Abstract
Hepcidin is the presumed negative regulator of systemic iron levels; its expression is induced in iron overload, infection, and inflammation, and by cytokines, but is suppressed in hypoxia and anemia. Although the gene is exquisitely sensitive to changes in iron status in vivo, its mRNA is devoid of prototypical iron-response elements, and it is therefore not obvious how it may be regulated by iron flux. The multiplicity of effectors of its expression also suggests that the transcriptional circuitry controlling the gene may be very complex indeed. In delineating enhancer elements within both the human and mouse hepcidin gene promoters, we show here that members of the basic helix-loop-helix leucine zipper (bHLH-ZIP) family of transcriptional regulators control hepcidin expression. The upstream stimulatory factor 2 (USF2), previously linked to hepcidin through gene ablation in inbred mice, appears to exert a polar or cis-acting effect, while USF1 may act in trans to control hepcidin expression. In mice, we found variation in expression of both hepcidin genes, driven by these transcription factors. In addition, c-Myc and Max synergize to control the expression of this hormone, supporting previous findings for the role of this couple in regulating iron metabolism. Transcriptional activation by both USF1/USF2 and c-Myc/Max heterodimers occurs through E-boxes within the promoter. Site-directed mutagenesis of these elements rendered the promoter unresponsive to USF1/USF2 or c-Myc/Max. Dominant-negative mutants of USF1 and USF2 reciprocally attenuated promoter transactivation by both wild-type USF1 and USF2. Promoter occupancy by the transcription factors was confirmed by DNA-binding and chromatin immunoprecipitation assays. Taken together, it would appear that synergy between these members of the bHLH-ZIP family of transcriptional regulators may subserve an important role in iron metabolism as well as other pathways in which hepcidin may be involved.
Introduction
Hepcidin is a small liver-derived acute-phase cationic peptide that is induced by iron overload, infection, and proinflammatory cytokines; on the other hand, its expression is attenuated in hypoxia and anemia.1-9 The relationship between this peptide hormone and iron is analogous to the link between blood glucose levels and insulin in that iron overload is accompanied by increased hepcidin expression by the liver, while the converse is true in iron deficiency. Initially isolated from urine and by subtraction cloning of its mRNA,1-3 this peptide has evolved as the primary regulator of iron homeostasis and a probable mediator of the anemia of chronic disease or inflammation. This role has been amply proven in a number of recent reports either by direct or indirect approaches. For example, in a study to define the role of upstream stimulatory factor 2 (USF2) in energy/glucose metabolism in mice,10,11 hepcidin was also surprisingly identified in association with an iron overload phenotype similar to hemochromatosis, as these mice produced diminished levels of hepcidin.12 On the other hand, transgenic mice that constitutively expressed hepcidin were severely anemic, thereby suggesting a role for this peptide in preventing iron overload.13,14 A frame-shift mutation identified in the human hepcidin gene has also been associated with a juvenile form of hemochromatosis.15 This peptide also appears to control iron trafficking by binding to and posttranslationally modulating ferroportin levels.16,17 Based on the foregoing, there is now almost incontrovertible evidence that this small peptide in some way communicates and modulates body iron levels, acting as the “gatekeeper” of iron trafficking. Intrinsic to its function as a modulator is a rapid18 (and possibly plastic) sensing mechanism that is still to be unraveled but is almost certain to involve a run-on surveillance mechanism; this is because hepcidin is de facto a defensin.1,2,5 Therefore, purely from first principles, the indications are that iron metabolism and innate immunity may be intimately linked to the role of hepcidin as a protective hormone. This therefore places it at a control nexus in whole-body homeostasis and defense mechanisms.
Although significant progress has been made with regard to the contribution of hepcidin in iron metabolism, we still do not know enough about how it is itself controlled transcriptionally or pre-/posttranslationally. It is clear from the variety of signals that induce hepcidin that the transcriptional controls underlying its expression may be varied and multifaceted. From the outset, its mRNA is devoid of the type of regulatory signature sequences, the iron-response elements, which are associated with key players in iron metabolism such as ferritin, ferroportin, and divalent metal iron transporter 1.19,20 On that basis, the link between iron metabolism and hepcidin is not immediately apparent. This indicates that its gene may be regulated by a different transcriptional and/or translational control circuitry. Although end-point assays (eg, Northern blotting or reverse transcriptase–polymerase chain reaction [RT-PCR]) show that the gene is induced by a number of effectors, it is not entirely clear which transcription factors are involved. To date, only C/EBPα has been shown to regulate the transcription of this gene,21 but it is almost certain that its control may be very complex indeed. Here we show that members of the basic helix-loop-helix leucine zipper (bHLH-ZIP) family of transcriptional regulators control hepcidin expression. By deletion mapping, transactivation and DNA-binding assays, we identified E-boxes within the hepcidin gene promoter and found that these enhancer elements could sufficiently mediate the transcriptional regulation of a reporter gene by USF1/USF2 and c-Myc/Max. We also establish that both mouse hepcidin genes are controlled via this transcription path. Thus the link between USF2 and hepcidin, although initially uncertain and serendipitous, is therefore functionally significant after all. This link is discussed in detail.
Materials and methods
Promoter and enhancer constructs
Approximately 1.1 kb of the human hepcidin gene, comprising 1026 bp of the promoter and nucleotides encoding the first 18 amino acids of prohepcidin, was amplified from (placental) genomic DNA with the following primers: sense, CAG GCTAGC CAT CGT ATA AAA TGT ACT CAT CGG; antisense, CAT CTCGAG CGA GGA GGA GGA GGA GCA (complementary to nucleotides 38-55 of the coding sequence); NheI and XhoI restriction sites are underlined. The cycling parameters were: 95°C for 5 minutes, then 95°C for 1 minute (denaturation), 60°C for 1 minute (annealing), and 72°C for 1 minute (extension); 35 cycles of PCR were performed with a final extension for 10 minutes at 72°C. The PCR product was subcloned into pGEM-T Easy vector (Promega, Southampton, United Kingdom) and sequenced for verification (MWG Biotech, Ebersberg, Germany). The construct was digested with NheI and XhoI (New England Biolabs, Hitchin, United Kingdom), and the insert was purified with Geneclean (BIO101; Anachem, Luton, United Kingdom) and ligated into the NheI and XhoI sites of pGL3Basic (Promega) to generate HepcP1.1-luc. To make deletion mutants, HepcP1.1-luc was digested using selected enzymes with unique sites within the promoter only (but not within the vector backbone). Following digestion, the products were resolved on a 1% agarose gel and purified with Geneclean. Compatible restriction ends of the linearized plasmids were religated directly; incompatible ends were polished with T4 DNA polymerase (New England Biolabs) and dNTPs for 20 minutes at 37°C. Ligations were performed using the Quick Ligation kit (New England Biolabs) and transformed into DH5α-competent cells (Invitrogen, Paisley, United Kingdom). Plasmid DNA was purified from isolated colonies using Nucleospin plasmid miniprep columns (Macherey-Nagel, Düren, Germany) and sequenced to verify that the intended deletions had occurred.
For mouse hepcidin gene promoter constructs, genomic DNA was isolated from liver tissue samples of male SWR mice (Harlan UK, Oxon, United Kingdom). Based on National Center for Biotechnology Information (NCBI) GenBank nucleotide accession numbers AC020841(mhepc1) and AC087143(mhepc2), the following primer pairs were designed and synthesized: mhepc1P sense, CAT AGCTAGCAG GCA GAA GGG GAA TCC AAC ATG AC; antisense, CAT GCT CGA GCG AGT GCT GAG TGC CAT CAT GCC TT; mhepc2P sense, CAT AGCTAGCGC TGC AGA AGT GAC TCC AAC ATG AG; and antisense, CAT GCTCGAGCG AGT GCT GAG TGC CAT CAT GCC TC (NheI and XhoI sites are underlined). Because of the high homology between the 2 genes, only primer sequences that differed by at least one nucleotide at the 3′ ends were selected, to avoid cross-amplification and chimerism. Thus, approximately 600 bp of both hepcidin gene promoters, as well as nucleotides encoding the first 6 amino acids of prohepcidin, were amplified by PCR as described. The PCR products were digested with NheI and XhoI, and directionally ligated into pGL3Basic. Both constructs were sequenced for authentication and labeled mhepc1P-luc and mhepc2P-luc, respectively.
To construct E-box plasmids, oligonucleotides modified by phosphorylation at their 5′ ends were synthesized (MWG Biotech). Their sequences were as follows. E-box1: sense, CTAGCG AAT TCC TGG GAA AAC ACC ACG TGC GGA TCG GGC ACA CGC; complement, TCGAGC GTG TGC CCG ATC CGC AC GTG G TGT TTT CCC AGG AAT TCG; E-box2: sense, CTAGCG AAT TCC CTG CCA GAA CCT ATG CAC GTG TGG TGA GAG CTC; complement, TCGAGA GCT CTC ACC ACA CGT GCA TAG GTT CTG GCA GGG AAT TCG; E-box3: sense, CTAGCG AAT TCA AAG GGC TCC CCA GAT GGC TGG TGA GCA GTG C; complement, TCGAGC ACT GCT CAC CAG CCA TCT GGG GAG CCC TTT GAA TTC G; and E-box4: sense, CTA GCG AAT TCT CCC AAG TAG CTG GGA CTA CAG ATG TGT GCC ACC ACG CCT GGC TC; complement, TCG AGA GCC AGG CGT GGT GGC ACA CAT CTG TAG TCC CAG CTA CTT GGG AGA ATT CG. To compare transcription from mouse and human hepcidin gene E-boxes we also subcloned the sequences CTAGCG AAT TCA GAA TCA GTA CTC ACT GCC ATG TGA AAC CAG TGT GC and TCGAGC ACA CTG GTT TCA CAT GGC AGT GAG TAC TGA TTC TGA ATT CG, containing an E-box conserved in both mhepc1 and mhepc2. (E-boxes are italicized in all cases.) Mutant E-box oligonucleotides (Emt's) with mutations (lowercase) within E-box1 were also synthesized as follows: sense, CTAGCG AAT TCC TGG GAA AAC ACC ACG gaC GGA TCG GGC ACA CGC; complement, TCGAGC CGT GTG CCC GAT CCG tcC GTG GTG TTT TCC AGG AAT TCG. A unique EcoRI site was included in the oligonucleotides to be used for linearization to ascertain oligonucleotide insertion into the vector. NheI and XhoI sites are underlined in all cases. Complementary oligonucleotides were combined in equimolar ratios (500 pmol each) in 1 × ligation buffer, incubated at 95°C for 3 to 5 minutes in a heating block, and annealed by slow cooling to below 30°C. All duplex oligonucleotides were directionally ligated into pGL3Promoter (Promega) and predigested with NheI and XhoI. The resulting plasmids were designated E-box1-luc, E-box2-luc, E-box3-luc, E-box4-luc, mhepcPE-luc (mouse enhancer construct), and E-null-luc, respectively. A construct (2xE-box-luc) encompassing the canonical E-boxes 1 and 2 (from –642 to –343; 300 bp), was derived by PCR amplification and subcloning of the 2 elements into pGL3Promoter, using the following primers: sense, CAT GCTAGC GCA TAA GCC ACT GTG CTG GGC C; antisense, CAT CTCGAG CTC ACT TCC CCA TCG CCT ACA TGC (NheI and XhoI sites are underlined). All constructs were verified by sequencing.
Site-directed mutagenesis of E-boxes
HepcP1.1-luc was subjected to site-directed mutagenesis using the QuikChange Multi Site-Directed Mutagenesis system (Strategene, Amsterdam, The Netherlands) as instructed by the manufacturer. Mutagenic primers (mutations in lowercase) were as follows: E-box1, CTG GGAAAA CAC CAC Gga CGG ATC GGG CAC ACG; E-box2, CCT GCC AGAACC TAT GCA CGg aTG GTG AGA GCT; E-box3, AAA GGG CTC CCC AGA gaG CTG GTG AGC AGT G; and E-box4, TCC CAA GTA GCT GGG ACT ACA GAg aTG TGC CAC CAC GCC TGG CT. After initial denaturation for 1 minute at 95°C, PCR cycling parameters were 95°C (1 minute), 55°C (1 minute), and 65°C (12 minutes), for a total of 30 cycles. Following DpnI digestion of wild-type HepcP1.1-luc, transformation of XL10 Gold cells with the mutagenesis reaction and selection on Luria agar/ampicillin plates, plasmid DNA was purified from overnight cultures of single colonies and restricted with PmlI (New England Biolabs), which cuts within the 2 canonical E-boxes (CAC ↓ GTG). Mutant promoter plasmids were identified as those that could not be cleaved by the enzyme due to loss of the restriction sites. Plasmids were sequenced to further ascertain mutagenesis of all E-boxes. The resulting mutant promoter construct was designated HepcP1.1-lucMtE4.
Cell culture, transfection, and reporter assays
All cell culture reagents were obtained from Invitrogen. HepG2 (obtained from the European Collection of Animal Cell Cultures, Porton Down, United Kingdom) and BHK cells (kindly provided by Jill Norman, University College London, United Kingdom) were cultured in minimal essential medium with Earle salts and supplemented with nonessential amino acids, 10% fetal bovine serum, and antibiotics; cells were grown under standard cell-culture conditions. Preliminary transfection assays with HepcP1.1-luc showed that BHK cells reported higher luciferase expression than HepG2 cells for reasons that are not apparent. Hence, for further transfections, these cells were seeded in Costar 24-well plates (Corning, Cambridge, MA) at densities of approximately 104 cells/well. Approximately 100 ng of the transactivation expression plasmids pCMV-USF1 and pCMV-USF2 (kindly provided by Michele Sawadogo, University of Texas, M.D. Anderson Cancer Center), and pTM3-His-cMyc and pTM3-Max (kindly provided by H. T. Marc Timmers, Utrecht University, The Netherlands) were diluted in OptiMEM 1 and cotransfected with 100 ng HepcP1.1-luc and its derivatives (deletion or site-directed mutants) as well as E-box enhancer constructs. All transfections were performed with Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer. A titration assay was also performed by cotransfecting different molar ratios of USF1 and USF2. As internal control, 50 ng pSVβgal vector (Promega) was included in all transfections to normalize transfection efficiencies. For trans-repression, various amounts of the USF1 dominant-negative mutant A-USF,22,23 (kindly provided by Charles Vinson, National Cancer Institute, National Institutes of Health, Bethesda, MD) and USF2 dominant-negative mutant USF2ΔB24,25 (kindly provided by L. Karl Olson, Michigan State University, East Lansing) were cotransfected with USF1 and USF2. Transfections with mhepc1P-luc, mhepc2P-luc, and mhepcPE-luc were similarly performed. Where necessary, pcDNA3.1 was added to equalize the total amount of DNA. Cells were harvested after 48 hours for reporter assays; luciferase activities were determined with the luciferase assay reagent and β-galactosidase (βgal) activity was measured using the Beta-Glo reagent (both from Promega). Luminescence was measured in a MicroLumatPlus LB96V microplate luminometer (Berthold Technologies, Baden-Wuerttenberg, Germany); luciferase levels were normalized with respect to βgal activity in the samples.
Preparation of nuclear extracts
HepG2 cells were harvested at log phase of growth by trypsinization, washed twice with phosphate-buffered saline, and resuspended in lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 1 mM DTT) supplemented with Complete protease inhibitor cocktail (Roche, Lewes, United Kingdom). They were incubated on ice for 30 minutes to lyse; to aid lysis, the cells were drawn 10 times through a 25 G needle. After centrifugation at 16 000g, 4°C for 20 minutes, the nuclear pellet was resuspended in an equal volume of extraction buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 25% glycerol, and 1 mM DTT) containing protease inhibitors. The nuclei were incubated on a rotary wheel at 4°C for 30 minutes and then centrifuged at 16 000g for 10 minutes at 4°C. Nuclear protein content was determined with the BCA reagent (Pierce, Rockford, IL) in microtiter plates (Nunc, Paisley, United Kingdom).
Electrophoretic mobility shift assay
For mobility shifts, E-box duplex oligonucleotides (100 pmol) were end-labeled with γ32P[ATP] (110 TBq/mmol; Amersham Biosciences, Bucks, United Kingdom) and T4 polynucleotide kinase (New England Biolabs) and diluted to 1 pmol/μL with Tris-EDTA (pH 8.0). Approximately 1 pmol of each probe was incubated with 10 μg nuclear extract in binding buffer (4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 10 mM HEPES pH 7.9, 50 mM NaCl, and 50 μg/mL poly [dI-dC] · poly[dI-dC]) in a total volume of 10 μL. For competitive inhibition, a 100-fold molar excess of the cold E-box oligonucleotide was added to the binding reaction 10 minutes before adding the labeled probe. Specific binding (supershift assay) was determined by incubating 10 μg nuclear extract with 4 μg USF1 (sc-8983X) or USF2 (sc-862X) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hour at room temperature. Thereafter, the radiolabeled E-box oligonucleotides were added, and after another 30 minutes at room temperature, the samples were resolved on a 6% nondenaturing polyacrylamide gel in 0.5 × TBE buffer. The gel was dried at 80°C for 2 hours and exposed to X-ray film for 6 hours at –80°C.
In a variation of the mobility shift assay, USF1, USF2, c-Myc, and Max were expressed by coupled in vitro transcription and translation using the T7 TNT Quick system (Promega) as instructed by the manufacturer. Equal amounts (3 μL each) of c-Myc and Max or USF1 and USF2 were mixed and incubated in binding buffer at room temperature for 30 minutes to form heterodimers; radiolabeled E-box oligonucleotide or the mutant E-box were then added. After incubating further for 30 minutes at room temperature, the samples were processed as described in the preceding paragraph.
ChIP assay
HepG2 cells were grown to 90% confluence. After formaldehyde cross-linking, chromatin was sonicated and precleared with protein A–sepharose/salmon sperm DNA. Immunoprecipitation of the hepcidin gene promoter was performed using a chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology, Lake Placid, NY) as instructed by the manufacturer, with 10 μg of antibodies to USF1, USF2, c-Myc (Sigma, Poole, United Kingdom), and Max (kindly provided by H.T. Marc Timmers). As a control, a nonspecific immunoglobulin G (IgG) of the same isotype was also used for ChIP. Chromatin immunoprecipitates were collected with fresh protein A–sepharose/salmon sperm DNA and washed sequentially in low-salt, high-salt, LiCl, and TE buffers. The immune complexes were eluted with 1% SDS and 0.1 M NaHCO3 at room temperature for 15 minutes. The eluates were incubated at 65°C overnight with NaCl (0.2 M final concentration) to reverse cross-linking and then with 10 mM EDTA, 40 mM Tris-HCl (pH 6.5), and 40 μg/mL proteinase K (Sigma) at 45°C for 1 hour. DNA was purified by phenol-chloroform extraction and precipitated with 2.5 vol ice-cold ethanol. PCR was performed with the primers used for amplification and subcloning of 2xE-box-luc; the latter was used as positive control for amplification. Amplification was performed for 35 cycles as described under “Promoter and enhancer constructs”; PCR products were resolved on a 2% agarose gel in 1 × TAE. The products were subcloned into pGEM-T Easy vector and sequenced to confirm their authenticity.
Statistical analysis
Data were analyzed and graphs were plotted with the GraphPad Prism software (GraphPad Software, San Diego, CA). Where appropriate, datasets were analyzed by the Mann-Whitney U test or the 1-way analysis of variance (ANOVA)/Student-Newman-Keuls multiple-comparisons test. Pairwise comparisons of control and test transfections or constructs and permutations thereof were performed from these analyses. Statistical significance was accepted at P values less than .05. Results were expressed as means ± SEM.
Results
Deletion mapping of the promoter and identification of enhancer elements
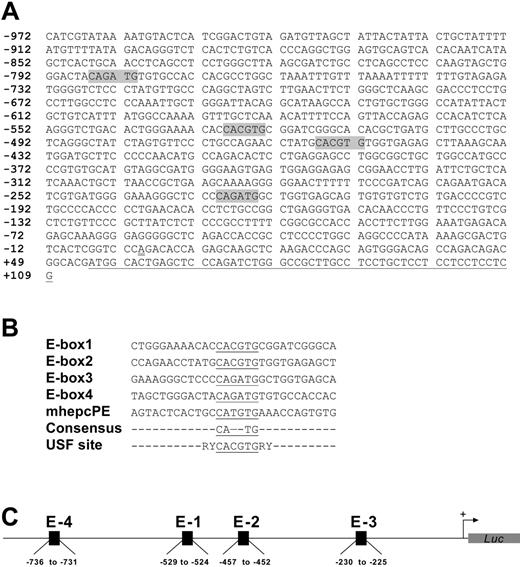
We cloned approximately 1.1 kb of the human hepcidin gene promoter as a transcriptional fusion with firefly luciferase to give HepcP1.1-luc; this construct also contained nucleotides encoding the first 18 amino acids of prohepcidin. Using unique restriction enzyme sites within the promoter only, we created a set of deletions (Figure 1A) to assess the basal activities of promoter segments. A comparison of luciferase activities reported by the various constructs showed that the wild-type promoter HepcP1.1-luc produced less luciferase than some deletion mutants (eg, ΔAP and ΔNA), possibly due to repressor elements within it. We delineated a minimal promoter of approximately 600 bp that was sufficient for optimal reporter activity (Figure 1B). Further deletions and close inspection of the sequence revealed enhancer activities associated with 2 canonical E-boxes at –529 to –524 (E-box1) and –457 to –452 (E-box2), each with the sequence CACGTG and separated by 66 nucleotides. Two identical consensus E-boxes with the sequences CAGATG were also identified at –230 to –225 (E-box3) and –786 to –781 (E-box4) (Figure 2A). A comparison of these boxes and juxtaposing nucleotides revealed little nucleotide conservation except for the core E-box consensus hexanucleotide CANNTG (Figure 2B) that is recognized by members of the bHLH-ZIP family of transcription factors, including USF.26,27 Incidentally, these elements lie within the region of the promoter (Figure 2C) that has been shown to be transactivated by C/EBPα.21 Deletions of E-box segments and transfection of the resulting plasmid constructs showed that basal promoter activity was partly dependent on these E-boxes. For example, the ΔNT deletion construct in which all 4 E-boxes had been excised showed diminished luciferase activity compared with constructs with the E-boxes.
Deletion mapping of the human hepcidin gene promoter. (A) HepcP1.1-luc (wild-type [WT]) was selectively restricted with enzymes to remove various segments. Constructs were generated by restriction with BalI(ΔBal; –598 to –379), PmlI(ΔPml; –526 to –454), NheI-AflII (ΔNA; –972 to –821), AflII-PmlI(ΔAP; –821 to –454), NheI-PmlI (ΔNP; –972 to –454), and NheI-TthIIII (ΔNT; –972 to –196). Nucleotide numbering is based on Courselaud et al.21 Note that the NheI restriction site was used for cloning purposes only and is not intrinsic to this segment of the promoter (PCR primers in “Materials and methods”). Also, although there are 2 TthIIII sites within the promoter, double-digestion with NheI removed all but approximately 200 bp of the promoter. (B) Basal transcriptional activity of promoter deletion constructs from panel A. Constructs (100 ng each) were cotransfected with pSVβgal (50 ng) into BHK cells for 48 hours, and reporter activities were measured as described above. Luciferase levels were normalized with respect to βgal activity. Fold activation was based on the activity of pGL3Basic, and assigned an arbitrary activation level of 1. Pairwise comparisons were made between WT and deletion constructs using the 1-way ANOVA/Student-Newman-Keuls test. **P < .005. Data are representative of 4 independent sets of transfections, shown as means ± SEM.
Deletion mapping of the human hepcidin gene promoter. (A) HepcP1.1-luc (wild-type [WT]) was selectively restricted with enzymes to remove various segments. Constructs were generated by restriction with BalI(ΔBal; –598 to –379), PmlI(ΔPml; –526 to –454), NheI-AflII (ΔNA; –972 to –821), AflII-PmlI(ΔAP; –821 to –454), NheI-PmlI (ΔNP; –972 to –454), and NheI-TthIIII (ΔNT; –972 to –196). Nucleotide numbering is based on Courselaud et al.21 Note that the NheI restriction site was used for cloning purposes only and is not intrinsic to this segment of the promoter (PCR primers in “Materials and methods”). Also, although there are 2 TthIIII sites within the promoter, double-digestion with NheI removed all but approximately 200 bp of the promoter. (B) Basal transcriptional activity of promoter deletion constructs from panel A. Constructs (100 ng each) were cotransfected with pSVβgal (50 ng) into BHK cells for 48 hours, and reporter activities were measured as described above. Luciferase levels were normalized with respect to βgal activity. Fold activation was based on the activity of pGL3Basic, and assigned an arbitrary activation level of 1. Pairwise comparisons were made between WT and deletion constructs using the 1-way ANOVA/Student-Newman-Keuls test. **P < .005. Data are representative of 4 independent sets of transfections, shown as means ± SEM.
Since the initial link between hepcidin and USF2 was established in mice, we compared the mouse and human hepcidin gene promoters for conserved E-boxes. We restricted comparisons to approximately 600 bp of both mouse gene promoters, equivalent to the minimal promoter identified for the human hepcidin gene promoter (Figure 1). Sequence analyses showed that unlike in the human ortholog, no canonical E-boxes were identified within the mouse sequence in a similar spatial arrangement. However, a number of consensus E-boxes punctuated the promoters of both mhepc1 and mhepc2 (data not shown). These putative E-boxes included 2 CAGGTG elements, CATATG and 2 overlapping elements, CACATGTG in the promoter of mhepc1 (underlined and italicized). The CATATG variant of the canonical E-box has been described as a high-affinity site within the mouse metallothionein gene promoter.28
Transcriptional control of hepcidin expression by USF1/USF2 and c-Myc/Max
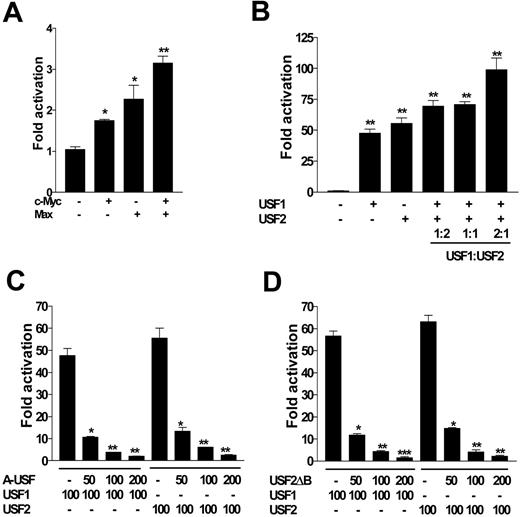
Transactivation assays showed that c-Myc and Max evoked a 2- to 4-fold increase in luciferase activity driven by HepcP1.1-luc (Figure 3A); this was modest compared with that of USF1 and USF2, which enhanced luciferase expression approximately 50- and 60-fold, respectively (Figure 3B). These differences may indicate that nucleotide specificities and preferences of c-Myc/Max and USF1/USF2 heterodimers around the E-box core may be different. In a titration assay using different USF1 and USF2 molar ratios, we found that within the context of these experiments, a USF1/USF2 ratio of 2:1 enhanced promoter activation 2-fold more than with either isoform alone (Figure 3B). To further confirm the specificity of USF1/USF2 in controlling hepcidin expression, we also used dominant-negative mutants of USF1 (A-USF) and USF2 (USF2ΔB) in cotransfection experiments. We found that both mutants significantly and reciprocally attenuated luciferase expression by HepcP1.1-luc in a dose-dependent manner when cotransfected with either (wild-type) USF1 or USF2 (Figure 3C-D).
To determine whether the mouse hepcidin genes could also be transactivated by USF1 and USF2, we cloned the respective promoters from the SWR mouse strain and used these in transactivation assays. In parallel, we performed trans-repression assays in cotransfections with the dominant-negative mutants. Alone, mhepc1P-luc reported 11-fold lower basal activity than mhepc2P-luc when transfected into cells. Upon cotransfection with USF1 or USF2, however, mhepc1P-luc was transactivated approximately 600-fold, 10-fold more than mhepc2P-luc by both transcription factors. While both promoters could be transactivated by USF1 and USF2 (Figure 4A), their respective dominant-negative mutants suppressed transactivation of both promoters by up to 2.5-fold, and in a dose-dependent manner (Figure 4B-C). The differences in transactivation between the 2 mouse hepcidin genes would suggest that regulatory polymorphisms in their cognate promoters might be important, particularly as we identified several single-nucleotide differences between these promoters (data not shown). We suggest that expression level polymorphisms in the 2 mouse genes may impact their responses to iron and/or infection, within and between inbred mice in general.
Organization of promoter and enhancer elements. (A) Spatial arrangement and nucleotide sequences of the E-boxes (shaded) within the cloned human hepcidin gene promoter. Numbers indicate nucleotide positions with respect to the putative transcription initiation site, which is double-underlined.21 The partial prohepcidin open-reading frame is single-underlined. (B) Sequences of E-boxes (underlined) and proximal nucleotides in HepcP1.1-luc compared with a canonical USF site; R indicates purine; and Y, pyrimidine. A representative mouse hepcidin gene E-box (mhepcPE) is included for comparison. (C) Schematic of spatial organization of canonical (E-1 and E-2) and consensus (E-3 and E-4) E-boxes in the context of the promoter.
Organization of promoter and enhancer elements. (A) Spatial arrangement and nucleotide sequences of the E-boxes (shaded) within the cloned human hepcidin gene promoter. Numbers indicate nucleotide positions with respect to the putative transcription initiation site, which is double-underlined.21 The partial prohepcidin open-reading frame is single-underlined. (B) Sequences of E-boxes (underlined) and proximal nucleotides in HepcP1.1-luc compared with a canonical USF site; R indicates purine; and Y, pyrimidine. A representative mouse hepcidin gene E-box (mhepcPE) is included for comparison. (C) Schematic of spatial organization of canonical (E-1 and E-2) and consensus (E-3 and E-4) E-boxes in the context of the promoter.
c-Myc/Max and USF1/USF2 differentially transactivate the promoter. (A) HepcP1.1-luc (100 ng) was transfected into BHK cells alone or with 100 ng c-Myc, Max, or c-Myc and Max. (B) HepcP1.1-luc was transfected with or without USF1, USF2 (100 ng each), or both. In the latter, various ratios of USF1 and USF2 were also cotransfected into BHK cells with HepcP1.1-luc. Fold activation in panels A and B was calculated with respect to the activity of HepcP1.1-luc alone. (C) HepcP1.1-luc (100 ng) was transfected into BHK cells alone or with 100 ng USF1 or USF2, and with increasing concentrations of dominant negative mutant USF1, A-USF, or (D) USF2 dominant-negative mutant, USF2ΔB. All transfections were performed in duplicate and included pSVβgal as internal control; luciferase levels were normalized to βgal activity. Fold repression by A-USF or USF2ΔB (C-D) was expressed with respect to the activity of HepcP1.1-luc in the presence of either USF1 or USF2. Data are representative of 3 independent experiments, and are plotted as means ± SEM. *P < .05; **P < .005; ***P < .001; determined by 1-way ANOVA and Student-Newman-Keuls multiple-comparisons test.
c-Myc/Max and USF1/USF2 differentially transactivate the promoter. (A) HepcP1.1-luc (100 ng) was transfected into BHK cells alone or with 100 ng c-Myc, Max, or c-Myc and Max. (B) HepcP1.1-luc was transfected with or without USF1, USF2 (100 ng each), or both. In the latter, various ratios of USF1 and USF2 were also cotransfected into BHK cells with HepcP1.1-luc. Fold activation in panels A and B was calculated with respect to the activity of HepcP1.1-luc alone. (C) HepcP1.1-luc (100 ng) was transfected into BHK cells alone or with 100 ng USF1 or USF2, and with increasing concentrations of dominant negative mutant USF1, A-USF, or (D) USF2 dominant-negative mutant, USF2ΔB. All transfections were performed in duplicate and included pSVβgal as internal control; luciferase levels were normalized to βgal activity. Fold repression by A-USF or USF2ΔB (C-D) was expressed with respect to the activity of HepcP1.1-luc in the presence of either USF1 or USF2. Data are representative of 3 independent experiments, and are plotted as means ± SEM. *P < .05; **P < .005; ***P < .001; determined by 1-way ANOVA and Student-Newman-Keuls multiple-comparisons test.
Next, we turned our attention to confirm the question of whether promoter transactivation was indeed mediated by the E-boxes. We concentrated particularly on the human hepcidin promoter, subcloning duplex oligonucleotides of each of its 4 E-boxes into the simian virus 40 (SV40) promoter vector pGL3Promoter (Figure 5A). In addition, we PCR-amplified and similarly subcloned the promoter segment containing the canonical E-boxes. For comparison, we also tested one of the E-boxes (CATGTG) conserved in both mouse hepcidin genes. We cotransfected these constructs with expression vectors for USF1, USF2, c-Myc, Max, and combinations thereof. Luciferase assays showed that both mouse and human hepcidin E-box constructs were functionally sufficient and equivalent in their ability to drive luciferase expression several-fold above background (Figure 5B); the mutant E-box construct E-null-luc reported only background activity (data not shown). This therefore suggests a common mechanism of hepcidin regulation in both mice and humans. Paradoxically, cotransfection of the E-boxes with combinations of USF1 and USF2 or c-Myc and Max showed that transactivation was not additive, possibly because of mass action or saturation effects. 2xE-box-luc surprisingly reported lower activity than expected compared with constructs with only one E-box. Due to the juxtaposition of the 2 boxes, it may indicate mutually competitive exclusion of either USF isoform by the other, or c-Myc exclusion by Max and vice versa. The higher levels of luciferase expression in the context of the entire promoter compared with the E-boxes alone strongly suggest that other unidentified transcription factors may synergize with USF1/USF2 and/or c-Myc/Max for hepcidin expression. Finally, we performed site-directed mutagenesis to further ascertain the role of the E-boxes for transactivation, mutating all 4 E-boxes in HepcP1.1-luc. We found that this diminished basal luciferase expression approximately 5-fold (Figure 6A). The mutant promoter (HepcP1.1-lucMtE4) was also 10-fold less responsive to transcriptional activation by USF1/USF2 than the wild-type promoter (Figure 6B), while transactivation by c-Myc/Max was reduced approximately 3-fold (Figure 6C).
Promoter occupancy by USF1/USF2 and c-Myc/Max
To ascertain that the E-boxes bound USF1/USF2 or c-Myc/Max, we performed electrophoretic mobility shift assays (EMSAs) with radiolabeled E-box oligonucleotides. We identified mobility shift profiles that were consistent with binding of USF1/USF2 and c-Myc/Max, possibly as homoand heterodimers.29-31 DNA binding could be competitively inhibited by a 100-fold molar excess of cold or unlabeled E-box oligonucleotide, further confirming that promoter occupancy by these transcription factors was specifically at the E-boxes. The mutant E-box Emt, in which the TG dinucleotide of the hexanucleotide CANNTG was changed to GA, failed to bind to the nuclear components (Figure 7A). To further ascertain binding specificity we incubated nuclear extracts with antibodies to USF1 or USF2 and observed supershifts consistent with USF1 or USF2 antibody complexes bound to the E-boxes (Figure 7B). E-box occupancy was also confirmed using in vitro–translated USF1 and USF2 or c-Myc and Max heterodimers; these proteins bound to the wild-type E-box oligonucleotides but not to Emt (Figure 7C).
To determine promoter occupancy by USF1/USF2 or c-Myc/Max in vivo, we performed ChIP assays. Using antibodies to USF1, USF2, and c-Myc/Max, we immunoprecipitated the target region of the promoter with the 2 canonical E-boxes (Figure 7D); the authenticity of the immunoprecipitated DNA was determined by sequencing. In contrast, we were unable to immunoprecipitate the promoter with a nonspecific IgG (Figure 7E). This assay therefore provided some proof for the recruitment of both USF1/USF2 and c-Myc/Max heterodimers to the promoter within its genomic context. The data also suggested that both sets of transcriptional regulators might be independently recruited to the E-boxes, although some competition for these sites cannot be ruled out.
Regulation of mouse hepcidin genes by USF1/USF2. (A) Approximately 600 bp of mhepc1 (mhepc1P-luc) and mhepc2 (mhepc2P-luc) promoters were cloned from the SWR mouse strain into pGL3Basic; BHK cells were cotransfected with 100 ng of each construct and with either USF1 or USF2 to determine transactivation. (B) Various concentrations of A-USF or (C) USF2ΔB were cotransfected into BHK cells with mhepc1P-luc or mhepc2P-luc together with 100 ng USF1 or USF2 plasmids to determine transcriptional repression. Data are representative of 2 independent assays (means ± SEM). *P < .05; **P < .005; ***P < .001 (determined by 1-way ANOVA/Student-Newman-Keuls comparisons test).
Regulation of mouse hepcidin genes by USF1/USF2. (A) Approximately 600 bp of mhepc1 (mhepc1P-luc) and mhepc2 (mhepc2P-luc) promoters were cloned from the SWR mouse strain into pGL3Basic; BHK cells were cotransfected with 100 ng of each construct and with either USF1 or USF2 to determine transactivation. (B) Various concentrations of A-USF or (C) USF2ΔB were cotransfected into BHK cells with mhepc1P-luc or mhepc2P-luc together with 100 ng USF1 or USF2 plasmids to determine transcriptional repression. Data are representative of 2 independent assays (means ± SEM). *P < .05; **P < .005; ***P < .001 (determined by 1-way ANOVA/Student-Newman-Keuls comparisons test).
Functional analyses of the putative enhancer elements. (A) Luciferase fusions of E-box duplex oligonucleotides (only partial sequences are shown) with 1 or 2 E-boxes (details in “Materials and methods”). A single E-box (mhepcPE) conserved in both mouse hepcidin genes was similarly subcloned. (B) E-box transactivation assays. Constructs in panel A, 100 ng each, were transfected into BHK cells alone or with 100 ng each of USF1, USF2, c-Myc, and Max expression vectors and combinations thereof. Fold transactivation by USF1, USF2, c-Myc, and Max was expressed with respect to the basal activity of each E-box construct alone. Data represent means of 3 independent experiments (± SEM). *P < .05; **P < .005; ***P < .001; determined by 1-way ANOVA and the Student-Newman-Keuls comparisons test.
Functional analyses of the putative enhancer elements. (A) Luciferase fusions of E-box duplex oligonucleotides (only partial sequences are shown) with 1 or 2 E-boxes (details in “Materials and methods”). A single E-box (mhepcPE) conserved in both mouse hepcidin genes was similarly subcloned. (B) E-box transactivation assays. Constructs in panel A, 100 ng each, were transfected into BHK cells alone or with 100 ng each of USF1, USF2, c-Myc, and Max expression vectors and combinations thereof. Fold transactivation by USF1, USF2, c-Myc, and Max was expressed with respect to the basal activity of each E-box construct alone. Data represent means of 3 independent experiments (± SEM). *P < .05; **P < .005; ***P < .001; determined by 1-way ANOVA and the Student-Newman-Keuls comparisons test.
Discussion
The interest in and importance of hepcidin is broad. To the physiologist, this hormone may be the answer to how the body controls and fine-tunes its iron requirements. To the clinician, hepcidin may be the panacea for iron overload disorders and might also provide clues for the treatment of the anemia of chronic disease. Yet, how this small peptide can provide all these answers remains undetermined, principally because we do not know enough about the regulatory mechanisms that underlie its expression. These mechanisms would help us understand how it can “switch” the body's iron supplies on and off. Recent data suggest an attractive feedback regulatory mechanism in which hepcidin may regulate iron absorption by binding to and controlling the constitutive levels and trafficking of ferroportin.17 However, the transcriptional regulators that govern hepcidin expression (with the exception of C/EBPα) are still unclear. On that basis we have identified members of the bHLH-ZIP family of transcriptional regulators as potentially significant modulators of hepcidin expression that consequently may serve an important auxiliary role an important role in iron homeostasis. Surprisingly, one of these transcription factors turned out to be USF2, which has been previously linked to iron metabolism through gene ablation in mice.
USF occurs in 2 isoforms of 43 kDa (USF1) and 44 kDa (USF2), encoded by 2 different genes.29-33 These transcription factors have a broad coverage of genes under their control, including glucose-responsive genes, the genes for ornithine decarboxylase, glycophorin B, transforming growth factor-β (TGF-β), vasopressin, actin, and cyclin B1, and genes that control the circadian rhythm (period, timeless, and clock); interestingly, β2-microglobulin, which plays a role in iron metabolism, is similarly regulated.34-44 The signature in all these genes is the E-box with the canonical sequence CACGTG (consensus, CANNTG). The nucleotide degeneracies within this element allow for combinatorial interactions between members of the bHLH-ZIP family of transcription factors. Consequently, this sequence is recognized by a large number of these proteins, including c-Myc, ARNT, Max, MyoD, AhR, Mxi1, TAL1, TFE3, and CLOCK.45-54 The binding specificities of these transcription factors to the cognate E-box are determined by other nucleotides juxtaposing the E-box core.26,27,55-57 Their ability to transactivate target genes is context dependent, and may also be tissue specific. In addition, their versatility enables cells to use different permutations of bHLH-ZIP members to control genes whose functions vary from cell-cycle control (eg, cyclin B1) to muscle cell differentiation (eg, MyoD) and cell death. While most of these transcription factors are tissue specific, others such as USF are ubiquitous.29 They also appear to function as heterodimers (eg, c-Myc/Max or AhR/ARNT). The association of (some) E-boxes with genes involved in rhythmicity is particularly intriguing, as it suggests that these genes may be transcribed in pulses. The glycolytic enzymes, circadian clock proteins, cyclin, and vasopressin, for instance, are under cyclical control and show robust expression in waves; in other words, these proteins are produced on demand. Our knowledge thus far of hepcidin expression indeed suggests that it may also be under pulsatile or rhythmic transcriptional control.
Site-directed mutagenesis of E-boxes in HepcP1.1-luc. All 4 E-boxes (Figure 2) were simultaneously mutated to produce HepcP1.1-lucMtE4. (A) Basal activities of both mutant and wild-type promoters were compared with the MannWhitney U test following transfection into BHK cells. ***P < .001. (B) Cotransfections of wild-type and mutant promoters with USF1/USF2 or (C) c-Myc/Max expression plasmids to determine transactivation levels. In panels B and C, ▦ and ▪ represent activation profiles for HepcP1.1-lucMtE4 and HepcP1.1-luc, respectively. Fold activation is a ratio of transactivation by USF1, USF2, c-Myc, and Max, and basal expression levels of the respective constructs. P values from pairwise comparison between transactivation of the wild-type or mutant promoters are indicated; these were determined by 1-way ANOVA/Student-Newman-Keuls test. Two independent experiments were performed; samples in each assay were run in duplicate. Data are presented as means ± SEM.
Site-directed mutagenesis of E-boxes in HepcP1.1-luc. All 4 E-boxes (Figure 2) were simultaneously mutated to produce HepcP1.1-lucMtE4. (A) Basal activities of both mutant and wild-type promoters were compared with the MannWhitney U test following transfection into BHK cells. ***P < .001. (B) Cotransfections of wild-type and mutant promoters with USF1/USF2 or (C) c-Myc/Max expression plasmids to determine transactivation levels. In panels B and C, ▦ and ▪ represent activation profiles for HepcP1.1-lucMtE4 and HepcP1.1-luc, respectively. Fold activation is a ratio of transactivation by USF1, USF2, c-Myc, and Max, and basal expression levels of the respective constructs. P values from pairwise comparison between transactivation of the wild-type or mutant promoters are indicated; these were determined by 1-way ANOVA/Student-Newman-Keuls test. Two independent experiments were performed; samples in each assay were run in duplicate. Data are presented as means ± SEM.
Promoter occupancy by c-Myc/Max and USF1/USF2 in vitro and in chromatin. (A) HepG2 cell nuclear extracts (10 μg) were incubated with radiolabeled E-boxes (including the mutant E-box, Emt); where indicated, excess unlabeled or cold competitor oligonucleotide (100 pmol) was added to the binding reactions. (B) Binding specificity was ascertained without or with USF1 and USF2 antibodies in a supershift assay. Arrows indicate E-box–nucleoprotein complexes; NS, nonspecific-binding nuclear components. **Supershifted complexes. (C) Binding of in vitro–translated USF1/USF2 and c-Myc/Max heterodimers to the E-boxes. L indicates E-box2 incubated with untranslated rabbit reticulocyte lysate as negative control; 1, E-box2 incubated with USF1/USF2 heterodimer; 2, E-box2 incubated with c-Myc/Max heterodimer; and 3, Emt incubated with c-Myc/Max heterodimer. Arrows show complexes of E-box and USF1/USF2 or c-Myc/Max heterodimers. (D) Arrangement of the 2 canonical E-boxes within ChIP target DNA; PCR primers (arrows) were chosen to amplify a 300-bp fragment of the promoter encompassing nucleotide sequences from –642 to –343. (E) ChIP assay. PCR was performed on whole chromatin without immunoprecipitation (Input) and on chromatin immunoprecipitated with either a nonspecific antibody (ns), or with (+) antibodies to c-Myc/Max together, USF1, and USF2. M indicates DNA molecular size marker. 2xE-box-luc was used as positive control (“C”) in PCR.
Promoter occupancy by c-Myc/Max and USF1/USF2 in vitro and in chromatin. (A) HepG2 cell nuclear extracts (10 μg) were incubated with radiolabeled E-boxes (including the mutant E-box, Emt); where indicated, excess unlabeled or cold competitor oligonucleotide (100 pmol) was added to the binding reactions. (B) Binding specificity was ascertained without or with USF1 and USF2 antibodies in a supershift assay. Arrows indicate E-box–nucleoprotein complexes; NS, nonspecific-binding nuclear components. **Supershifted complexes. (C) Binding of in vitro–translated USF1/USF2 and c-Myc/Max heterodimers to the E-boxes. L indicates E-box2 incubated with untranslated rabbit reticulocyte lysate as negative control; 1, E-box2 incubated with USF1/USF2 heterodimer; 2, E-box2 incubated with c-Myc/Max heterodimer; and 3, Emt incubated with c-Myc/Max heterodimer. Arrows show complexes of E-box and USF1/USF2 or c-Myc/Max heterodimers. (D) Arrangement of the 2 canonical E-boxes within ChIP target DNA; PCR primers (arrows) were chosen to amplify a 300-bp fragment of the promoter encompassing nucleotide sequences from –642 to –343. (E) ChIP assay. PCR was performed on whole chromatin without immunoprecipitation (Input) and on chromatin immunoprecipitated with either a nonspecific antibody (ns), or with (+) antibodies to c-Myc/Max together, USF1, and USF2. M indicates DNA molecular size marker. 2xE-box-luc was used as positive control (“C”) in PCR.
Our in vitro data strongly suggested that both mouse and human hepcidin genes are regulated by USF1/USF2 and c-Myc/Max heterodimers, and we envisage that this may also apply in vivo. In our detailed analyses of the human hepcidin gene promoter we found that transactivation from the E-boxes by either c-Myc/Max or USF1/USF2 heterodimers was nonadditive (ie, these enhancer elements are functionally redundant). Although the transfection data showed that each transcription factor (eg, USF1 or c-Myc) could transactivate the promoter independently of either member of the couple (ie, USF2 or Max), it is more likely that they operate as heterodimers within the cell. For example, constitutively expressed or endogenous USF2 may dimerize with ectopically expressed USF1, and vice versa. This may be true for a number of reasons. Deletion mapping showed that the E-boxes coincided with that portion of the promoter with maximal basal activity (ie, even when neither USF isoform was cotransfected with the promoter construct). Furthermore, site-directed mutagenesis of all the E-boxes caused a significant decrease in basal and USF1/USF2 or c-Myc/Max transactivation of the promoter. These observations therefore suggest some contribution from endogenous USF1/USF2 or c-Myc/Max in promoter transactivation.
Results accrued from the study of Usf knockout mice also suggest a complex relationship between the 2 isoforms in vivo in a manner that may or may not affect iron metabolism. Some of these mice (referred to as the Paris Usf2–/– mice) did not express hepcidin, and consequently exhibited extensive iron overload even though both hepcidin genes were intact. These mice were generated with a Usf2 allele in which nucleotides (in exon 7) encoding the basic DNA-binding domain had been disrupted, but this allele still retained the region encoding the HLH-ZIP domain, which is required for heterodimer formation.10,12 However, in a different Usf2 knockout mouse model, the Houston Usf2–/– mouse, iron metabolism was apparently normal. In this model a null Usf2 allele was generated by deleting all of the exons and some of the 5′ noncoding regions of Usf2; interestingly, these mice concomitantly expressed attenuated levels of Usf1. In contrast, in the same study it was found that Usf1 knockout mice expressed increased levels of Usf2 to compensate for Usf1 loss.58 Another Usf1 knockout mouse model was generated by deleting exons 4 to 9 together with part of exon 10 (ie, the DNA binding and heterodimerization [HLH-ZIP] as well as activation domains of Usf1 were deleted)11 ; iron metabolism was apparently normal in those mice. Taken together, these observations suggest 2 points: (1) that there may be epistatic interactions between Usf1 and Usf2; and (2) depending on how these genes are targeted and ablated, they may become null or dominant-negative inhibitors. This is particularly important since our data suggests a cis-acting effect of Usf2 on hepcidin expression. Because the hepcidin genes and Usf2 juxtapose each other (the 3′ untranslated region of Usf2 overlaps the promoter region of both mouse as well as human hepcidin genes), targeting the mouse Usf2 locus would consequently have a polar or positional effect on hepcidin expression. For example, chromatin remodeling and changes in nucleosome positioning as a result of the mutant Usf2 transgene integration may inhibit hepcidin expression by occluding important transcription factor–binding sites that are required for basal expression of the gene.
However, our study suggests an alternative and plausible explanation: that dominant-negative Usf2 (and Usf1) mutants may inhibit run-on transcription of the hepcidin gene to precipitate the deranged iron metabolism observed in some Usf2 knockout mice. This assertion may be true because we found that at least in vitro, a dominant-negative USF2 mutant, USF2ΔB, abrogated both mouse and human hepcidin gene promoter transactivation by USF1 and USF2; the dominant-negative USF1 mutant A-USF had similar reciprocal effects. USF2ΔB lacks the DNA-binding domain encoded by exon 7 (Figure 8A) but can heterodimerize with other USF isoforms through its (intact) HLH-ZIP domain. Similarly, A-USF has a defective DNA-binding domain because the basic region of USF1 has been replaced with an acidic motif that confers increased stability between this mutant and endogenous USF1 and USF2.22 As more than 66% of USF1 and USF2 exists in vivo as heterodimers,32 it is possible that they may be sequestered and transcriptionally crippled by endogenous dominant-negative mutants were these to be expressed. This proposed mechanism of transcriptional repression of hepcidin expression (Figure 8B, bottom panel) may be supported not only by our in vitro data but also by previous work that showed that the deletion or interruption of the DNA-binding domain (exon 7) of Usf2 converted it into a dominant-negative mutant.22-25 Our data may also partially unify conflicting observations of differences in iron metabolism in the various Usf2–/– mice described earlier in this section. Although this proposition is attractive, it is highly unlikely that these transcription factors have an overarching control on hepcidin expression since a number of disparate effectors or stimuli influence hepcidin levels.
Proposed mechanisms of transcriptional activation or repression of the hepcidin gene by USF1/USF2 and their dominant-negative mutants. (A) A simplified depiction (not to scale) of the exon-intron arrangement of Usf2 and its deletion mutants. E1 to E10 are the 10 exons of the gene. BR indicates basic DNA-binding domain; HLH, helix-loop-helix domain; and LL, leucine zipper domain. The DNA-binding domain of Usf2 encoded by E7 has been interrupted (▾) in the Paris Usf2 knockout mouse and in the dominant-negative mutant USF2ΔB. (B) Model of transactivation or repression. In transactivation, USF2 (expressed upstream of hepcidin) forms heterodimers with USF1 through their HLH-ZIP domains. Following recruitment of coactivator complexes (eg, the adenovirus E1A-binding protein p300/CREB-binding protein [CBP]), the heterodimer binds to the E-boxes (designated E-1 to E-4) to drive hepcidin transcription (top panel). However, in the presence of a dominant-negative mutant (A-USF or USF2ΔB), USF2 (or USF1) may form stable but transcriptionally crippled heterodimers (shown with a clip) that are unable to recruit coactivator complexes; hepcidin transcription is therefore stalled (bottom panel). *USF2 stop codon.
Proposed mechanisms of transcriptional activation or repression of the hepcidin gene by USF1/USF2 and their dominant-negative mutants. (A) A simplified depiction (not to scale) of the exon-intron arrangement of Usf2 and its deletion mutants. E1 to E10 are the 10 exons of the gene. BR indicates basic DNA-binding domain; HLH, helix-loop-helix domain; and LL, leucine zipper domain. The DNA-binding domain of Usf2 encoded by E7 has been interrupted (▾) in the Paris Usf2 knockout mouse and in the dominant-negative mutant USF2ΔB. (B) Model of transactivation or repression. In transactivation, USF2 (expressed upstream of hepcidin) forms heterodimers with USF1 through their HLH-ZIP domains. Following recruitment of coactivator complexes (eg, the adenovirus E1A-binding protein p300/CREB-binding protein [CBP]), the heterodimer binds to the E-boxes (designated E-1 to E-4) to drive hepcidin transcription (top panel). However, in the presence of a dominant-negative mutant (A-USF or USF2ΔB), USF2 (or USF1) may form stable but transcriptionally crippled heterodimers (shown with a clip) that are unable to recruit coactivator complexes; hepcidin transcription is therefore stalled (bottom panel). *USF2 stop codon.
Diseases associated with USF1 or USF2 have been limited, but recent data identified USF1 as the prime candidate for susceptibility to familial combined hyperlipidemia, an inherited disorder that presents with abnormal blood lipid levels and type 2 diabetes.59 Thus, USF1 has also been suggested to contribute to the metabolic syndrome by virtue of its transcriptional control of genes that are associated with hypertension and diabetes.60 Our identification of this transcription factor as an integral part of the transcriptional circuitry controlling iron flux through hepcidin may therefore also add a new perspective to its role in cellular (patho)physiology. That the c-Myc/Max transcription couple is a coregulator of hepcidin transcription, albeit modest in comparison with USF1 and USF2, is also consistent with the role of c-Myc in controlling iron levels by coordinating the expression of iron-regulatory protein 2 and ferritin heavy chain.61 Our data may therefore provide some insight into how this couple (ie, c-Myc/Max) may control cell-cycle progression, growth, and transformation by regulating cellular iron pools through hepcidin. Taken together, these observations show a direct role for these members of the bHLH-ZIP family of transcription factors in regulating hepcidin expression and, by default, iron metabolism and other pathways in which hepcidin may play a part such as in inflammation and cellular defense. We suggest that cross talk (or defects thereof) between these transcriptional regulators and possibly with others, such as C/EBPα, may underlie the many functions of hepcidin. This cross talk may involve transcriptional synergy, autoregulatory feedback loops, or antagonism between the various transactivators. Intriguingly C/EBPα has been shown to up-regulate USF1, which in turn transactivates C/EBPα.62 It would therefore appear that transcriptional and posttranslational feedback loops17 may be the defining mechanism(s) by which hepcidin controls iron flux.
Authorship
Contribution: H.K.B. designed research, performed experiments, analyzed the data, and wrote the paper; H.M. contributed vital new reagents and analytical tools; and S.K.S.S. designed research, analyzed the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, 8/10/2006; DOI 10.1182/blood-2005-07-027037.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Michele Sawadogo for USF1 and USF2 expression plasmids, H. T. Marc Timmers for pTM3-His-cMyc and pTM3-Max, Charles Vinson for A-USF, and L. Karl Olson for USF2ΔB. We also thank H. T. Marc Timmers for an anti-Max antibody.
This work was supported by the Wellcome Trust, the Medical Research Council, and the Charles Wolfson Trust (H.K.B.).

![Figure 1. Deletion mapping of the human hepcidin gene promoter. (A) HepcP1.1-luc (wild-type [WT]) was selectively restricted with enzymes to remove various segments. Constructs were generated by restriction with BalI(ΔBal; –598 to –379), PmlI(ΔPml; –526 to –454), NheI-AflII (ΔNA; –972 to –821), AflII-PmlI(ΔAP; –821 to –454), NheI-PmlI (ΔNP; –972 to –454), and NheI-TthIIII (ΔNT; –972 to –196). Nucleotide numbering is based on Courselaud et al.21 Note that the NheI restriction site was used for cloning purposes only and is not intrinsic to this segment of the promoter (PCR primers in “Materials and methods”). Also, although there are 2 TthIIII sites within the promoter, double-digestion with NheI removed all but approximately 200 bp of the promoter. (B) Basal transcriptional activity of promoter deletion constructs from panel A. Constructs (100 ng each) were cotransfected with pSVβgal (50 ng) into BHK cells for 48 hours, and reporter activities were measured as described above. Luciferase levels were normalized with respect to βgal activity. Fold activation was based on the activity of pGL3Basic, and assigned an arbitrary activation level of 1. Pairwise comparisons were made between WT and deletion constructs using the 1-way ANOVA/Student-Newman-Keuls test. **P < .005. Data are representative of 4 independent sets of transfections, shown as means ± SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2005-07-027037/8/m_zh80240605120001.jpeg?Expires=1765960156&Signature=CEmrMTmEM35cSh63iecMn3Tqtmxu5JXTuEYten6fQ3NjcLMLDWoEwxlIvv6qkCxTcaaG3Hl~3OQnvMvngPM9i1F~wn5yVarR7vDAsW5ixj6P853zMLO2HpfDupZo0PMc3uWCOiq~S5JX8Hb6Vf4Ge5qUvJbLDa8NpgAAhq1kb6S~fHfkpapRxwy2DAB043ufzvs0YeCQZtxbi1KZwUA85MQTL48pC69lbq5cI0RHguCojBYUaUAL89YnZrrlDaO7f6QZ2DlQoxv8qyGyz07bRAVyUm~e46KdHOcBJj0PxocRNmzLPfT6xbbM9~TyYB2k1NFdNWfbn7gWX-Y1St2GAQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 8. Proposed mechanisms of transcriptional activation or repression of the hepcidin gene by USF1/USF2 and their dominant-negative mutants. (A) A simplified depiction (not to scale) of the exon-intron arrangement of Usf2 and its deletion mutants. E1 to E10 are the 10 exons of the gene. BR indicates basic DNA-binding domain; HLH, helix-loop-helix domain; and LL, leucine zipper domain. The DNA-binding domain of Usf2 encoded by E7 has been interrupted (▾) in the Paris Usf2 knockout mouse and in the dominant-negative mutant USF2ΔB. (B) Model of transactivation or repression. In transactivation, USF2 (expressed upstream of hepcidin) forms heterodimers with USF1 through their HLH-ZIP domains. Following recruitment of coactivator complexes (eg, the adenovirus E1A-binding protein p300/CREB-binding protein [CBP]), the heterodimer binds to the E-boxes (designated E-1 to E-4) to drive hepcidin transcription (top panel). However, in the presence of a dominant-negative mutant (A-USF or USF2ΔB), USF2 (or USF1) may form stable but transcriptionally crippled heterodimers (shown with a clip) that are unable to recruit coactivator complexes; hepcidin transcription is therefore stalled (bottom panel). *USF2 stop codon.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2005-07-027037/8/m_zh80240605120008.jpeg?Expires=1765960156&Signature=kgJio4w4BgUBc~BDMHz4IxA-~Rt2Yb1kRncoE0UIdsaMloM6xg8UGQbLrg0nB1CgP3kViviy5OlcT7D3nOrEpG7-31rjMFXYkpyYJ02rkXGEa9nmfS27KLDMfHa20B1qtEVtABGyRN9D3LxlCrIOfCPtN1NED51iUfSiH~VRHBBqPKTFsWCqoLHy~SazSAOMY2e9mCXf3LzT73mw7xgx7OCjlkt26knuFkpZvhcYDFoFI03WON0RuYWNla4GOYrZiDmA4h2chE0pBNNIaK6~Il1wa9lH4W6BYUGECL8RAvgpOP-sW8dVS9C92Fmpdolt0T1DaSy~6EBV1y5glcTytg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal