Abstract
An activating JAK2 mutation (JAK2 V617F) is present in the chronic myeloproliferative disorders (MPDs), polycythemia vera (PV), idiopathic myelofibrosis (IMF), and essential thrombocytosis (ET). JAK2 is also a chaperone for Mpl and responsible for its cell-surface expression. We observed a reciprocal relationship between neutrophil JAK2 V617F allele percentage and platelet Mpl expression in JAK2 V617F–positive PV, IMF, and ET patients. However, severely impaired platelet Mpl expression was present in JAK2 V617F–negative MPD patients. While JAK2 V617F allele status did not necessarily correlate with the clinical MPD phenotype, the degree of impaired platelet Mpl expression did. We conclude that multiple molecular abnormalities are involved in the pathogenesis of the MPDs and that aberrant Mpl expression may be a common denominator of aberrant signaling in both the JAK2 V617F–positive and JAK2 V617F–negative MPDs.
Introduction
Polycythemia vera (PV), idiopathic myelofibrosis (IMF), and essential thrombocytosis (ET) share their origin in a multipotent hematopoietic progenitor cell1 ; cytogenetic abnormalities of chromosomes 1, 8, 9, 13, and 202 ; growth factor–independent in vitro hematopoietic colony formation3 ; and molecular abnormalities, including increased granulocyte PRV-1 mRNA expression,4 impaired megakaryocyte and platelet thrombopoietin receptor (Mpl) expression,5 and the acquisition of JAK2 V617F.6-9 Because JAK2 has important biologic associations with regard to Mpl expression as well as signal transduction,10 we examined the relationship between JAK2 V617F and platelet Mpl expression in PV, IMF, and ET.
Patients, materials, and methods
The study was approved by our institutional review board and written informed consent was obtained from all patients, in accordance with the Declaration of Helsinki. Diagnoses of PV and ET were based on the Polycythemia Vera Study Group criteria11 ; the diagnosis of IMF was based on the Italian Consensus Conference.12 Genomic DNA was prepared from density gradient–purified granulocytes using the Puregene Cell kit (Gentra Systems, Minneapolis, MN). RNA was prepared from platelets isolated by differential centrifugation using the RNAeasy kit (Qiagen, Valencia, CA). Complementary DNA was prepared from platelet RNA using the TaqMan Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA) and random hexamers.
Platelet Mpl densitometry
Platelet lysate (20 μg) was applied to 10% precast gradient gels (Bio-Rad, Hercules, CA), transferred, and immunoblotted with a rabbit polyclonal antiserum to the C-terminal of the Mpl protein and anti–glycoprotein IIB antiserum as described previously.13 Quantitative densitometry was performed using ImageJ software (National Institutes of Health, Bethesda, MD) by a blinded observer; Mpl normalized to IIB was expressed as the percent of 2 averaged controls on each blot. Sixteen secondary erythrocytosis patients defined the normal range, in which the mean was 98% and the range was 52% to 149%. The intra-assay correlation coefficient for replicates was 0.99, and the interassay coefficients of variation for the normal and low controls were 5% and 22%, respectively.
JAK2 V617F determination
Custom TaqMan SNP Genotyping Assays (Applied Biosystems) for JAK V617F with templates from either genomic DNA or cDNA consisting of real-time polymerase chain reaction (PCR) primers flanking the mutant region plus 2 TaqMan MGB real-time PCR probes specific for the normal or mutant sequence (JAK2 genomic assay: forward primer, AAGCTTTCTCACAAGCATTTGGTTT and reverse primer, AGAAAGGCATTAGAAAGCCTGTAGTT; JAK2 cDNA assay: forward primer, AAGCTTTCTCACAAGCATTTGGTTT and reverse primer, CCAAATTTTACAAACTCCTGAACCAGAA; JAK2 genomic assay: probes, VIC-TCTCCACAGACACATAC and FAM-TCCACAGAAACATAC; JAK2 cDNA assay: probes, VIC-CTCCACAGACACATAC and FAM-TCCACAGAAACATAC) were performed on an Applied Biosystems Prism 7700 sequence detection system using the default PCR conditions. DNA mixtures corresponding to V617F allele percentages of 5%, 10%, 25%, 50%, 75%, and 90% were assay reference standards. Intra-assay replicates did not vary more than 5%, and interassay replicates did not vary more than 12%.
Statistical analysis
Statistical calculations were performed using SigmaStat (STSTAT Software, Point Richmond, CA).
Results and discussion
Our study population consisted of a clinically well-defined group of PV, IMF, and ET patients followed for up to 30 years. In agreement with previous studies, we detected JAK2 V617F in neutrophil genomic DNA in 92% of PV (n = 92), 45% of ET (n = 84), and 42% of IMF (n = 19) patients.6,7,14,15 There were no significant differences in age, disease duration, sex, or degree of splenomegaly between the JAK2 V617F–positive and –negative patients.
The percent of JAK2 alleles that are JAK2 V617F is related both to the extent of dominance of the JAK2 V617F–positive clone16 and also to the clone's JAK2 V617F genotype, which can be heterozygous or homozygous due to a mitotic recombination event.17 Therefore, we measured the percentage of JAK2 V617F alleles in PV, IMF, and ET neutrophils using a quantitative, allele-specific assay sensitive to the presence of 5% JAK2 V617F. We found that the median neutrophil JAK2 V617F allele percentage was greater in PV (67%; range, 35%-100%) than in ET (47%; range, 10%-63%; P < .001), and that allele percentages more than 63% were restricted to PV and IMF, a finding similar to some18 but not all19 studies. When stratified by disease duration, 100% neutrophil JAK2 V617F allele percentages were present in only 15% of PV patients less than 3 years from diagnosis compared with 40% of PV patients 3 to 10 years and 40% of PV patients between 10 and 26 years since diagnosis, although these differences were not significant (Figure 1A).
In PV patients, there was no difference between the median JAK2 V617F percentage in platelet cDNA and neutrophil genomic DNA (65% versus 61%, respectively) (Figure 1B). However, in ET, the median JAK2 V617F percentage was significantly higher in platelet cDNA than in neutrophil genomic DNA (42% versus 22%, respectively; P = .001) (Figure 1B). In 3 of 11 ET patients, JAK2 V617F was not detected in neutrophil genomic DNA but was easily detected in platelet cDNA. The differences in neutrophil JAK2 V617F allele percentage in ET compared with PV and the observation that JAK2 V617F can be detected in platelets only in ET suggest that factors such as the level of stem-cell commitment upon acquisition of JAK2 V617F or the degree of retained polyclonal hematopoiesis are related to differences in clinical phenotypes in JAK2 V617F–positive PV and ET.
JAK2 VG17F allele percentages in the MPDs. (A) Neutrophil JAK2 V617F allele percentages in the MPDs. Thirty-six ET and 77 PV patients were stratified by neutrophil JAK2 V617F allele percentage and disease duration. Boxes indicate the quartiles of the distribution; medians are indicated by black lines. All of the PV median neutrophil allele percentages were significantly greater than all of the ET median neutrophil allele percentages, regardless of disease duration (P = .008). Within ET or PV, the differences in allele percentages as a function of disease duration were not statistically significant. The JAK2 V617F allele percentage in the 8 IMF patients (median of 100, range of 48-100) could not be compared with the other groups because of the small sample size. (B) JAK2 V617F percentages in PV and ET neutrophils and platelets. Neutrophil genomic DNA and platelet cDNA were isolated simultaneously from blood samples in 23 PV and 13 ET patients. Circles indicate PV samples; triangles, ET. Median neutrophil JAK2 V617F allele percentage in PV (66%, gray bar) was greater than in ET (22%, black bar) (P < .001); median platelet JAK2 V617F allele percentage in PV (65%, gray bar) was greater than in ET (43%, black bar) (P = .002). In PV, the median neutrophil JAK2 V617F allele percentage was not significantly different (ns) from the platelet JAK2 V617F allele percentage, whereas in ET the platelet JAK2 V617F allele percentage was significantly greater than the neutrophil JAK2 V617F allele percentage (P = .001). Lines indicate cases in which JAK2 V617F was detected in platelet cDNA only. Neutrophil cDNA analysis yielded similar results (data not shown).
JAK2 VG17F allele percentages in the MPDs. (A) Neutrophil JAK2 V617F allele percentages in the MPDs. Thirty-six ET and 77 PV patients were stratified by neutrophil JAK2 V617F allele percentage and disease duration. Boxes indicate the quartiles of the distribution; medians are indicated by black lines. All of the PV median neutrophil allele percentages were significantly greater than all of the ET median neutrophil allele percentages, regardless of disease duration (P = .008). Within ET or PV, the differences in allele percentages as a function of disease duration were not statistically significant. The JAK2 V617F allele percentage in the 8 IMF patients (median of 100, range of 48-100) could not be compared with the other groups because of the small sample size. (B) JAK2 V617F percentages in PV and ET neutrophils and platelets. Neutrophil genomic DNA and platelet cDNA were isolated simultaneously from blood samples in 23 PV and 13 ET patients. Circles indicate PV samples; triangles, ET. Median neutrophil JAK2 V617F allele percentage in PV (66%, gray bar) was greater than in ET (22%, black bar) (P < .001); median platelet JAK2 V617F allele percentage in PV (65%, gray bar) was greater than in ET (43%, black bar) (P = .002). In PV, the median neutrophil JAK2 V617F allele percentage was not significantly different (ns) from the platelet JAK2 V617F allele percentage, whereas in ET the platelet JAK2 V617F allele percentage was significantly greater than the neutrophil JAK2 V617F allele percentage (P = .001). Lines indicate cases in which JAK2 V617F was detected in platelet cDNA only. Neutrophil cDNA analysis yielded similar results (data not shown).
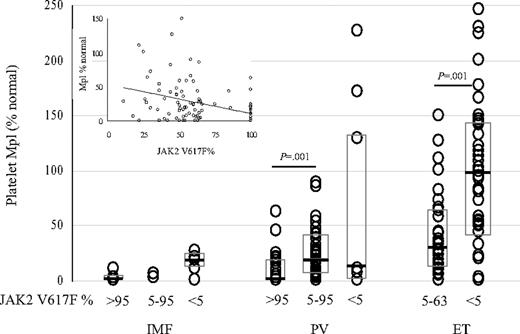
Quantitative platelet Mpl protein expression in the MPDs stratified by disease and neutrophil JAK2 V617F status. Platelet Mpl protein expression and neutrophil JAK2 V617F allele percentages were determined in 70 PV, 69 ET, and 13 IMF patients. Patients in each disease category are stratified by neutrophil JAK2 V617F allele percentage of more than 95%, between 5% to 95%, and less than 5%. Boxes indicate the quartiles of the distribution; medians are indicated by black lines. Linear regression analysis of neutrophil JAK2 V617F allele percentage and platelet Mpl expression in 101 PV, ET, and IMF patients was significant (P < .001) with a correlation coefficient of –0.390 (inset).
Quantitative platelet Mpl protein expression in the MPDs stratified by disease and neutrophil JAK2 V617F status. Platelet Mpl protein expression and neutrophil JAK2 V617F allele percentages were determined in 70 PV, 69 ET, and 13 IMF patients. Patients in each disease category are stratified by neutrophil JAK2 V617F allele percentage of more than 95%, between 5% to 95%, and less than 5%. Boxes indicate the quartiles of the distribution; medians are indicated by black lines. Linear regression analysis of neutrophil JAK2 V617F allele percentage and platelet Mpl expression in 101 PV, ET, and IMF patients was significant (P < .001) with a correlation coefficient of –0.390 (inset).
JAK2 is the cognate tyrosine kinase of Mpl, and impaired megakaryocyte and platelet Mpl expression have been documented in many laboratories in the myeloproliferative disorders (MPDs).20-22 Therefore, we examined the concordance of impaired platelet Mpl expression with disease phenotype and JAK2 V617F status in 152 MPD patients. Platelet Mpl expression was significantly different among the MPDs, with a median of 55% in ET (range, 1-246; reduced in 43%), 16% in PV (range, 1-227; reduced in 89%; P < .001 with respect to ET), and 5% in IMF patients (range, 1-22; reduced in 100%; P = .076 with respect to PV). In PV, median platelet Mpl expression was lower in patients with only the JAK2 V617F allele compared with patients with both the wild-type and mutant alleles (P = .001) (Figure 2). The median platelet Mpl expression of 98% in JAK2 V617F–negative ET patients was higher than the median of 28% in JAK2 V617F–positive ET patients (P = .001). Linear regression analysis of neutrophil JAK2 V617F allele percentage and platelet Mpl expression in the JAK2 V617F–positive cases demonstrated a significant inverse relationship (r =–0.36, P < .001) (Figure 2). However, while a negative correlation between neutrophil JAK2 V617F allele percentage and platelet Mpl expression existed, the JAK2 V617F allele percentage did not completely account for the variability of platelet Mpl expression. Of importance, the same association of impaired Mpl expression with an MPD phenotype occurred in JAK2 V617F–negative patients. This suggests that the same phenotype may develop as a consequence of different molecular mechanisms.
A previous study did not find a strong association between JAK2 V617F and impaired Mpl expression23 ; however, the study was limited by a small number of Mpl measurements (n = 25), all of which were in ET patients, and neither neutrophil JAK2 V617F nor platelet Mpl expression was measured quantitatively. Impaired Mpl expression is in part related to defective Mpl processing, which may lead to retention in the Golgi or aberrant localization in the cytoplasm.13 Constitutively activated JAK2 may also reduce Mpl protein expression by causing receptor down-regulation. Indeed, the inverse correlation between neutrophil JAK2 V617F allele percentage and platelet Mpl reduction suggests that these are related abnormalities. Identification of other lesions in Mpl and the JAK signaling pathways in the JAK2 V617F–negative MPDs will further define the significance of aberrant Mpl protein expression to the molecular pathogenesis of the MPDs.
Authorship
A.R.M. and D.M.W. designed and performed research and wrote the paper; O.R. designed and performed the research; J.L.S. designed the research and wrote the paper.
The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 15, 2006; DOI 10.1182/blood-2006-03-008805.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Drs Sophie M. Lanzkron and Michael B. Streiff for patient recruitment.
This work was supported by the Myeloproliferative Disorders Foundation and the National Institutes of Health (RO1-HL082995).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal