The 2 most frequent human MLL hematopoietic malignancies involve either AF4 or AF9 as fusion partners; each has distinct biology but the role of the fusion partner is not clear. We produced Mll-AF4 knock-in (KI) mice by homologous recombination in embryonic stem cells and compared them with Mll-AF9 KI mice. Young Mll-AF4 mice had lymphoid and myeloid deregulation manifest by increased lymphoid and myeloid cells in hematopoietic organs. In vitro, bone marrow cells from young mice formed unique mixed pro-B lymphoid (B220+CD19+CD43+sIgM–, PAX5+, TdT+, IgH rearranged)/myeloid (CD11b/Mac1+, c-fms+, lysozyme+) colonies when grown in IL-7– and Flt3 ligand-containing media. Mixed lymphoid/myeloid hyperplasia and hematologic malignancies (most frequently B-cell lymphomas) developed in Mll-AF4 mice after prolonged latency; long latency to malignancy indicates that Mll-AF4–induced lymphoid/myeloid deregulation alone is insufficient to produce malignancy. In contrast, young Mll-AF9 mice had predominately myeloid deregulation in vivo and in vitro and developed myeloid malignancies. The early onset of distinct mixed lymphoid/myeloid lineage deregulation in Mll-AF4 mice shows evidence for both “instructive” and “noninstructive” roles for AF4 and AF9 as partners in MLL fusion genes. The molecular basis for “instruction” and secondary cooperating mutations can now be studied in our Mll-AF4 model.
Introduction
The myeloid/mixed lymphoid leukemia gene (MLL) on human chromosome 11 was first described from a cell line derived from a patient with a hematologic malignancy that resulted from a reciprocal translocation involving chromosome 4.1 MLL was subsequently shown to partner with many other genes to result in hematologic malignancy.2 The fusion of MLL to AF4 family members, LAF4 and AF5, results in malignancies that are the most common and unique among the MLL fusion gene malignancies in that they are generally lymphoid or lymphoid/myeloid in type but rarely purely myeloid. They are also unique because of the high frequency in infants, extensive spread beyond the hematopoietic compartment, and a poor outcome with treatment.3-10 In contrast to MLL-AF4, the fusion of MLL with most other partners, including the second most common partner AF9,8 results in myeloid malignancies.
To date, no murine model of MLL-AF4 translocation has been reported and thus neither the premalignant early events nor the eventual malignancies have been defined. In this study, we produced Mll-AF4 knock-in (KI) mice and compare them with Mll-AF9 mice developed previously.11 The KI models, which have the advantage of having a single copy of the fusion gene in all stem/progenitor cells, permit control of bias introduced by a variable number of gene copies in the various progenitor populations in other models.
The mechanisms for the association between the MLL partner gene and type of malignancy have not been elucidated. The MLL partner may be “instructive” in directing the selective expansion and transformation of cells toward fusion gene-specific lineages.12 Some evidence derives from studies in which MLL-AF4 and MLL-AF9 had differing effects when introduced into Drosophila13 or a human cell line.14 Instructed multipotent stem/progenitor cells may have the capacity for either lymphoid or myeloid expansion depending on the MLL fusion gene partner. However, it is also likely that some of the effects of the MLL fusion genes are partner independent.
Our new Mll-AF4 model produced deregulation of mixed-lineage progenitor cells (in vitro) and lymphoid and myeloid proliferation (in vivo) in young mice. In contrast, myeloid deregulation predominated in Mll-AF9 mice. After a long latency period, Mll-AF4 mice developed mixed lymphoid/myeloid hyperplasia and hematologic malignancies (most frequently B-cell lymphomas), whereas myeloid malignancies predominated in Mll-AF9 mice.
Materials and methods
Construction of Mll-AF4–targeting vector and generation of targeted ES cells
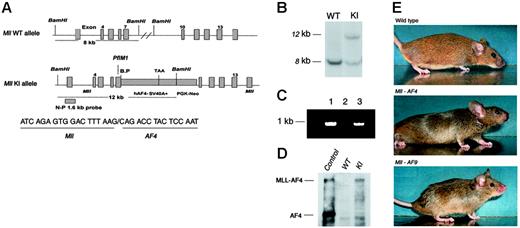
Mouse gDNA containing the Mll locus was isolated from a P1 library by polymerase chain reaction (PCR) screening (Genome Systems, St Louis, MO). Human MLL-AF4 cDNA (GenBank accession no. L22179) was cloned using standard methods.15,16 Because the exon 7 of mouse Mll gene shares 100% homology with the exon 7 of human MLL between the PflM1 site and the breakpoint in RS4;11 cells, the PflM1 site in Mll exon 7 was used to generate the in-frame fusion of mouse Mll with human AF4 cDNA. The KI construct contains a 5′ and a 3′ arm from Mll, the fragment of human AF4 cDNA from the breakpoint to the 3′ end, a 953-bp fragment of SV40 with stop signals, and a PGK-Neo cassette (Figure 1A). The targeting vector was linearized by XhoI, electroporated into passage 10 of CJ7 embryonic stem (ES) cells (derived from 129/sv mouse strain) obtained from the Mouse Genetics Laboratory, University of Minnesota, Cancer Center, and selected by G418. gDNA of ES cells was purified and digested with BamHI. The N-P1.6 probe used for Southern blotting detects only an 8-kb fragment in wild-type ES cells but both the 12-kb and 8-kb fragment in ES cells carrying the Mll-AF4 fusion gene.
Generation of heterozygous Mll-AF4+/– mice
Two correctly targeted ES-cell clones were used for injection into C57Bl/6 blastocysts to produce chimeric mice. Before injections, a complete cytogenetic analysis showed a normal karyotype of the ES cells. Male chimeric mice were bred to C57Bl/6 females. Heterozygous offspring were selected by PCR (Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online article). These germline offspring were bred on a C57Bl/6 background for 2 generations and then bred to FVB mice to improve fertility and viability of offspring.
The Mll-AF9 mice were originally produced in the laboratory of Dr Terence Rabbitts (Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom)11 using the same homologous recombination approach. CCB ES cells (derived from mouse strain of 129/sv) were targeted to produce Mll-AF9 mice that comprise a KI of the human AF9 short form (breakpoint to 3′ end) into exon 8 of the mouse Mll gene. In summary, both CJ7 ES cells used for Mll-AF4 and CCB ES cells used for Mll-AF9 were derived from 129/sv strain. As with the Mll-AF4 mice, the Mll-AF9 mice originally were on a C57Bl/6 background. To permit Mll-AF4 versus Mll-AF9 experiments controlled for strain differences, we recently crossed Mll-AF9 C57Bl/6 with FVB mice. There were no significant differences in hematologic analysis (including white blood cell [WBC] count, absolute numbers of immature cells, monocytes, neutrophils, and lymphocytes in blood), colony numbers, and percentage of different type of colonies in colony-forming assays, or malignancies; thus the results reported were combined from C57Bl/6 and C57Bl/FVB hybrid mice.
Construction of the Mll-AF4 KI allele and characterization of targeted ES cells. (A) A diagrammatic description of the KI allele. The top map represents the Mll wild-type allele, indicating the 8-kb fragment detected by N-P1.6 probe. The bottom map represents the Mll-exon7-AF4–targeting allele, indicating the position of the Mll-AF4 breakpoint, PGK-Neo cassette, and a 12-kb fragment detected by N-P1.6 probe in Southern blotting. The Mll-AF4 breakpoint and junction sequence are also shown. (B) Southern blotting analysis of wild-type and targeted ES cells. The 8-kb wild-type (WT) band and 12-kb KI band are indicated. (C) Detection of Mll-AF4 fusion gene by PCR. PCR was performed with gDNA from ES cells with KI allele (lane 1), wild-type ES cells (lane 2), and a representative Mll-AF4 mouse (lane 3). The 5′ primer of Mll exon 6 and 3′ primer of human AF4 amplified a 930-bp Mll-AF4 fragment in lanes 1 and 3. (D) Expression of Mll-AF4 fusion protein in targeted ES cells detected by Western blotting. The 240-kDa Mll-AF4 fusion protein was detected in targeted ES cells. A cell line transfected with MLL-AF4 DNA was used as positive control. (E) Mll-AF4, Mll-AF9, and wild-type mouse at the age of 5 weeks. Mll-AF4 mice have a shortened face and large ears. Mll-AF9 mice have a more pointed face and large ears.
Construction of the Mll-AF4 KI allele and characterization of targeted ES cells. (A) A diagrammatic description of the KI allele. The top map represents the Mll wild-type allele, indicating the 8-kb fragment detected by N-P1.6 probe. The bottom map represents the Mll-exon7-AF4–targeting allele, indicating the position of the Mll-AF4 breakpoint, PGK-Neo cassette, and a 12-kb fragment detected by N-P1.6 probe in Southern blotting. The Mll-AF4 breakpoint and junction sequence are also shown. (B) Southern blotting analysis of wild-type and targeted ES cells. The 8-kb wild-type (WT) band and 12-kb KI band are indicated. (C) Detection of Mll-AF4 fusion gene by PCR. PCR was performed with gDNA from ES cells with KI allele (lane 1), wild-type ES cells (lane 2), and a representative Mll-AF4 mouse (lane 3). The 5′ primer of Mll exon 6 and 3′ primer of human AF4 amplified a 930-bp Mll-AF4 fragment in lanes 1 and 3. (D) Expression of Mll-AF4 fusion protein in targeted ES cells detected by Western blotting. The 240-kDa Mll-AF4 fusion protein was detected in targeted ES cells. A cell line transfected with MLL-AF4 DNA was used as positive control. (E) Mll-AF4, Mll-AF9, and wild-type mouse at the age of 5 weeks. Mll-AF4 mice have a shortened face and large ears. Mll-AF9 mice have a more pointed face and large ears.
Hematologic analysis
Young Mll-AF4 and Mll-AF9 mice (5-8 weeks old) and their wild-type littermates were humanely killed. The total WBC counts in peripheral blood were determined by Unopette Micro Collection System (BD Biosciences, San Diego, CA). Blood and bone marrow smears were stained with Wright-Giemsa stain. Differential counts were determined by counting 200 nucleated cells under a microscope. Statistical significance between means was determined by the Student t test.
Bone marrow cell cultures
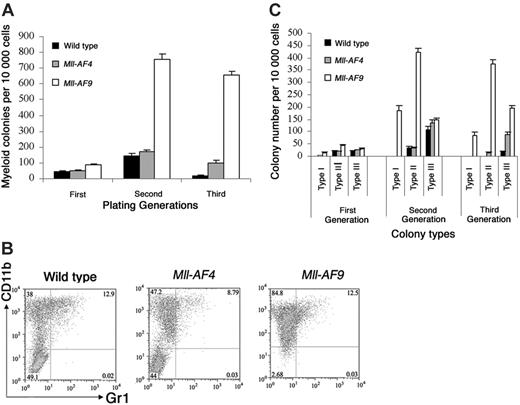
Single bone marrow cell suspension was cultured in methylcellulose medium under either lymphoid (pro-B) conditions using Methocult 3630 (StemCell Technologies, Vancouver, BC, Canada) supplemented with 50 ng/mL flt3 ligand (R&D Systems, Minneapolis, MN) for the growth of pro-B colonies, or myeloid conditions using Methocult 3534 (StemCell Technologies) supplemented with 10 ng/mL GM-CSF (R&D Systems) for the growth of myeloid colonies. The cells were cultured in triplicate for 3 consecutive generations of 7 to 8 days each. The myeloid colonies were counted and classified into 3 types (I, II, and III), as previously described.17 Briefly, type I colonies are compact colonies with a dense center and a smooth edge, type II colonies have a compact center and a halo of loose cells, and type III colonies have dispersed cells but no center. Type I colonies are composed mostly of immature myeloid precursors, whereas the cells in type II and type III colonies are progressively more differentiated. The colony containing more than 50 cells was scored. The total number of pro-B colonies was scored and counted as previously reported.18
Histopathology and multiparameter FACS analysis
For histopathologic analysis, mice that were terminally ill were humanely killed. Tissues were fixed with 10% formalin. Hematoxylin and eosin (H&E) staining and immunochemistry were performed according to standard protocols. Primary antibodies used here were antimyeloperoxidase (DakoCytomation, Carpinteria, CA), anti-B220/CD45R, anti-CD19 (PharMingen, San Diego, CA), anti-IgM (BioGenex, San Ramon, CA), and anti-PAX5 and anti–Bcl-6 (Santa Cruz Biotechnology, Santa Cruz, CA). Images were processed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA). After staining with primary antibodies, sections were incubated with biotinylated secondary antibodies and the streptavidin HRP enzyme conjugate (DakoCytomation) or the alkaline phosphatase enzyme system (Biogenex), followed by 3,3′-diaminobenzidine tetrahydrochloride (Dakocytomation), and were counterstained with Mayer hematoxylin. Photomicrographs were taken using a SPOT Insight digital camera along with SPOT Advanced software version 4.0.9 (Diagnostic Instruments, Sterling Heights, MI) under a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan). Nikon Plan 2 ×/0.08 numeric aperture (NA) and Plan 40 ×/0.75 NA objectives were used for Figures 7A and 7B-H, respectively.
For flow cytometric analysis, cells were stained with FITC- and PE-conjugated antibodies to CD11b/Mac1, Gr-1, CD43, B220/CD45R, CD19, IgM, and CD3 (PharMingen). Stained samples were acquired by fluorescence-activated cell sorting with a BD FACSCalibur using Cell Quest Pro software. Multiparameter analysis of the data was done with FloJo software (Tree Star, San Carlos, CA).
Gene expression by RT-PCR
RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA). cDNA was synthesized with SuperSpript first-strand synthesis system for reverse transcription-PCR kit (RT-PCR; Invitrogen) according to the manufacturer's instruction. PCR primers are described in Table S1.
Mll-AF4 and Mll-AF9 expression by real-time quantitative RT-PCR
Quantitative RT-PCR was performed to detect the expression of Mll-AF4 and Mll-AF9 gene in young and old diseased mice. Total RNA was isolated from bone marrow cells using the RNeasy kit (Qiagen, Valencia, CA), incorporating on-column DNase digestion. For real-time quantitative RT-PCR, TaqMan primer and probe sets were obtained from Applied Biosystems (ABI, Foster City, CA). The probes spanned the Mll-AF4 and Mll-AF9 junctions. Quantitative PCR was performed using an ABI prism 7500 sequence detection system. Quantitative analysis was performed using REST (Relative Expression Software Tool).19
Immunoglobulin heavy-chain rearrangements by PCR or Southern blotting
DJH rearrangements of the immunoglobulin heavy chain in the cultured cells from colonies under pro-B condition were determined by PCR using 2 upstream degenerate primers (DFL/DSP and DQ52) and one reverse primer. All 3 primers were used in a single PCR to detect rearrangements from DJH1 to DJH4.20
Immunoglobulin heavy-chain rearrangements in the organs from sick Mll-AF4 mice were determined by Southern blotting. gDNA was digested with EcoRI. Plasmid pBR322 containing mouse IgH J1-J4 sequences was used as the probe.21 gDNA was also digested with HindIII followed by Southern blotting to confirm IgH rearrangements.
All the experiments using mice were conducted after approval by the Institutional Animal Care and Research Committee, University of Minnesota.
Results
Generation of Mll-AF4 mice
We constructed a KI vector to fuse the 5′ portion of the murine Mll with the 3′ portion of the human AF4. Blastocyst injection with targeted ES cells resulted in 14 Mll-AF4 chimeric mice with more than 70% chimerism as judged by fur color. Two founders (A and D1) showed germline transmission. However, offspring of the founder D1 were not fertile; therefore, all Mll-AF4 germline mice used in this study were derived from founder A. The organization of the wild-type Mll allele and the Mll-AF4 KI allele were shown (Figure 1A). The breakpoint and junction sequence was confirmed by DNA sequencing (Figure 1A). Southern analysis showed a specific 12-kb band derived from the Mll-AF4 gene in KI ES clones, different from the 8-kb band from wild-type Mll (Figure 1B). The Mll-AF4 fusion gene was also detected by PCR (Figure 1C). The 240-kDa Mll-AF4 fusion protein was detected in the KI ES cells, as shown by Western blotting (Figure 1D), indicating expression of full-length Mll-AF4 protein.22
Expression of the Mll-AF4 or Mll-AF9 fusion gene in bone marrow from randomly selected young and old diseased mice was tested using real time quantitative RT-PCR. As expected, all samples tested showed expression of the appropriate fusion gene (Table S2). In both Mll-AF4 and Mll-AF9 mice that were older and diseased with lymphoma and myeloid hyperplasia, relative expression levels were lower than in young mice (P < .01). The difference was larger among Mll-AF4 and smaller among Mll-AF9 mice. Overall, there are no significance differences between Mll-AF4 and Mll-AF9 (P > .05). However, differences between Mll-AF4 and Mll-AF9 may be masked by the “interaction” between age/disease and type of gene as described in Table S2. Additional studies using purified cell populations and larger sample sizes will be necessary to further evaluate potential differences in detail.
Young Mll-AF4 and Mll-AF9 mice have distinctive phenotypic features and different lymphoid and myeloid expansion in vivo
Mll-AF4 and Mll-AF9 mice were small at birth compared to wild-type littermates. At 5 weeks of age, Mll-AF4 mice weighed a mean of 13.0 g, Mll-AF9 mice 13.6 g, and wild-type littermates 17.4 g. Mll-AF4 mice had a distinctive appearance, including a shortened blunt face, large ears, and a high-set tail (Figure 1E). Mll-AF9 mice had faces that were also shortened but pointed rather than square. The ears were also enlarged but less pronounced than those of the Mll-AF4 animals.
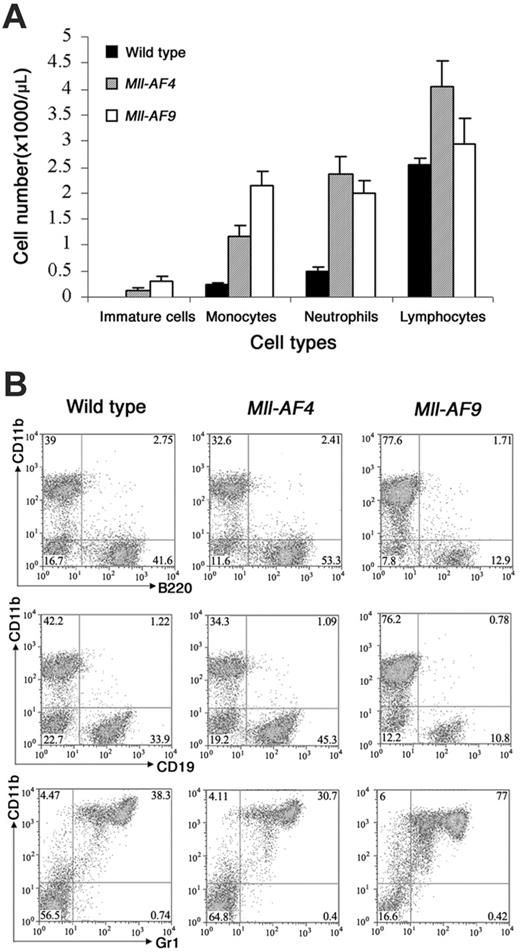
We evaluated the effects of the 2 fusion genes on hematopoietic differentiation in young (5–8-week-old) mice. Elevated WBC counts were found in both Mll-AF4 mice (7.2 × 103 ± 1.2 × 103/μL; n = 10) and Mll-AF9 (7.3 × 103 ± 1.3 × 103/μL; n = 8) as compared to wild-type controls (3.9 × 103 ± 1.0 × 103/μL; n = 9). Mll-AF4 and Mll-AF9 mice, but not wild-type mice, showed immature cells in blood (P < .001; Figure 2A). Notably, Mll-AF4 mice showed the highest total number of lymphocytes (Mll-AF4 versus wild type, P < .01). A statistically insignificant increase also observed in Mll-AF9. The highest number of monocytes was found in Mll-AF9 mice, although Mll-AF4 mice also showed an increase compared to wild-type mice (P < .01 for all the compared groups). Neutrophils were increased in both Mll-AF9 and Mll-AF4 (P < .001) but differences between Mll-AF4 and Mll-AF9 were not significant.
To further define the role of the Mll fusion partners, AF4 and AF9, in hematopoietic differentiation, multiparameter FACS analysis was used to characterize the leukocytes in bone marrow (Figure 2B). The highest percentage of B220+ or CD19+ B-lymphoid lineage cells was found in Mll-AF4 marrow (P < .01, Mll-AF4 versus wild type), with intermediate level in wild-type and lowest in Mll-AF9 marrow (P < .001, Mll-AF9 versus wild type). In contrast, CD11b+Gr1+ cells predominated in Mll-AF9 marrow (P < .001, Mll-AF9 versus wild type).
Cell types in blood and bone marrow of young wild-type, Mll-AF4, and Mll-AF9 mice. (A) The total numbers of immature cells, monocytes, neutrophils, and lymphocytes in the blood of young wild-type, Mll-AF4, and Mll-AF9 mice. The numbers were calculated by multiplying total WBC counts with percentage of each cell type and expressed as cell numbers per microliter peripheral blood. The error bars represent SEMs. (B) Immunophenotype of nucleated cells in the bone marrow of young Mll-AF4, Mll-AF9, and wild-type mice. Phenotypic analysis of bone marrow cells from representative 5- to 8-week-old Mll-AF4, Mll-AF9, and wild-type mice was performed by FACS.
Cell types in blood and bone marrow of young wild-type, Mll-AF4, and Mll-AF9 mice. (A) The total numbers of immature cells, monocytes, neutrophils, and lymphocytes in the blood of young wild-type, Mll-AF4, and Mll-AF9 mice. The numbers were calculated by multiplying total WBC counts with percentage of each cell type and expressed as cell numbers per microliter peripheral blood. The error bars represent SEMs. (B) Immunophenotype of nucleated cells in the bone marrow of young Mll-AF4, Mll-AF9, and wild-type mice. Phenotypic analysis of bone marrow cells from representative 5- to 8-week-old Mll-AF4, Mll-AF9, and wild-type mice was performed by FACS.
Mll-AF4 bone marrow from young mice shows lymphoid progenitor deregulation in vitro
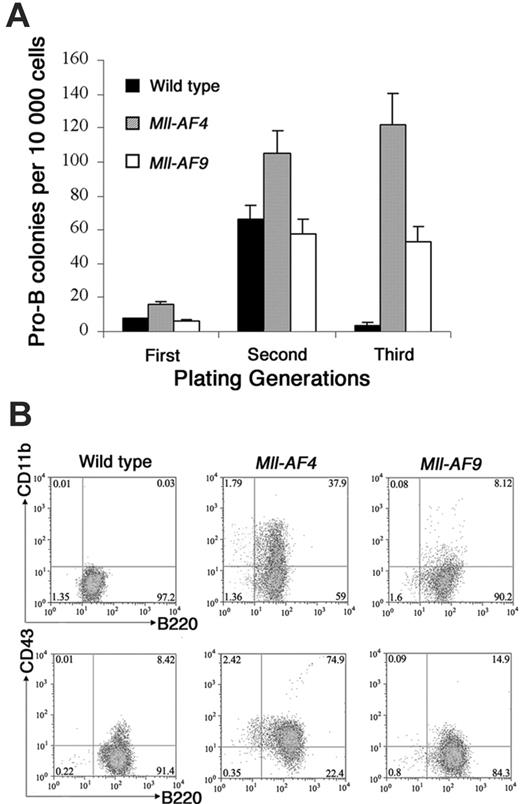
Colony assays, which allowed direct observation of cells that have undergone deregulation based on plating abilities over several generations, were performed to evaluate potential differences in young Mll-AF4 and Mll-AF9 compared to wild-type mice. Pro-B colonies grown in methylcellulose containing Flt3 ligand and IL-723,24 were studied. Results are shown in Figure 3A. Pro-B colony-forming units (CFUs) from Mll-AF4 mice were significantly higher than those from wild-type and Mll-AF9 mice beginning in the first generation of plating (P < .01). As expected, pro-B colonies were almost undetectable in wild-type mice by the third generation. However, significant numbers of Mll-AF4 and fewer Mll-AF9 pro-B colonies were found in the third generation of plating. As shown, the number of pro-B CFUs of the Mll-AF4 mice was always significantly higher than the one of Mll-AF9 mice (Figure 3A).
Mixed pro-B lymphoid (B220+ CD19+CD43+sIgM–, PAX5 +, TdT+, IgH rearranged)/myeloid (CD11b/Mac1+, c-fms+, lysozyme+) progenitor cells are produced from marrow culture of young Mll-AF4 mice
Multiparameter FACS analysis was carried out on pooled cells from the methylcellulose culture of the third generation grown under pro-B lymphoid conditions. Cells from Mll-AF4 colonies were B220+CD43+ and coexpressed lymphoid and myeloid-associated markers, B220+CD11b+ (a representative experiment is shown in Figure 3B). The percentage of mixed-lineage B220+ CD43+ CD11b+ cells in the Mll-AF4 colonies was reproducibly higher than wild-type and Mll-AF9 mice.
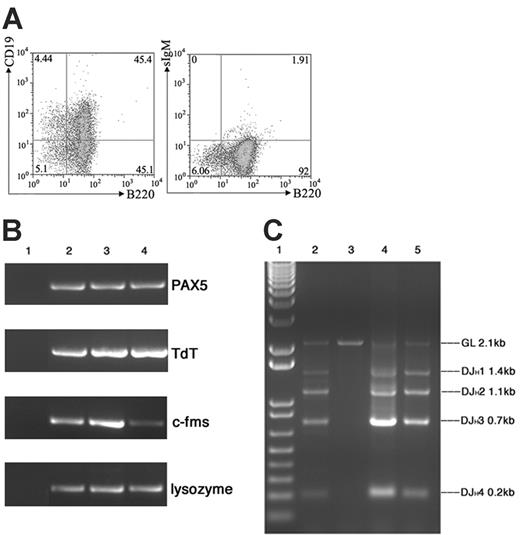
To further characterize the B220+CD43+CD11b+ population from the bone marrow culture of Mll-AF4 mice, the B-cell–associated markers CD19 and sIgM were also examined by FACS. The result indicated that these cultured bone marrow cells were sIgM negative and about 50% of them were B220+CD19+ (Figure 4A). RT-PCR was conducted to detect the expression of B-cell markers (PAX5, TdT) and myeloid markers (c-fms, lysozyme). As shown in Figure 4B, these cultured cells were positive for PAX5, TdT, c-fms, and lysozyme. DJH1-DJH4 rearrangements of immunoglobulin heavy chain were also found in this population (Figure 4C). All of these data indicated that this expanded population from Mll-AF4 bone marrow grown under pro-B condition was positive for both pro–B cell markers (B220+ CD19+CD43+sIgM–, PAX5 +, TdT+, IgH rearranged) and myeloid cell markers (CD11b/Mac1+, c-fms+, lysozyme+).
Cells from the third-generation culture were subsequently put in IMDM supplemented with fetal bovine serum, Flt3 ligand, and IL-7 to evaluate the growth potential of the mixed-lineage Mll-AF4 cells. None of the cells from beyond the third-generation Mll-AF4 lymphoid cultures could grow for more than 2 months, suggesting that these cells had not attained a fully malignant phenotype. Similarly, no cells from Mll-AF9 or wild-type mice survived under these lymphoid growth conditions.
Mll-AF4 marrow progenitor cells expressed myeloid and lymphoid markers (B220, CD43, and CD11b) when grown under pro-B lymphoid conditions. (A) Colonies from young (5- to 8-week-old) mice. Bone marrow cells from wild-type (n = 8), Mll-AF4 (n = 7), and Mll-AF9 (n = 8) mice were plated in methylcellulose medium under lymphoid conditions for 3 generations. The error bars represent SEMs. (B) Representative results of multiparameter FACS analysis on pooled cells from the methylcellulose culture of the third generation under lymphoid conditions.
Mll-AF4 marrow progenitor cells expressed myeloid and lymphoid markers (B220, CD43, and CD11b) when grown under pro-B lymphoid conditions. (A) Colonies from young (5- to 8-week-old) mice. Bone marrow cells from wild-type (n = 8), Mll-AF4 (n = 7), and Mll-AF9 (n = 8) mice were plated in methylcellulose medium under lymphoid conditions for 3 generations. The error bars represent SEMs. (B) Representative results of multiparameter FACS analysis on pooled cells from the methylcellulose culture of the third generation under lymphoid conditions.
Marrow cells from Mll-AF4 mice expressed additional lymphoid (CD19, PAX5, TdT, DH-JH but not sIgM) and myeloid markers (c-fms and lysozyme) when cultured under pro-B culture conditions. (A) FACS analysis of B220/CD19 and B220/sIgM. (B) PAX5, TdT, c-fms, and lysozyme expression detected by RT-PCR. Lane 1, H2O control; lane 2, mouse spleen cells as control; lanes 3-4, bone marrow cells from 2 Mll-AF4 mice cultured for 21 days. (C) Immunoglobulin DH-JH heavy-chain rearrangement detected by PCR. Lane 1, molecular weight marker; lane 2, mouse spleen DNA as control; lane 3, mouse kidney DNA as control; and lanes 4-5, DNA of bone marrow cells from 2 Mll-AF4 mice cultured for 21 days.
Marrow cells from Mll-AF4 mice expressed additional lymphoid (CD19, PAX5, TdT, DH-JH but not sIgM) and myeloid markers (c-fms and lysozyme) when cultured under pro-B culture conditions. (A) FACS analysis of B220/CD19 and B220/sIgM. (B) PAX5, TdT, c-fms, and lysozyme expression detected by RT-PCR. Lane 1, H2O control; lane 2, mouse spleen cells as control; lanes 3-4, bone marrow cells from 2 Mll-AF4 mice cultured for 21 days. (C) Immunoglobulin DH-JH heavy-chain rearrangement detected by PCR. Lane 1, molecular weight marker; lane 2, mouse spleen DNA as control; lane 3, mouse kidney DNA as control; and lanes 4-5, DNA of bone marrow cells from 2 Mll-AF4 mice cultured for 21 days.
In contrast to Mll-AF4, Mll-AF9 bone marrow shows predominately myeloid progenitor cell expansion in colony assays
We also studied myeloid colony formation of young mouse marrow in methylcellulose supplemented with GM-CSF, IL-3, SCF, and IL-6 (Figure 5A). Myeloid CFUs from Mll-AF9 marrow were significantly higher in number than from wild-type and Mll-AF4 mice beginning in the first generation (P < .01). We observed no increase in Mll-AF4 myeloid colonies in the first generation compared to wild-type mice. As expected, few myeloid colonies persisted in control cultures by the third generation. However, significant expansion of myeloid CFUs persisting through 3 generations was clearly evident in the Mll-AF9 cultures. Some evidence of myeloid deregulation of Mll-AF4 marrow was found but much less than that of Mll-AF9 marrow.
Bone marrow cultures under myeloid growth conditions. (A) Myeloid colonies from young mice. Cells from wild-type (n = 6), Mll-AF4 (n = 7), and Mll-AF9 (n = 12) mice were plated in methylcellulose medium under myeloid conditions for 3 generations. (B) Multiparameter FACS analysis on pooled cells from the methylcellulose culture of third-generation cells grown under myeloid conditions. (C) Colony distribution of murine bone marrow cultures under myeloid growth conditions. The frequency of myeloid type I, II, and III colonies was determined from young wild-type, Mll-AF4, and Mll-AF9 mice for 3 generations. The error bars represent SEMs.
Bone marrow cultures under myeloid growth conditions. (A) Myeloid colonies from young mice. Cells from wild-type (n = 6), Mll-AF4 (n = 7), and Mll-AF9 (n = 12) mice were plated in methylcellulose medium under myeloid conditions for 3 generations. (B) Multiparameter FACS analysis on pooled cells from the methylcellulose culture of third-generation cells grown under myeloid conditions. (C) Colony distribution of murine bone marrow cultures under myeloid growth conditions. The frequency of myeloid type I, II, and III colonies was determined from young wild-type, Mll-AF4, and Mll-AF9 mice for 3 generations. The error bars represent SEMs.
To further characterize cells from myeloid cultures, multiparameter FACS analysis of third-generation cells was carried out (Figure 5B). Mll-AF9 colonies contained the highest number of CD11b+. Minorities of the cells were also Gr1+. As expected, cells from myeloid cultures were B220 negative in all 3 types of mice (data not shown).
Type I myeloid colonies are found predominately in Mll-AF9 mice
Under myeloid conditions Mll fusion gene–expressing marrow cells form 3 types of distinct colonies as described previously.17,25,26 The colony distribution of bone marrow cultures from young Mll-AF4, Mll-AF9, and wild-type mice under myeloid conditions was compared (Figure 5C). Type I colonies were found in the culture of Mll-AF9 marrow and these colonies persisted through all generations. The distribution of Mll-AF4 colonies was similar to wild-type mice in 3 plating generations. Thus, although Mll-AF4 had some ability to deregulate myeloid cells, the type I colonies, which were primarily composed of immature myeloid cells, were found only in Mll-AF9 mice.
Mll-AF4 mice develop a complex and variable mix of lymphoid and myeloid hyperplasia, B-cell lymphoma, and myeloid malignancies
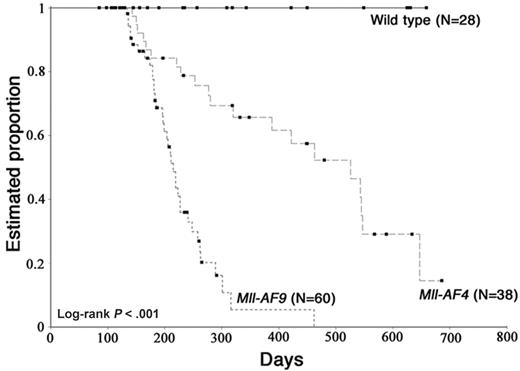
We observed Mll-AF4 mice up to 22 months for development of malignancy. Moribund Mll-AF4 mice showed typical features of severe hematologic disease, rough coat, hunched posture, and abnormal behaviors. Mice that were terminally ill were humanely killed. Kaplan-Meier analysis of the proportion of animals alive and free of disease are shown (Figure 6). The median time to development of hematologic malignancy was 520 days (17 months) for Mll-AF4 and 220 days (7 months) for Mll-AF9 mice. No wild-type littermate mice died during the 22-month observation.
Kaplan-Meier analysis of the estimated proportion of animals alive and free of hematologic malignancy. The analysis was done on Mll-AF4 (n = 38), Mll-AF9 animals (n = 60), and WT (n = 28). Overall log-rank P < .001.
Kaplan-Meier analysis of the estimated proportion of animals alive and free of hematologic malignancy. The analysis was done on Mll-AF4 (n = 38), Mll-AF9 animals (n = 60), and WT (n = 28). Overall log-rank P < .001.
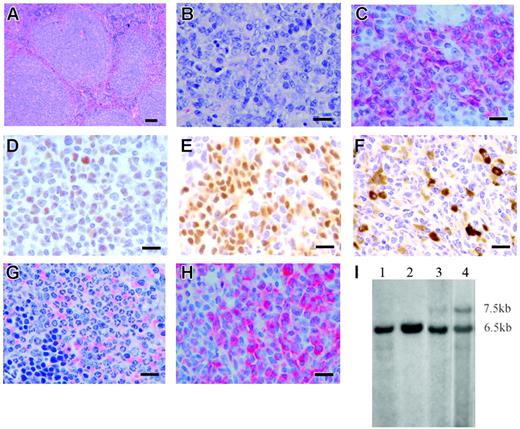
A total of 20 of the 38 Mll-AF4 mice became sick and died or were killed. WBC counts, cell types in blood, spleen weights, FACS analysis of spleen, and immunohistochemistry were evaluated. Histopathology was reviewed independently by 3 experts in murine hematopathology. Immunoglobulin gene rearrangements and Pax5 expressions were also tested. The Bethesda classification was used for diagnosis in all cases.27,28 Results of each mouse, including the final diagnosis, are shown in Table S3. The mean WBC count of Mll-AF4 mice with malignancy was higher than the one of wild-type mice (10.4 × 103 ± 0.6 × 103/μL [n = 15] versus 2.4 × 103 ± 0.2 × 103/μL [n = 10]; P = .01). When compared to wild-type mice, Mll-AF4 mice notably showed the presence of blasts and increases in the total numbers of monocytes and neutrophils in blood. All 20 mice had evidence of “benign” lymphoid and/or myeloid hyperplasia in spleen and bone marrow. Spleens were significantly enlarged with a mean 7.3-fold increase in size (means of Mll-AF4 = 0.51 g compared to 0.07 g in wild-type mice). The histopathology and immunohistochemistry of the spleen and lymph nodes were abnormal. These animals had lymphoid tumors arising from follicular centers (Table S3). Spleen histopathology from one mouse with lymphoid malignancy is shown (Figure 7A-B). The lymphoid malignancy affected spleen, lymph nodes, and often liver, lung, intestine, or kidney but not blood; cells from the spleen and other sites studied by immunohistochemistry had a follicular center B-cell phenotype: B220+ (Figure 7C), Bcl-6+ (Figure 7D), Pax5+ (Figure 7E), sIgM+ (Figure 7F), CD19+. Monoclonal or oligoclonal IgH rearrangements were detected in mice with lymphoma. The rearrangement in the spleen and lymph node from one representative mouse is shown in Figure 7I. The IgH rearrangements were also confirmed by HindIII digestion followed by Southern blotting (data not shown). The morphologic features combined with the immunophenotypic characteristics of blood and hematopoietic organs and immunoglobulin gene rearrangements resulted in the diagnosis of follicular B-cell lymphoma.28 Erythroid leukemia was present in one animal (FM1, Table S3). Spleen from mouse ChG2 is shown in H&E-stained (Figure 7G) and myeloperoxidase-stained sections (Figure 7H). This MPO+ myeloid malignancy and that of ChB were classified as MPD-like myeloid leukemia.29 Myeloid hyperplasia with no evidence of frank malignancy was present in 3 mice (ChD2, FM6m, and FM105). In summary, FACS of splenic cells, WBCs, cell morphology, and histopathology of different organs showed alterations in both lymphoid and myeloid cells in most of the 20 animals that developed fatal disease, indicating these Mll-AF4 mice had a complex, variable mixed lymphoid (most frequently B-cell lymphoma) and myeloid disease.
Histopathology and immunohistochemistry of Mll-AF4 mice with leukemia or lymphoma. (A) Spleen section from a representative Mll-AF4 mouse with lymphoma stained with H&E and shown at low magnification demonstrates loss of normal architecture resulting from follicular B-cell lymphoma. Bar represents 200 μm. (B) The same spleen section is shown at high magnification. Bar represents 50 μm. (C) Liver section indicates B220+ lymphoma cell infiltration. Bar represents 50 μm. (D) Liver section from the same specimen shows Bcl-6+ lymphoma. Bar represents 50 μm. (E) Spleen section shows PAX5+ lymphoma cells. Bar represents 50 μm. (F) Small intestine section shows IgM+ lymphoma cells. Bar represents 50 μm. (G) Spleen section from an Mll-AF4 mouse with MPD-like myeloid leukemia stained with H&E indicates loss of normal architecture and heavy infiltration of myeloid cells at all stage of maturation. Bar represents 50 μm. (H) Spleen section from a Mll-AF4 mouse with MPD-like myeloid leukemia stained with myeloperoxidase. Bar represents 50 μm. (I) IgH rearrangements in the spleen and lymph nodes of an Mll-AF4 mouse with follicular B-cell lymphoma. Lane 1, spleen from a wild-type mouse; lane 2, kidney from the same wild-type mouse; lane 3, spleen from an Mll-AF4 mouse; lane 4, lymph node from the same mouse. gDNA was digested by EcoRI. Germline band, 6.5 kb; additional band in lane 2 and lane 3, 7.5 kb.
Histopathology and immunohistochemistry of Mll-AF4 mice with leukemia or lymphoma. (A) Spleen section from a representative Mll-AF4 mouse with lymphoma stained with H&E and shown at low magnification demonstrates loss of normal architecture resulting from follicular B-cell lymphoma. Bar represents 200 μm. (B) The same spleen section is shown at high magnification. Bar represents 50 μm. (C) Liver section indicates B220+ lymphoma cell infiltration. Bar represents 50 μm. (D) Liver section from the same specimen shows Bcl-6+ lymphoma. Bar represents 50 μm. (E) Spleen section shows PAX5+ lymphoma cells. Bar represents 50 μm. (F) Small intestine section shows IgM+ lymphoma cells. Bar represents 50 μm. (G) Spleen section from an Mll-AF4 mouse with MPD-like myeloid leukemia stained with H&E indicates loss of normal architecture and heavy infiltration of myeloid cells at all stage of maturation. Bar represents 50 μm. (H) Spleen section from a Mll-AF4 mouse with MPD-like myeloid leukemia stained with myeloperoxidase. Bar represents 50 μm. (I) IgH rearrangements in the spleen and lymph nodes of an Mll-AF4 mouse with follicular B-cell lymphoma. Lane 1, spleen from a wild-type mouse; lane 2, kidney from the same wild-type mouse; lane 3, spleen from an Mll-AF4 mouse; lane 4, lymph node from the same mouse. gDNA was digested by EcoRI. Germline band, 6.5 kb; additional band in lane 2 and lane 3, 7.5 kb.
Transplantation of spleens and lymph nodes from Mll-AF4 mice with lymphoma and myeloid hyperplasia resulted in 2 distinct types of fatal malignancy. One was B-cell lymphoma in hematopoietic organs with IgH rearrangements, identical to that in donor Mll-AF4 mice with lymphoma. The second was fatal myeloid disease, characterized by increased number of CD11b+ Gr1+ F4/80+ myeloid cells in hematopoietic organs.
Mll-AF9 mice develop predominately myeloid malignancies
Thirty-seven Mll-AF9 mice developed hematologic malignancies at a median of 7 months (Figure 6). These malignancies were MPD-like myeloid leukemias in 36 mice, similar to earlier reports,11,25,30 and follicular B-cell lymphoma with MPD in one mouse. The mean WBC count of Mll-AF9 mice with myeloid disease was dramatically increased compared to wild-type littermates (225.1 × 103 ± 42.4 × 103/μL [n = 17] versus 2.4 × 103 ± 0.2 × 103/μL [n = 10]; P < .01) and higher than Mll-AF4 mice. Mll-AF9 mice also had significant leukemic infiltration of bone marrow, spleen, liver, lymph nodes, and Peyer patches. Malignant cells showed variable degrees of differentiation, ranging from blasts to cells with maturation into segmented neutrophils. Macrophages with a striated eosinophilic cytoplasm were frequently noted. Tumors were immunopositive for myeloperoxidase. FACS analysis demonstrated these malignancies to be CD11b+Gr1+B220– as reported earlier.17,30 Because the number of immature/blast cells were increased, but less than 10%, these malignancies, previously termed Gr1+ myeloid leukemias,17 were reclassified as MPD-like myeloid leukemia.29
Discussion
In this report, we describe the first model of murine Mll-AF4 disease. Mll-AF4 mice had abnormalities in the hematopoietic and skeletal systems. Skeletal and hematologic changes included distinctive craniofacial features that differed from both Mll-AF9 and wild-type mice. Mll-AF4 mice were small with reduced fertility. The effects of Mll-AF4 on lymphohematopoiesis were found early after birth and progressed to a fatal, complex, and variable mixed lymphoid (most frequently B-cell lymphoma)/myeloid disease in most mice after a long latency period.
MLL is known to be important in mammalian embryonic skeletal and hematopoietic development.31,32 AF4 is expressed in brain and multiple other organs.21,33 AF4-deficient mice have defective B-lymphoid development,34 and LAF4 is highest in B-lymphoid progenitor cells.35 In humans, MLL fusions with AF4 family members (AF4, LAF4, and AF5q31) result in lymphoid or mixed/multilineage malignancies.3,4,6,8-10 MLL-AF9 fusions most often result in myeloid malignancies.5,36 Our data showing preferential effects of Mll-AF4 on lymphoid and mixed-lineage cells are consistent with the patterns of MLL-AF4.
Our results showed distinctive effects of Mll-AF4 on lymphoid and myeloid populations of young premalignant mice. These differences were evident both in culture systems and in mice. Mixed-lineage pro-B (B220+CD19+CD43+sIgM–, PAX5+, TdT+, IgH rearranged)/myeloid (CD11b/Mac1+, c-fms+, lysozyme+) colonies were frequent in Mll-AF4 bone marrow grown under lymphoid conditions (Figures 3B and 4), which may represent multilineage capability possibly contributing to both lymphoid and myeloid proliferation in vivo. The existence of cells with mixed-lineage markers and multipotential for growth in normal marrow has been reported previously in normal marrow.37 Previously, murine marrow, retrovirally transfected with Mll-ENL or Mll-GAS7 and cultured under the similar conditions, grew mixed-lineage/multipotential cells.20,24 Depending on their self-renewal potential, increased progenitor cells in our cultures are also likely to increase the number of cellular targets at risk for secondary mutations. The increased numbers of premalignant Mll-AF4 progenitor cells could also be the result of reduced apoptosis. Previous studies have demonstrated resistance to apoptosis in MLL-AF4 leukemia38 or in cells transfected with MLL-AF4.14
Our Mll-AF4 KI murine model, in its current form, results in a long latency period before fatal lymphoid or myeloid disease rather than B-lineage leukemias found in humans. Thus, the model is not fully penetrant and indicates probably replicating the first does not replicate later steps in evolution of human Mll-AF4 disease. Whereas rarely are MLL-AF4 lymphomas found in humans,39 most human MLL-AF4 malignances are pro-B or pro-B/myeloid mixed-lineage leukemias. The most likely explanation for the differences with human disease follows from an absence of secondary mutations that are present in human Mll-AF4 disease. There may also be species differences, chromatin-limited accessibility of mutable genes, and lack of haploinsufficiency in the murine model or other to-be-identified factors. In other models of Mll fusion genes, including Mll-ENL and Mll-CBP, cooperating secondary mutations have been suggested to play a key role in development of frank malignancy, although in these models, as well as the Mll-AF4 model, these mutations remain largely undefined.40,41 The development of a fully penetrant model through induction of important additional cooperating mutations in the Mll-AF4 model is now a high priority for our laboratory. As in humans with MLL fusion gene leukemias, Flt3 might be one of a number of candidates for secondary mutations in transgenic mice with Mll fusions genes.3,42
In the Mll-AF4 and Mll-AF9 models, all hematopoietic progenitors and stem cells contain the fusion gene and yet different cellular effects are observed. The results show that both Mll-AF4 and Mll-AF9 fusion partners are active in deregulating hematopoiesis in young mice long before the development of hematologic malignancy. Our data provide evidence that some of cell type–preferential downstream effects are fusion partner dependent. Our findings showing that at least some of the effects are partner dependent suggest that the “hits” resulting from chromosomal translocation are not required to be in different cell types to obtain differing effects on cellular proliferation and differentiation.
Is Mll partner-dependent “instruction” a consequence of varying levels of expression of the fusion oncogene? We studied the expression of the Mll-AF4 or Mll-AF9 gene in selected bone marrows of young and older diseased mice using quantitative RT-PCR. All mice had expression of the appropriate gene. To date, no significant overall differences have been seen when Mll-AF4 and Mll-AF9 mice are compared. This result might be expected in the KI model where the 5′ region of Mll of Mll-AF4 or Mll-AF9 is not disrupted and under the control of the endogenous promoter. However, there are likely to be cell type–specific differences in expression levels. This possibility is suggested from the result of mice tested with differing age and disease status. Older diseased mice had lower levels of gene expression than young mice in both the Mll-AF4 and Mll-AF9 groups. Additional studies are under way to study these differences in purified populations of cells.
Mll fusion partner-independent effects were also found in our experiments with lymphoid and myeloid abnormalities in both Mll-AF4 and Mll-AF9 mice. Thus, while preferential, the effects of AF4 and AF9 were partially overlapping to some extent. Myeloid and lymphoid expansions were enhanced by both fusion genes. This crossover suggests that there are common features to both fusion genes that could result from effects of shared functional domains either in MLL or in the partners or both. Gain-of-function models proposed by many investigators for MLL fusion oncogenes have mostly focused on “partner-independent” characteristics.2,43
How do Mll-AF4 and Mll-AF9 produce preferential “instructive” expansion of mixed-lineage or myeloid cells, respectively, and set in motion the processes that eventually result in leukemia? Both AF4 and AF9 contain critical 3′ transactivation sequences that are retained in the fusion genes.2,41-43 The 5′ DNA-binding sequences of Mll, when fused to the 3′ portion of AF4 or AF9, may change the normal transcriptional regulation of downstream genes, such as Hox genes.17,20 Hox genes have been identified as the downstream targets of the MLL gene in both Drosophila and mammals.30,31,44,45 Up-regulation of Hox genes has been reported in both MLL-AF4 and MLL-AF9 leukemia3,46,47 in human and Mll-AF9 mice.17 Hox genes can alter the phenotype of leukemias but there is apparent redundancy in Mll fusion gene leukemogenesis.17,48 The differing abnormalities of facial morphology in Mll-AF4 and Mll-AF9 mice, which results in part from skeletal defects, may be the results of altered expression patterns of Hox genes during early development. Further studies of Hox and other genes in these mice are under way in our laboratory.
Partner-dependent “instructive” expression of other genes is likely to be important in the differing effects of Mll-AF4 and Mll-AF9. Milne et al49 have shown that the cyclin-dependent kinase inhibitor, p27kip1, is regulated by MLL through binding to the p27 promoter. Xia et al50 showed that MLL-AF4 binds to the CDKN1B promoter and p27 is highly expressed in MLL-AF4 but not MLL-AF9 leukemic cell lines. Menin is likely to play an important role in the effects of MLL fusion genes, as Yokoyama et al51 showed Menin to be an essential oncogenic cofactor in MLL fusion gene leukemia.
The fact that Mll-AF4 malignancies differ in phenotype from most other Mll fusion-induced malignancies in both mice and humans suggests that the “downstream” cells responsible for development of malignancy may be different in Mll-AF4 family member malignancies compared to most other MLL malignancies. Currently, the phenotype of the malignancy-initiating stem cells52 in Mll-AF4 versus Mll-AF9 malignancies remains to be determined. These important cells may differ from the bulk of cells found in the diverse malignancies that range from follicular B-cell to immature myeloid and mixed lineage in our Mll-AF4 transgenic animals. Additional studies of the progression and evolution of these important malignancy-inducing stem cells are now possible with the Mll-AF4 KI mice.
The Mll-AF4 KI model will also provide mice and cells for molecular studies of the AF4 partner in Mll fusion gene-induced lymphoid/myeloid deregulation and the later development of malignancy. Molecular studies of promoter transactivation, protein-protein interactions, and signal transduction pathways now can be done with this model.
Prepublished online as Blood First Edition Paper, March 21, 2006; DOI 10.1182/blood-2005-08-3498.
The research was supported in part by grant R01 CA087053 (J.H.K.) from the National Institutes of Health, and the Children's Cancer Research Fund.
J.H.K. designed and directed the research; W.C. was involved in experimental design, colony studies, FACS, and RT-PCR; Q.L. developed the Mll-AF4 knock-in ES cells and detected IgH rearrangements with PCR and Southern blotting; W.A.H. was involved in mouse genotyping, necropsy, and FACS; A.K. performed real-time quantitative RT-PCR; N.K. conducted pathologic analysis; W.C., Q.L., and W.A.H. performed all the other experiments; and J.H.K., W.C., and Q.L. analyzed data. All the authors were involved in manuscript writing and checking.
W.C. and Q.L. contributed equally to this study.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Bin Liu for construction of Mll-AF4 KI vector, Dr Stanley Korsmeyer for providing human MLL-AF4 cDNA, Drs Ilze Matise and Cathy Carlson for preparing histopathologic and immunohistochemical images, Dr Terence H. Rabbitts for providing Mll-AF9 mice, Drs Jerrold M. Ward and Torgny N. Fredrickson for review of necropsy and histopathologic materials, the University of Minnesota Mouse Genetics Laboratory for blastocyst injections, and Dr David Largaespada for helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal