of relevance to our study is the report by Steinbach and colleagues, who found significantly higher median ABCG2 gene expression levels in 24 pediatric AML patients who failed to achieve remission after initial induction chemotherapy compared with the 21 patients who achieved remission.73 Similar and discrepant results have been reported by others using varying and sometimes discordant analytic methods and study designs.72,74,75
The significant ABCG2 overexpression among members of our high induction failure cluster, B, supports a role for ABCG2 in chemoresistance, possibly in combination with MDR1.76,77 Permeability glycoprotein (MDR1, P-gp, or ABCB1) was concurrently overexpressed among patients in cluster B, but MDR1 alone did not significantly differentiate this group from the other AML patients. Drug-sensitive cells transfected with ABCG2 become resistant to mitoxantrone, doxorubicin, daunorubicin, and topotecan,70 while ABCG2-expressing cells from AML patients are resistant to daunorubicin in vitro.78 Therefore, treatment methods circumventing ABCG2-mediated multidrug resistance should be considered for evaluation in future patients with gene profiles similar to cluster B members. These include the use of ABCG2 inhibitors and antineoplastic agents that show poor ABCG2-mediated efflux (ie, idarubicin or newer agents in development).79
The other poor-outcome cluster, C, had the highest rate of CR to induction therapy (58%), with only 16% of the 31 patients showing initially resistant disease. However these CRs were comparatively short lived: In the analysis of DFS, cluster C had the largest hazard ratio of the 6 clusters. Many high-ranking genes defining this cluster were down-regulated (54% of the top 50 genes). Among the significantly overexpressed genes were genes involved in immunoregulation, including interferon regulatory factor-4 (IRF4), a gene regulated by NF-κB member c-rel80 ; IL-10RA, a member of the interferon receptor family; and MALT1, a factor required for NF-κB activation. Immune-mediated antitumor effects may have played a role in the initial therapeutic responses in this ultimately poor-outcome group.
Cluster D had the largest membership (n = 42), the highest prevalence of karyotypic abnormalities (76% of members), and a low prevalence of NPM1 mutations (5% of members). This cytogenetically diverse group contained most of the patients in the favorable cytogenetic risk group (10 of 12; 83%; including all t(8;21)) as well as the largest percentage of patients with unfavorable karyotypes (15 of 44, 34%; 44% of members). A study of 116 adult AML patients initially using an unsupervised clustering approach also found karyotypic diversity within cluster groups.35 Analogous to their findings, primary translocating events may be less important in the transformation to leukemia than the overall dysregulation of signaling pathways or other genetic events better reflected in gene expression profiles. In cluster D, the “high proliferative activity” gene signature dominated and may have obscured gene signatures more specific to the divergent karyotypes. Most of the top-ranked genes in cluster D were associated with DNA proliferation and repair. It is unclear whether the “high proliferative” signature led to an increase in detectable cytogenetic abnormalities, because in vitro proliferation is required to detect such karyotypic abnormalities, or whether the cytogenetic abnormalities led to the increase in proliferative genes. Notable in this cluster was the low expression of class I homeobox A genes (HOXA9, HOXA10). HOXA gene expression has been shown to be lower among AMLs with favorable karyotypes compared with unfavorable33,81,82 and higher among patients with NPM1 mutations,39 normal karyotypes, or subsets of patients with intermediate-risk karyotypes.33,83 Down-regulation of HOXA9 and HOXA10 in cluster D may reflect the lower proportion of patients with NPM1 mutations or decreased normal karyotypes (24%) relative to the other clusters (42%-75%).
Cluster E represented a small group of patients (n = 18), many of whom were registered to SWOG protocol S9500 for the treatment of younger adults (younger than 56 years). Cluster F (n = 33) was defined by AML with monocytic differentiation (97% of members), the highest pretreatment WBC counts, and a high percentage of NPM1 mutations (61%). NPM1 mutations have been shown to be increased among monocytic leukemias, and the gene expression profiles contained many genes pertinent to monocyte function.25-27 When our gene lists were compared with those in the study by Valk et al,34 23% of the top 40 genes defining cluster F were similar to those seen in the cluster of Valk et al34 containing monocytic leukemias (see Table S16).44 Similarly, when significant cluster-defining genes in our study were analyzed using datasets of Valk et al34 and Bullinger et al,35 the strongest gene expression relationships were found among the monocytic groups in all the studies (see Figures S2-S8).44 This confirms the importance of monocyte morphology on AML gene expression signatures and raises the question of whether the strong “monocyte signature” masks other genes of potential biologic significance in these groups.35 We are currently evaluating whether the outcome of monocytic leukemias with NPM1 mutations and a “monocytic gene signature” differs significantly from AMLs with a “stem-cell molecular signature” seen in cluster A and reported by Alcalay et al.39
This gene expression profiling study highlights the divergent mechanisms and pathways of leukemic transformation that are not appreciated by current methods of AML diagnosis, classification, and risk assignment. No bias was induced during cluster selection in this analysis, and therefore these subsets represent true reflections of the intrinsic biology in this cohort of patients. For example, the significance of NPM1 mutations in AML was unknown at the initiation of this study, yet the gene expression profiles clustered groups of patients together with this unique genetic abnormality. Additional studies will be important to determine whether the improved survivals in cluster A with increased NPM1 mutations relate to the gene expression signatures displayed by these cases, regardless of the FLT3 mutational status. We are now evaluating the relative significance of these genes in predictive models of outcome using supervised learning methods in this same cohort. It is hoped that the gene signatures identified in this study will provide clues to new therapeutic interventions for older AML patients who have historically done poorly with current treatment regimens. Confirmatory studies and prospective validation of our results are required to continue to understand the significance of our clusters of patients, such as cluster B with increased RD. These analyses are important to enhance risk classification and the identification of individual genes and pathways that can be exploited for improved therapeutic interventions.
Prepublished online as Blood First Edition Paper, April 4, 2006; DOI 10.1182/blood-2004-12-4633.
Supported in part by Department of Health and Human Services, National Institutes of Health grants NCI CA88361 and NCI CA32102, the W. M. Keck Foundation, the Dedicated Health Research Fund of the State of New Mexico, and the University of New Mexico Cancer Center Genomics, Biostatistics, and Biocomputing Shared Facilities. Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the United States Department of Energy under contract DE-AC04-94AL85000.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Julia H. Engel for excellent technical assistance.
Supplemental data
Genes predictive of the designated VxInsight clusters are listed in the columns. The rows represent the gene expression for each AML patient in the dataset of Bullinger et al8 relative to the other patients. The patients in this study formed 2 main clusters along with discrete subclusters. The data are limited by the presence of only 85 matching genes in the cDNA platform for analysis. In addition, some patients had unusable signal data for particular genes, which are colored gray. Despite these limitations, overlap of gene expression signatures is noted among the monocytic leukemias in the dataset of Bullinger et al8 and cluster F.REFERENCES1. Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733-3739.2. Stirewalt DL, Mashinchi S, Kussick SJ, et al. Novel FLT3 point mutations within exon 14 found in patients with acute myeloid leukaemia. Br J Haematol. 2004;124:481-484.3. Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001;97:2434-2439.4. Gene Expression in Human Leukemia Experimental & Computational Methodology website. Available at: http://hsc.unm.edu/crtc/willmanresearch/Pages/UNMHSC_HPC_SNL_Methodology.htm. Accessed May 6, 2006.5. Valk PJM, Verhaak RGW, Beijen AM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617-1628.6. Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747-3754.7. Affymetrix. Available at: http://www.affymetrix.com. Accessed May 6, 2006.8. Bullinger L, Dohner K, Bair E, et al. Use of gene expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605-1616.9. Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763-774.10. Alcalay M, Tiacci E, Bergomas R, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPM1c+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899-905.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal