Abstract
Somatic mutation of PIGA in hematopoietic stem cells causes deficiency of glycosyl phosphatidylinositol–anchored proteins in paroxysmal nocturnal hemoglobinuria (PNH) that underlies the intravascular hemolysis but does not account for expansion of the PNH clone. Immune mechanisms may mediate clonal selection but appear insufficient to account for the clonal dominance necessary for PNH to become clinically apparent. Herein, we report 2 patients with PNH whose PIGA-mutant cells had a concurrent, acquired rearrangement of chromosome 12. In both cases, der(12) had a break within the 3′ untranslated region of HMGA2, the architectural transcription factor gene deregulated in many benign mesenchymal tumors, that caused ectopic expression of HMGA2 in the bone marrow. These observations suggest that aberrant HMGA2 expression, in concert with mutant PIGA, accounts for clonal hematopoiesis in these 2 patients and suggest the concept of PNH as a benign tumor of the bone marrow.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is a consequence of nonmalignant clonal expansion of hematopoietic stem cells with somatic mutation of PIGA.1 Mutant PIGA2 explains the deficiency of glycosyl phosphatidylinositol–anchored proteins (GPI-APs) that underlies the intravascular hemolysis of PNH.3 However, PIGA-mutant stem cells have no intrinsic proliferative advantage,4,5 suggesting a 2-step model of pathogenesis.
Step 1 of this model, clonal selection,6,7 is envisioned as a conditional survival advantage that depends on deficiency of 1 or more GPI-APs. The close association of PNH with aplastic anemia, suggests that the selection pressure is immune mediated.6,7 But, although 60% to 70% of patients with aplastic anemia have small, subclinical populations of GPI-AP– hematopoietic cells at diagnosis,8 only 10% to 15% subsequently develop clinically apparent PNH.9 In the remainder, GPI-AP– cells persist subclinically or disappear,8 suggesting that mutant PIGA (and the consequent deficiency of GPI-APs) is necessary for clonal selection but is insufficient to account for the clonal expansion required for clinical manifestations of PNH to become apparent.
Clonal expansion, step 2 of the PNH pathogenesis model, is envisioned as a consequence of clonal evolution in which a second somatic mutation bestows on the PIGA-mutant stem cell a proliferative advantage.10 Herein, we present evidence supporting this 2-step model by showing a concurrent, acquired genetic abnormality in the PIGA-mutant cells of 2 patients that establishes a novel mechanism for the nonmalignant clonal hematopoieis characteristic of PNH.
Patients, materials, and methods
Patients
Informed consent was obtained from patients J20 and US1 according to protocols approved by the Institutional Review Boards of Osaka University Hospital (Osaka, Japan) and the University of Utah School of Medicine (Salt Lake City, UT), respectively.
Hybrid cell lines
Monocytes derived from J20 or US1 were fused with the hypoxanthine phosphoribosyltransferase–negative mouse myeloma cell line, P3-X63-Ag8.653, as previously described.11 Lines carrying human chromosome 12 were selected by analyses of expression of both CD9 and polymorphic markers D12S77 and D12S78. The B-lymphoblastoid cell line JY2512 was used as a control in some experiments.
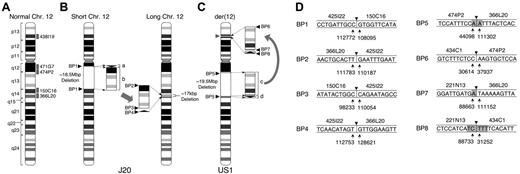
Chromosomal abnormalities in 2 patients with PNH. (A) Idiogram of normal chromosome 12 (Chr 12) modified from the NCBI Map viewer. Labeled gray boxes indicate the positions of the designated BAC clones used for FISH analysis. (B) J20. The karyotypic abnormality identified in the GPI-AP– bone marrow cells of J20 was defined as an interchromosomal insertion. An 18.5M-bp region from q12 to q14 (surrounded by broken lines) is deleted in short chromosome 12 as defined by characterization of BP1. The 2.7-kbp small fragment (bracketed arrowhead, labeled a) and the 18.5-Mbp large fragment (bracketed rectangle, labeled b) deleted from short chromosome 12 are inserted inversely and directly, respectively, into the 12q14 region of long chromosome 12 generating BP2, BP3, and BP4. The deleted region into which the 2 fragments are inserted lacks 17 kbp of sequence (broken lines). BP1, BP2, BP3, and BP4 indicate the breakpoint junctions generated by the chromosomal abnormality. (C) US1. The karyotypic abnormality identified in the bone marrow cells of US1 was defined as an intrachromosomal insertion. The large fragment (19.5 Mbp, labeled c) and the small fragment (300 kbp, labeled d) are inserted into the TEL locus (gray arrowhead) on 12p13. BP5 is generated by the deleted region. BP6, BP7, and BP8 are generated by rearranged fragments c and d. (D) Sequences of BP junctions in J20 and US1. The sequences around BP junctions 1 to 8 are shown. BAC clones containing the sequence are denoted above the lines. Arrows indicate the nucleotide numbers of the BAC clones. Arrowheads indicate one of the candidate breakpoints, and gray regions indicate ambiguous sequences shared between the 2 BAC clones at the site of the breakpoint.
Chromosomal abnormalities in 2 patients with PNH. (A) Idiogram of normal chromosome 12 (Chr 12) modified from the NCBI Map viewer. Labeled gray boxes indicate the positions of the designated BAC clones used for FISH analysis. (B) J20. The karyotypic abnormality identified in the GPI-AP– bone marrow cells of J20 was defined as an interchromosomal insertion. An 18.5M-bp region from q12 to q14 (surrounded by broken lines) is deleted in short chromosome 12 as defined by characterization of BP1. The 2.7-kbp small fragment (bracketed arrowhead, labeled a) and the 18.5-Mbp large fragment (bracketed rectangle, labeled b) deleted from short chromosome 12 are inserted inversely and directly, respectively, into the 12q14 region of long chromosome 12 generating BP2, BP3, and BP4. The deleted region into which the 2 fragments are inserted lacks 17 kbp of sequence (broken lines). BP1, BP2, BP3, and BP4 indicate the breakpoint junctions generated by the chromosomal abnormality. (C) US1. The karyotypic abnormality identified in the bone marrow cells of US1 was defined as an intrachromosomal insertion. The large fragment (19.5 Mbp, labeled c) and the small fragment (300 kbp, labeled d) are inserted into the TEL locus (gray arrowhead) on 12p13. BP5 is generated by the deleted region. BP6, BP7, and BP8 are generated by rearranged fragments c and d. (D) Sequences of BP junctions in J20 and US1. The sequences around BP junctions 1 to 8 are shown. BAC clones containing the sequence are denoted above the lines. Arrows indicate the nucleotide numbers of the BAC clones. Arrowheads indicate one of the candidate breakpoints, and gray regions indicate ambiguous sequences shared between the 2 BAC clones at the site of the breakpoint.
Determination of chromosomal breakpoints
Initial mapping of breakpoints required a combination of polymerase chain reaction (PCR), inverse PCR, and Southern blotting. For fine mapping, sequence-tagged site markers were generated by PCR using primers based on sequences of ends of bacterial artificial chromosome (BAC) clones or on data in the human genome database of the National Center for Biotechnical Information. GenBank accession numbers of BAC clones (CHORI BACPAC Resource Center) are as follows: RP11-425I22 (425I22), AC074030; RP11-471G7 (471G7), AC024935; RP11-150C16 (150C16), AC046129; RP11-366L20 (366L20), AC090673; RP11-474P2, AC025031; RP11-221N13, AC090023; RP11-434C1, AC007450; RP11-438I19 (438I19), N0438I19.
PCR primers used to amplify breakpoints
PCR primers used to amplify junction sequences of the breakpoints in J20 were as follows: breakpoint (BP) 1, CTTATGTCTCACTTGGGCAC (108462-108443 in 150C16) and CCTTCACTTCACTTGTTAGC (113076-113057 in 425I22); BP2, TTCCTACAGAGCCAAAATGCCA (111355-111376 in 366L20) and ACTGCAACACCTCTCTAGCAG (109879-109899 in 425I22); BP3, TTGAACCCTTTGCCATTACGT (111047-111027 in 425I22) and TATTTAACACCTATCTGACTCC (97995-98016 in 150C16). Primers, GTGCCCAAGTGAGACATAAG (108443-108462 in 150C16) and TGTTGACTGAGCCCCATGAT (108598-108579 in 150C16), were used for positive control PCR.
Junction sequences of the breakpoints in US1 were confirmed by PCR with the following primer mixture to amplify both normal and abnormal alleles at the same time: BP5, CCAAAAGTGGGCTTACACATAAAA (44148-44125 in 474P2), TTCGCTCCTCCCACCTCATA (primer A, 111517-111498 in 366L20), and ACTCCCTGTAGTGAATCCTCTGTTTAGA (primer B, 111043-111070 in 366L20); BP6, GCCCGGGTTAATGTCGCTGAAT (37462-37483 in 474P2), GTTGGGGGTGGGGACAAAATG (primer C, 30520-30540 in 434C1), and CGTTGGCAAAGCAGGGTTCCT (primer D, 31301-31281 in 434C1); BP7, GAAGCTCCAACTTTGCCCTCTG (88565-88586 in 221N13), primer B and primer A; and BP8, GGGCAGTTGGAATTGGGGAGAT (89202-89181 in 221N13), primer D and primer C.
Fluorescence in situ hybridization (FISH)
BAC clones 471G7, 150C16, and 366L20 were used as probes against hybrid cell lines derived from J20 and 438I19; 474P2 and 366L20 were used against bone marrow cells from US1. Signals were detected with biotinylated BAC probes and fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories, Burlingame, CA) and amplified by using biotinylated antiavidin antibody (Vector Laboratories) and FITC-conjugated avidin. Chromosomes were stained with propidium iodide and mounted with glycerol-based medium containing 1,4-diazabicyclo(2,2,2)octane antifade. The chromosomal localization was captured through a PlanApochromat objective lens (63×/1.4 NA oil objective) using a Zeiss laser-scanning LSM510 microscope (Carl Zeiss, Jena, Germany) for J20 samples and through a Leica HCS PL FLUOTAR objective lens (100×/0.6-1.3 NA oil objective) using a Leica fluoromicroscope DM RXA2 with a Leica DC350F digital camera (Leica Microsystems, Wetzlar, Germany) for US1. Images were analyzed using Zeiss LSM510 software or Leica QFluoro software, respectively, and were processed with Adobe Photoshop (Adobe Systems, San Jose, CA).
Real-time PCR
Marrow samples were collected from J20 and US1, and control samples from healthy donors were purchased from a commercial vendor (Cambrex, Walkersville, MD). CD59– cells were isolated from the J20 sample by cell sorting (Becton Dickinson, San Jose, CA). Random hexamer-primed RNA from these samples was reverse transcribed using Superscript III (Invitrogen, Carlsbad, CA).
On the basis of the sequence of HMGA2, the following PCR primers and TaqMan MGB probe were designed: forward primer, 5′-TTCAGCCCAGGGACAACCT (located in exon 1); reverse primer, 5′-TCTTGTTTTTGCTGCCTTTGG (located in exon 2); TaqMan MGB probe, 6-carboxyfluorescein (FAM)–AGCAAGAACCAACCGGT–non-fluorescent quencher–Minor groove binder (MGB) (the sequence of the probe spanned the boundary between exons 1 and 2) (Applied Biosystems, Foster City, CA). The method for amplifying β-glucuronidase cDNA as an internal control has been published.13 PCR was performed using QuantiTect Multiplex PCR Kit (QIAGEN, Valencia, CA) on an ABI 7900HT sequence detection system according to the manufacturer's instructions (Applied Biosystems).
Measurement of allele frequency
Real-time PCR was used to measure the ratio of der(12) to normal chromosome 12 in hematopoietic cells of US1. Primers were designed around one of the breakpoints for detection of der(12). For detection of the wild-type allele, a region deleted in der(12) was analyzed. To measure the frequency of mutant PIGA, exon 2 was amplified, and PCR products were cloned into pGEM-Teasy vector (Promega, Madison, WI) and sequenced.
Analysis of allele-specific expression of HMGA2
The previously described polymorphic marker in the 5′ UTR of HMGA2 was used to determine allele-specific gene expression in bone marrow cells14 using the following primers: 5′-GACCCTATCCCGGCGGAGTCTC and 5′-TTGAAATGTTAGGCGGGGAAAGAA. DNA from hybridoma cell lines carrying one chromosome 12 was used to determine the origin of each allele. PCR fragments were separated by 15% to 25% gradient polyacrylamide gel electrophoresis (PAGmini; Daiichi Pure Chemicals, Tokyo, Japan).
Results
Patients
At age 33, J20 presented with pancytopenia and a hypocellular marrow without karyotypic abnormalities.15 Five months later, an abnormal karyotype, 46, XX, t(12;12)(q13;q15), was reported in 3 (14%) of 21 metaphase cells. One year after diagnosis, blood counts were essentially normal, and marrow analysis showed normal cellularity with mild erythroid dysplasia. Cytogenetic analysis indicated expansion of the mutant clone with 10 (50%) of 20 metaphase cells having the abnormal karyotype. Laboratory evidence of hemolysis was noted, and flow cytometry showed 55% to 60% GPI-AP– erythrocytes. A mutation in exon 2 of PIGA (G715A) was shown in patient neutrophils.15 Marrow mononuclear cells were separated into GPI-AP+ and GPI-AP– populations, and the abnormal karyotype was found only in GPI-AP– cells.15 These findings established the somatic nature of both the PIGA mutation and the chromosome 12 rearrangement and suggested that both genetic abnormalities were involved in the pathogenesis of clonal hematopoiesis.
US1 was 31 years old at presentation with complaints of fatigue and dark urine. Her white blood cell count was 4.1 × 109/L (4100/μL), hemoglobin level was 38 g/L (3.8 g/dL), and platelet count was 171 × 109/L (171 000/μL). Laboratory studies indicated intravascular hemolysis, and flow cytometry showed 88% GPI-AP– neutrophils. Marrow analysis revealed normal cellularity with erythroid hyperplasia without dysplasia and a karyotype of 46, XX, ins(12)(p12∼13q13q12) in 20 (100%) of 20 metaphases. FISH showed the insertion split the TEL locus at 12p13. The abnormal karyotype was identified in 23% of mitogen-stimulated lymphocytes. A 14-bp deletion in the 3′ end of exon 2 of PIGA (693-706) was identified in neutrophil DNA. These findings confirmed the somatic nature of both the der(12) and mutant PIGA in US1.
Chromosomal abnormalities
Hybrid cell lines between patient monocytes and mouse myeloma cells were established. From J20, 2 lines (S1 and S2) carrying short chromosome 12 and 2 lines (L1 and L2) carrying long chromosome 12 (Figure 1B) were developed.11 From US1, hybrid cells carrying normal chromosome 12 (US1W) or der(12) (US1M) were developed (Figure 1A,C). Chromosomal abnormalities (Figure 1) were delineated by using a combination of PCR analysis based on sequence-tagged site markers, FISH (Figure 2), Southern blotting, and inverse PCR (not shown).
For J20, the abnormality was defined as insertion of an 18.5-Mbp fragment derived from one chromosome 12 (short chromosome 12) into the other (long chromosome 12) (Figure 1B). The small fragment (a) and the large fragment (b) derived from the deleted region of short chromosome 12 were inversely and directly, respectively, inserted into 12q14 of long chromosome 12, generating BP2, BP3, and BP4 (Figure 1B).
For US1, the abnormality was an intrachromosomal insertion. The large fragment (19.5 Mbp, labeled c) and the small fragment (300 kbp, labeled d) derived from a region deleted from 12q13q14 are inserted inversely and directly, respectively, into the TEL locus on 12p13 (Figure 1C).
A combination of Southern blotting (not shown), PCR (Figure 2A-B), and FISH (Figure 2C-D) were used to confirm that breakpoints that defined the karyotypic abnormalities were present in peripheral blood and bone marrow of J20 and US1.
For J20, only GPI– cells had the chromosome 12 abnormality.15 To investigate whether cells of US1 were also double mutants, the percentage of marrow cells with der(12) and mutant PIGA was quantitated. Mutant PIGA and der(12) were found in 91.8% and 93.9%, respectively, of bone marrow cells, indicating that the 2 mutations coexisted in the same cells. Together, these observations show that clonal hematopoiesis in these 2 patients is derived from a hematopoietic stem cell with somatic mutations of both PIGA and chromosome 12.
Effects of chromosome 12 abnormalities
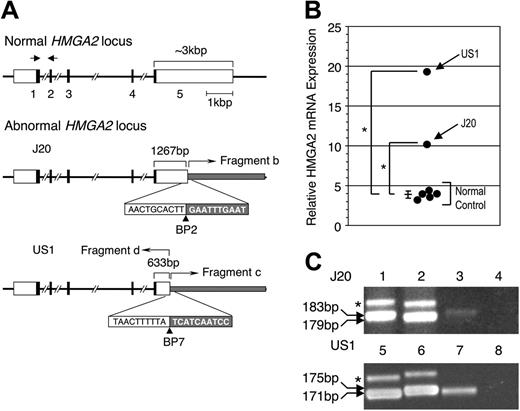
Although the molecular details are different (Figure 1), the result of the chromosome 12 rearrangements is almost identical for the 2 patients, because in both cases, HMGA2 is disrupted in the 3′ UTR of exon 5 (Figure 3A). No other effects of chromosome 12 rearrangement were detected for either J20 or US1. For US1, chimeric transcripts derived from TEL and HMGA2 were not detected, and TEL transcripts appeared normal both quantitatively and qualitatively (not shown).
Real-time PCR showed that relative expression of HMGA2 in bone marrow cells of both J20 and US1 was greater than normal (Figure 3B). In addition to rearranged HMGA2, both J20 and US1 have one intact HMGA2 locus (Figure 1). To determine the allelic origin of HMGA2 expression, a polymorphic region in the 5′ UTR14 was analyzed. For both patients, HMGA2 expression was derived almost exclusively from the rearranged locus (Figure 3C).
Discussion
These studies showed, in PIGA-mutant cells of 2 patients, rearrangement of chromosome 12 (Figures 1, 2) that resulted in ectopic expression of HMGA2 (Figure 3). The findings identify for the first time a molecular mechanism for clonal expansion of hematopoiesis in PNH.
HMGA2 is a member of the high-mobility group of proteins (HMGA1a, HMGA1b, HMGA2) that function as architectural transcription factors.16-18 HMG members possess no intrinsic transcriptional activity. Instead, these nonhistone proteins orchestrate assembly of stereospecific transcriptional regulatory proteins into enhanceosomes.18,19 The cellular targets of HMGA2 are incompletely defined but appear to include cyclin A.19
Molecular studies established a causal role for HMGA2 in benign mesenchymal tumors.17,20 Rearrangement of 12q13-15 is observed in these neoplasms, but tumorigenesis does not depend on generation of chimeric proteins derived from fusion of HMGA2 with specific translocation partners. Rather, clonal expansion induced by HMGA2 appears to result from deregulated expression of a truncated version of the protein.21-23 For the 2 patients with PNH, ectopic expression appears to be a consequence of gain-of-function mutational events (Figure 3B) caused by disruption of the 3′ UTR (Figure 3A) shown to contain elements that negatively regulate HMGA2 transcription.24 This hypothesis is supported by experiments showing that HMGA2 transcripts from marrow of J20 and US1 are derived almost exclusively from the rearranged alleles (Figure 3C). Additional studies will be required to determine whether aberrant expression of HMGA2 underlies clonal expansion in patients with PNH without structural abnormalities of 12q13-15.
PNH manifests many of the characteristics of a benign tumor because there is limited expansion of PIGA mutant clones (the peripheral blood of patients is a relatively stable mosaic of normal and abnormal cells), PIGA-mutant cells respect tissue boundaries (there is no invasion of nonhematopoietic tissues), PIGA-mutant cells respond appropriately to signals that normally regulate hematopoiesis (function is not autonomous) and transformation into acute leukemia occurs rarely (PNH is not a premalignant condition).25 Our studies suggest the concept of PNH as a benign tumor of the bone marrow with aberrant expression of HMGA2 acting in concert with mutant PIGA (and the consequent deficiency of GPI-APs) to produce the proliferative phenotype that underlies clonal expansion. However, our studies neither establish the sequence of events that culminated in the clonal outgrowth of the double mutant cells nor define how the aberrant expression of HMGA2 works additively or synergistically with mutant PIGA to produce the proliferative phenotype. This latter issue will be the subject of future studies.
Confirmation of breakpoint junctions generated by the chromosomal abnormalities. (A) Confirmation of the breakpoints in J20 by PCR. Genomic DNA from L1 (lane 1), L2 (lane 2), S1 (lane 3), S2 (lane 4), JY25 (the wild-type control) (lane 5), white blood cells (WBCs) from J20 (lane 6), WBCs of a healthy volunteer (lane 7), or no DNA template (negative control, lane 8) was used for PCR analysis using primer sets designed according to the flanking regions of BP1, BP2, and BP3 (illustrated in Figure 1). Both the integrity and quantity of the DNA templates were confirmed by PCR using control primers (labeled Control). That appropriate-sized PCR products were amplified using DNA derived from the circulating WBCs of J20 (lane 6) shows that both the S1 and L1 versions of chromosome 12 were present in vivo. (B) Confirmation of the breakpoints in US1 by PCR. Genomic DNA from WBCs of a healthy volunteer (lane 1), bone marrow cells of US1 (lane 2), PMN of US1 (lane 3), US1W (the hybrid cell line containing wild-type chromosome 12) (lane 4), or US1M [the hybrid cell line containing (der(12)] (lane 5), or no DNA template (negative control, lane 6) were used for PCR analysis using primer sets designed according to the sequence of flanking regions of the BP5, BP6, BP7, and BP8 (illustrated in Figure 1). These experiments show that both wild-type chromosome 12 and der(12) were present in the peripheral blood and bone marrow of US1. (C) Metaphase FISH showing the chromosomal abnormality in J20. BAC probes 471G7, 150C16, and 366L20 were hybridized against chromosomal specimens derived from the cell line (L1) that contains only long chromosome 12. Two hybridization signals (arrows) were detected with all 3 BAC probes, confirming that long chromosome 12 contained the inserted material deleted from short chromosome 12. (D) Metaphase FISH showing the chromosomal abnormality in US1. BAC probes 366L20, 438I19, and 474P2 (illustrated in Figure 1) were hybridized with chromosomal specimens derived from bone marrow cells of US1. Each sample contained a der(12) (indicated by 2 hybridization signals on the same chromosome, double arrows) and a wild-type chromosome 12 (indicated by one hybridization signal, arrow). The intrachromosomal insertion splits the signal on 12p generated by hybridization of probe 438l19, whereas signals are generated on 12q and 12p when probes that overlap BP5 on the centromeric (474P2) and telomeric (366L20) ends are used.
Confirmation of breakpoint junctions generated by the chromosomal abnormalities. (A) Confirmation of the breakpoints in J20 by PCR. Genomic DNA from L1 (lane 1), L2 (lane 2), S1 (lane 3), S2 (lane 4), JY25 (the wild-type control) (lane 5), white blood cells (WBCs) from J20 (lane 6), WBCs of a healthy volunteer (lane 7), or no DNA template (negative control, lane 8) was used for PCR analysis using primer sets designed according to the flanking regions of BP1, BP2, and BP3 (illustrated in Figure 1). Both the integrity and quantity of the DNA templates were confirmed by PCR using control primers (labeled Control). That appropriate-sized PCR products were amplified using DNA derived from the circulating WBCs of J20 (lane 6) shows that both the S1 and L1 versions of chromosome 12 were present in vivo. (B) Confirmation of the breakpoints in US1 by PCR. Genomic DNA from WBCs of a healthy volunteer (lane 1), bone marrow cells of US1 (lane 2), PMN of US1 (lane 3), US1W (the hybrid cell line containing wild-type chromosome 12) (lane 4), or US1M [the hybrid cell line containing (der(12)] (lane 5), or no DNA template (negative control, lane 6) were used for PCR analysis using primer sets designed according to the sequence of flanking regions of the BP5, BP6, BP7, and BP8 (illustrated in Figure 1). These experiments show that both wild-type chromosome 12 and der(12) were present in the peripheral blood and bone marrow of US1. (C) Metaphase FISH showing the chromosomal abnormality in J20. BAC probes 471G7, 150C16, and 366L20 were hybridized against chromosomal specimens derived from the cell line (L1) that contains only long chromosome 12. Two hybridization signals (arrows) were detected with all 3 BAC probes, confirming that long chromosome 12 contained the inserted material deleted from short chromosome 12. (D) Metaphase FISH showing the chromosomal abnormality in US1. BAC probes 366L20, 438I19, and 474P2 (illustrated in Figure 1) were hybridized with chromosomal specimens derived from bone marrow cells of US1. Each sample contained a der(12) (indicated by 2 hybridization signals on the same chromosome, double arrows) and a wild-type chromosome 12 (indicated by one hybridization signal, arrow). The intrachromosomal insertion splits the signal on 12p generated by hybridization of probe 438l19, whereas signals are generated on 12q and 12p when probes that overlap BP5 on the centromeric (474P2) and telomeric (366L20) ends are used.
Effects of the chromosome 12 abnormalities in 2 patients with PNH. (A) Structure of normal and abnormal HMGA2 locus in J20 and US1. White, black, and gray boxes indicate UTRs, coding regions, and abnormally fused fragments, respectively. The exon numbers of HMGA2 are shown below the boxes. The nucleotide sequences on both sides of BP2 and BP7 (arrowheads) are shown in the white and gray boxes. The truncated HMGA2 exon 5 of J20 and US1 are indicated by the brackets with the size (bp) shown above the brackets. The bent arrows indicate the fused fragments. The 3′ UTR of exon 5 of HMGA2 on long chromosome 12 is disrupted as a result of insertion of fragment b (the 12q12q14 fragment from short chromosome 12; see Figure 1). In the case of US1, a similar disruption of exon 5 resulted from the rearrangement that occurred when material deleted from 12q (fragments c and d) was inserted into 12p (see Figure 1). (B) Real-time PCR analysis of HMGA2 transcripts in J20 and US1. The amount of HMGA2 transcripts in bone marrow cells of 5 healthy individuals and of patients J20 and US1 were quantitated by using the TaqMan MGB PCR method. The positions of the forward (right-facing arrow) and reverse (left-facing arrow) PCR primers are indicated above the normal HMGA2 locus shown in panel A. The relative expression of HMGA2 transcripts is normalized to expression of β-glucuronidase transcripts. Each value of the relative expression indicates the average of triplicate measurements. Expression of HMGA2 was greater than normal (mean ± SD, 3.87 ± 0.45) for both J20 (mean, 10.23) and US1 (mean, 19.34) (*P < .01). The long and short horizontal bars indicate average and standard deviation (SD) in healthy individuals, respectively. (C) Allele-specific expression of HMGA2. A polymorphic region (based on TC repeats) in the 5′ UTR of HMGA2 was amplified by PCR and analyzed by polyacrylamide gel electrophoresis. The products were also cloned and sequenced to characterize the polymorphisms. (Top panel) A 183-bp product (containing 29 TC repeats) was generated from the J20-derived hybrid cell line containing long chromosome 12 (lane 1), whereas a 179-bp product (containing 27 TC repeats) was generated from the cell line containing short chromosome 12 (lane 2). Analysis of the PCR product generated by amplification of cDNA derived from GPI-AP– bone marrow cells of J20 revealed only the 183-bp product (lane 3). No PCR products were visualized when the PCR template was prepared without reverse transcriptase (lane 4). (Bottom panel) A 171-bp product (containing 23 TC repeats) was generated from the US1 hybrid cell line containing the der(12) (lane 5), whereas a 175-bp product (containing 25 TC repeats) resulted from amplification of DNA from the cell line containing normal chromosome 12 (lane 6). Analysis of the PCR product generated by amplification of cDNA derived from unfractionated bone marrow cells of US1 revealed only the 171-bp product (lane 7). No PCR products were visualized when the PCR template was prepared without reverse transcriptase (lane 8). The asterisk (left of each panel) indicates the position of an uncharacterized PCR product. The abnormal allele-specific expression of HMGA2 in the bone marrow cells of US1 was confirmed by using a genetic analyzer (3100-Avant; Applied Biosystems) (not shown). For both J20 and US1, HMGA2 expression appears to be derived exclusively from the mutant allele.
Effects of the chromosome 12 abnormalities in 2 patients with PNH. (A) Structure of normal and abnormal HMGA2 locus in J20 and US1. White, black, and gray boxes indicate UTRs, coding regions, and abnormally fused fragments, respectively. The exon numbers of HMGA2 are shown below the boxes. The nucleotide sequences on both sides of BP2 and BP7 (arrowheads) are shown in the white and gray boxes. The truncated HMGA2 exon 5 of J20 and US1 are indicated by the brackets with the size (bp) shown above the brackets. The bent arrows indicate the fused fragments. The 3′ UTR of exon 5 of HMGA2 on long chromosome 12 is disrupted as a result of insertion of fragment b (the 12q12q14 fragment from short chromosome 12; see Figure 1). In the case of US1, a similar disruption of exon 5 resulted from the rearrangement that occurred when material deleted from 12q (fragments c and d) was inserted into 12p (see Figure 1). (B) Real-time PCR analysis of HMGA2 transcripts in J20 and US1. The amount of HMGA2 transcripts in bone marrow cells of 5 healthy individuals and of patients J20 and US1 were quantitated by using the TaqMan MGB PCR method. The positions of the forward (right-facing arrow) and reverse (left-facing arrow) PCR primers are indicated above the normal HMGA2 locus shown in panel A. The relative expression of HMGA2 transcripts is normalized to expression of β-glucuronidase transcripts. Each value of the relative expression indicates the average of triplicate measurements. Expression of HMGA2 was greater than normal (mean ± SD, 3.87 ± 0.45) for both J20 (mean, 10.23) and US1 (mean, 19.34) (*P < .01). The long and short horizontal bars indicate average and standard deviation (SD) in healthy individuals, respectively. (C) Allele-specific expression of HMGA2. A polymorphic region (based on TC repeats) in the 5′ UTR of HMGA2 was amplified by PCR and analyzed by polyacrylamide gel electrophoresis. The products were also cloned and sequenced to characterize the polymorphisms. (Top panel) A 183-bp product (containing 29 TC repeats) was generated from the J20-derived hybrid cell line containing long chromosome 12 (lane 1), whereas a 179-bp product (containing 27 TC repeats) was generated from the cell line containing short chromosome 12 (lane 2). Analysis of the PCR product generated by amplification of cDNA derived from GPI-AP– bone marrow cells of J20 revealed only the 183-bp product (lane 3). No PCR products were visualized when the PCR template was prepared without reverse transcriptase (lane 4). (Bottom panel) A 171-bp product (containing 23 TC repeats) was generated from the US1 hybrid cell line containing the der(12) (lane 5), whereas a 175-bp product (containing 25 TC repeats) resulted from amplification of DNA from the cell line containing normal chromosome 12 (lane 6). Analysis of the PCR product generated by amplification of cDNA derived from unfractionated bone marrow cells of US1 revealed only the 171-bp product (lane 7). No PCR products were visualized when the PCR template was prepared without reverse transcriptase (lane 8). The asterisk (left of each panel) indicates the position of an uncharacterized PCR product. The abnormal allele-specific expression of HMGA2 in the bone marrow cells of US1 was confirmed by using a genetic analyzer (3100-Avant; Applied Biosystems) (not shown). For both J20 and US1, HMGA2 expression appears to be derived exclusively from the mutant allele.
Findings reported herein provide new insights into the cause of the nonmalignant clonal hematopoiesis of PNH. Together with observations of others,6,7 our studies support a 2-step process consisting of clonal immunoselection based on phenotype (ie, GPI-AP deficiency resulting from mutant PIGA) and clonal expansion as a consequence of a second somatic mutation that bestows the proliferative advantage. Clonal immunoselection may induce exit of PIGA-mutant stem cells from a dormant state,4 thereby favoring acquisition of the mutation that underlies clonal expansion. But the benign nature of PNH suggests that genes involved in clonal expansion of PIGA-mutant stem cells are different from those that underlie malignant clonal diseases such as acute leukemia. Characterizing the molecular basis of benign clonal hematopoiesis is important not only for understanding the pathobiology of PNH but also for developing novel strategies for treatment of bone marrow failure and for enhancing stem cell function for both transplantation and gene therapy.
Authorship
Contribution: N.I. designed research, performed research, and wrote the paper; T.I.-S. designed and performed research; Y.M., Y.E., and J.-I.N. performed research; K.K. contributed bioinformatics expertise; M.K., H.S., T.M., and Y.K. collected data; G.M. performed research; C.W. designed research; Z.C. performed research; W.B. and D.F.-L. provided essential material; and C.J.P. and T.K. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
N.I. and T.I.-S. contributed equally to this study.
Prepublished online as Blood First Edition Paper, August 29, 2006; DOI 10.1182/blood-2006-05-025148.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Andrew Zinn (The University of Texas Southwestern Medical School at Dallas) and Kiran Chada (Robert Wood Johnson Medical School, University of Medicine and Dentistry of New Jersey) for helpful discussions, and Kiyo Kawata, Fumiko Ishii-Mori, and Keiko Kinoshita for technical assistance.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Ministry of Health, Labour, and Welfare of Japan (T.K. and N.I.); the Osaka Medical Research Foundation for Incurable Diseases (N.I.); the Mochida Memorial Foundation for Medical Pharmaceutical Research (N.I.); the Japan Health Sciences Foundation (J.-I.N.); and the National Institutes of Health (grant K23 RR020043) (G.M. and C.J.P.).

![Figure 2. Confirmation of breakpoint junctions generated by the chromosomal abnormalities. (A) Confirmation of the breakpoints in J20 by PCR. Genomic DNA from L1 (lane 1), L2 (lane 2), S1 (lane 3), S2 (lane 4), JY25 (the wild-type control) (lane 5), white blood cells (WBCs) from J20 (lane 6), WBCs of a healthy volunteer (lane 7), or no DNA template (negative control, lane 8) was used for PCR analysis using primer sets designed according to the flanking regions of BP1, BP2, and BP3 (illustrated in Figure 1). Both the integrity and quantity of the DNA templates were confirmed by PCR using control primers (labeled Control). That appropriate-sized PCR products were amplified using DNA derived from the circulating WBCs of J20 (lane 6) shows that both the S1 and L1 versions of chromosome 12 were present in vivo. (B) Confirmation of the breakpoints in US1 by PCR. Genomic DNA from WBCs of a healthy volunteer (lane 1), bone marrow cells of US1 (lane 2), PMN of US1 (lane 3), US1W (the hybrid cell line containing wild-type chromosome 12) (lane 4), or US1M [the hybrid cell line containing (der(12)] (lane 5), or no DNA template (negative control, lane 6) were used for PCR analysis using primer sets designed according to the sequence of flanking regions of the BP5, BP6, BP7, and BP8 (illustrated in Figure 1). These experiments show that both wild-type chromosome 12 and der(12) were present in the peripheral blood and bone marrow of US1. (C) Metaphase FISH showing the chromosomal abnormality in J20. BAC probes 471G7, 150C16, and 366L20 were hybridized against chromosomal specimens derived from the cell line (L1) that contains only long chromosome 12. Two hybridization signals (arrows) were detected with all 3 BAC probes, confirming that long chromosome 12 contained the inserted material deleted from short chromosome 12. (D) Metaphase FISH showing the chromosomal abnormality in US1. BAC probes 366L20, 438I19, and 474P2 (illustrated in Figure 1) were hybridized with chromosomal specimens derived from bone marrow cells of US1. Each sample contained a der(12) (indicated by 2 hybridization signals on the same chromosome, double arrows) and a wild-type chromosome 12 (indicated by one hybridization signal, arrow). The intrachromosomal insertion splits the signal on 12p generated by hybridization of probe 438l19, whereas signals are generated on 12q and 12p when probes that overlap BP5 on the centromeric (474P2) and telomeric (366L20) ends are used.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2006-05-025148/7/m_zh80240605040002.jpeg?Expires=1767710266&Signature=QjHuPqY5q9j58ZFrGyfoWKIrKdk0uy26BIryC-z3e0Fb61meezVIYgM30aInwNNSZYfxG4pQ200O668gNRjasP9O8kFc2JjBnPfHd-2pjhI87LrFO6pewBG2N0dc2v2ze1xBV6BTLHksdthkmrQZXQ9eI0r1qoBXHAXYPyjPu-XSbzvvOy~MpskchmNqKnYhAMEzueSmuWR3uqz9ctIhJzvwDxmzBb8NGHTpvxlrNQxoQMQQMgkDJTPxtmHftE6hdRJA~RAlkOtrbpEKnENestQaDkohk0TcJqYUVmq2wQl02dKf5FL9yRCG6oIcPQ8jrbjj953qri9nCCy9sthi5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal