Abstract
Nearly 15 years have elapsed since the US Food and Drug Administration last approved a major new hematopoietic cytokine. Promiscuous binding to multiple receptors, or to receptors expressed by multiple tissues, reduces growth factor specificity and promotes side effects. Here we show that hematopoiesis can be differentially regulated using receptors rather than ligands. Conditional derivatives of both fibroblast growth factor receptor-1 (F36VFGFR1) and the thrombopoietin receptor (F36VMpl) induced a sustained expansion of mouse marrow cells ex vivo, and erythroid cells in vivo. Only F36VFGFR1 could support the ex vivo expansion of short-term repopulating hematopoietic stem cells (HSCs), the ex vivo survival of long-term repopulating HSCs, and the prolonged in vivo expansion of granulocytes, monocytes, and platelets. Only F36VMpl induced a response sufficiently rapid to accelerate recovery from radiation-induced anemia. These results establish receptors as a new class of hematopoietic regulators possessing activities unobtainable with growth factors.
Introduction
Hematopoietic stem cells (HSCs) are defined by their ability to self-renew and to differentiate into most or all cells present in blood.1,2 In choosing between self-renewal versus differentiation, HSCs must balance the immediate demands of hematopoiesis with the longer-range goal of maintaining blood cell production for the lifetime of an individual. In striking this balance a mitotic HSC has 3 options: to symmetrically self-renew, yielding a pair of HSCs; to differentiate, yielding committed progeny that lack the full potential and/or staying power of the parent; or to asymmetrically divide, yielding 1 HSC and 1 committed daughter.
Developing ways to manipulate the behavior of hematopoietic cells in vitro or, more desirably, in vivo, may have many applications in clinical medicine.3 Relevant to this effort is a longstanding debate regarding the extent to which HSC self-renewal, lineage choice, and differentiation are intrinsically determined, versus being subject to external control.4,5 A strict intrinsic (or “stochastic”6 ) view holds that self-renewal or lineage commitment ensue upon attaining an appropriate complement of intracellular factors, and this process cannot be influenced by exogenous growth factors.7 In this model, growth factor receptors merely allow for cell survival. A competing instructive view postulates that the probability of attaining the necessary threshold of transcription factors to permit self-renewal or lineage choice is subject, at least partially, to the directive influence of growth factors.4 In this model, signaling by different growth factor receptors is expected to engender different responses in HSCs and multipotential progenitors.
One clinically applicable method for regulating the proliferation of transplanted cells uses chemical inducers of dimerization (CIDs)8 to activate engineered signaling proteins.8,9 In mouse marrow cells engineered to express the transgene in all lineages, a derivative of the thrombopoietin receptor (F36VMpl) induced an exponential, CID-dependent expansion of megakaryocytes and multipotent progenitors (but not HSCs) in culture.10-14 When administered in vivo during steady-state hematopoiesis, CID-triggered F36VMpl signaling expanded red blood cells, but had modest effects on platelets, and negligible effects on neutrophils.14-16 In mice given transplants of marrow cells containing a CID-activated derivative of Janus kinase 2 (Jak2), the CID response was restricted to red blood cells.17 A pragmatic question arising from these findings is whether CID-regulated proliferation can be used in hematopoiesis for anything other than regulating transduced red cells.
The fibroblast growth factor (FGF) family comprises at least 23 ligands that are involved in critical biological processes such as cell proliferation, differentiation, migration, morphogenesis, and angiogenesis.18,19 While a physiologic role of FGFs in adult hematopoiesis has not been defined, homozygous deletion of FGFR1 in mouse embryonic stem (ES) cells severely reduces hematopoietic differentiation in vitro,20 and 5 different translocation partners that result in constitutive activation of FGFR1 have been identified in myeloproliferative and or T lymphoma syndromes.21,22 Recently, primitive mouse marrow cells that express transcripts for FGFR1, FGFR3, and FGFR4 were found to expand markedly in cultures containing FGF-1,23 a ligand capable of activating all FGFRs.24 Here we show that F36VFGFR1 induces hematologic effects distinct from those obtained using F36VMpl, and highlight the potential of using receptors as regulators of hematopoiesis.
Materials and methods
Mice
Eight- to 12-week-old female C57BL/Ly5.1(B/6.SJL-CD45a-Pep3b)(CD45.1+) and C57BL/6-Ly5.2(CD45.2+) mice, purchased from The Jackson Laboratory (Bar Harbor, ME) were used in these experiments. STAT 5a/bΔNΔN mice25 were kindly provided by Evan Parganas and James Ihle at St Jude Children's Hospital (Memphis, TN). All mice were housed in specific pathogen–free facilities at the University of Washington. All procedures were approved by the University of Washington's Animal Care Committee.
Retroviral vectors
Retroviral transduction
Female C57BL/6J mice were intraperitoneally injected with 150 mg/kg 5-fluorouracil. After 48 hours, marrow cells were harvested and cultured in Dulbecco modified Eagle medium (DMEM) containing 16% fetal calf serum (FCS; Millipore, Billerica, MA), 5% interleukin-3 (IL-3)–conditioned medium (Stem Cell Technologies, Vancouver, BC, Canada; or BD Biosciences, San Jose, CA), 100 ng/mL recombinant human IL-6, 50 ng/mL recombinant murine stem cell factor (rhIL-6 and rmSCF, respectively; both from Peprotech, Rocky Hill, NJ), 50 U/mL penicillin, and 50 mg/mL streptomycin (Pen/Strep) in a 37°C, 5% CO2 incubator. After 48 hours, cells were transferred onto irradiated (1500 cGy) producer cells and cocultivated using identical growth conditions except for the addition of polybrene (8 μg/mL). Marrow cells were harvested after 48 hours of cocultivation. Transduction efficiency was determined by culturing cells in Iscove modified Dulbecco medium (IMDM) containing 10% FCS, Pen/Strep with 50 ng/mL murine stem cell factor, and 100 ng/mL human flt-3 ligand for an additional 48 hours, then by determining the percentage of cells expressing green fluorescent protein (GFP) by flow cytometry.
Colony assays
Colony assays were performed as described previously10 using methylcellulose (Stem Cell Technologies), 5% murine IL-3 culture supplement, 100 ng/mL rhIL-6, and 50 ng/mL rmSCF.
Suspension cultures
Following retroviral transduction, marrow cells were cultured in IMDM containing 10% FCS and Pen/Strep plus AP20187 (100 nM) in a 37°C, 5% CO2 incubator. Cell numbers were determined using a hemocytometer and trypan blue exclusion.
Bone marrow transplantation
F36VFGFR1- or F36VMpl-transduced bone marrow cells were injected via the tail vein into recipients receiving a 1050-cGy radiation dose from a dual-cesium Cs137 gamma source (GammaCell 40; AEC, Kanata, ON, Canada).
In vivo AP20187 administration
Lyophilized AP20187 (ARIAD Pharmaceuticals, Cambridge, MA) was solubilized in 100% ethanol to produce a 5-mg/mL stock that was stored at –20°C and diluted freshly on the day of injection. The final solution for injection contained 10% polyethylene glycol (PEG) 400 and 1.4% Tween 80. AP20187 was administered by daily intraperitoneal injection in a total volume of 400 μL per injection.
Hematologic analysis
Blood (50 μL) was collected using a retroorbital technique in tubes with EDTA (Microtainer; Becton Dickinson, San Jose, CA). Red cell counts were performed using 4 μL of fresh blood diluted 1:50 in phosphate-buffered saline (PBS) without Ca2+/Mg2+. A portion (10 μL) of this solution was added to 10 mL of Streck-Diluent III (Streck Labs, Omaha, NE) in Dilu-Vial (Fisher Scientific, Hampton, NH) to achieve a final dilution of 1:50 000. For white blood cell (WBC) counts 10 μL of fresh blood was added to 10 mL of Streck-Diluent III in Dilu-Vial to achieve a final dilution of 1:1000. Three drops of Zap-oglobin II Lytic Reagent (Beckman Coulter, Fullerton, CA) was added to lyse red cells. Counts were determined using a Coulter Z series particle count and size analyzer model Z1 dual (Coulter, Miami, FL) with a lower size limit of 20 for red cells and 25 for white cells. Normal ranges were obtained from Russell and Bernstein.27 Platelet counts were determined from nonheparinized blood diluted 1:200 in Rees-Ecker solution (3.8 g sodium citrate, 0.2 mL 37% formaldelyde, and 0.1 mL brilliant cresyl blue in 100 mL water). All marrow and spleen histologic sections were examined in conjunction with a veterinary pathologist (H.D.L.).
For flow cytometry, 2 μL whole blood was diluted in PBS without Ca2+/Mg2+ and analyzed for GFP-positive red cells and platelets based on standard forward–side scatter parameters by FACSCAN (BD Biosciences, San Jose, CA). Remaining blood was lysed (PharM Lyse; BD Biosciences) and divided between 1 tube containing PBS with 1% bovine serum albumin (BSA) and phycoerythrin (PE)–conjugated αGr-1 antibody plus αCD11b-PerCP-Cy5.5, and another tube with αThy1.2-PE and PE-Cy5–conjugated αCD45R/B220 (all from BD Biosciences). Stained cells were washed, resuspended in PBS with 1% BSA, and analyzed by FACSCAN.
Intracellular flow cytometry
Anti–phospho-Erk1/2 staining. F36VFGFR1- and F36VMpl-transduced mouse bone marrow cells expanded for 5 weeks in IMDM containing 10% FBS and 100 nM AP20187 were washed with PBS containing 2% FBS, then starved overnight in IMDM containing 0.5% BSA. The following day, a portion of cells was stimulated with AP20187 (100 nM) for 45 minutes at 37°C. Stimulated and unstimulated cells were then fixed in 2% paraformaldehyde (37°C for 15 minutes), permeabilized in 90% methanol (30 minutes on ice), washed twice in PBS containing 2% FBS, and stained with Alexa Fluor 647–conjugated anti–phospho-Erk1/2 antibody (catalog no. 612593; BD Pharmingen, San Diego, CA), and analyzed on an LSR flow cytometer (BD Biosciences).
Anti-HA staining. Mice that received transplants of freshly transduced marrow cells (either F36VMpl or F36VFGFR1) were killed 4 months after transplantation (without exposure to CID). Marrow cells were collected, lineage-depleted using a biotinylated cocktail of lineage-specific antibodies and avidin-coated magnetic beads (Miltenyi, Auburn, CA), and stained with c-kit PE-Cy5 (eBioscience, San Diego, CA), Sca-1 PE-Cy7 (BD Pharmingen), and anti-biotin APC (Miltenyi) to identify residual lin+ cells. Following fixation and permeabilization (Fix&Perm kit; Caltag Laboratories, Burlingame, CA), cells were stained with a PE-conjugated antihemagglutinin (HA) tag antibody (Miltenyi), resuspended in PBS, 2% FBS, and 0.1% sodium azide, and analyzed using an LSRII (BD Biosciences) with FlowJo software (TreeStar, Ashland, OR). Expression using the HA antibody was compared with an isotype control antibody.
Transplantation of ex vivo–expanded cells
Following transduction with either F36VMpl or F36VFGFR1, 2.5 × 106 bone marrow cells were expanded in the presence of AP20187 (100 nM) for 28 days. Ex vivo–expanded cells (8 × 106) were resuspended in PBS then injected via tail vein into irradiated (1050 cGy) C57BL/6 recipients. Where indicated, cultured cells were cotransplanted concurrently with 100 000 fresh marrow cells.
LAM-PCR
Insertion site analysis was performed using a 3-arm modification of linear amplification–mediated polymerase chain reaction (LAM-PCR)28 (M.A.H., Rajinder Kaul, Michael A. Jacobs, Peter Kurre, Don Bovee, Ruth Levy, and C.A.B., manuscript in preparation). Reactions were conducted with 50 to 100 ng of column-purified DNA. All amplifications were done with Advantage II Polymerase Mix and buffer system (BD Biosciences Clontech, Palo Alto, CA), using the following cycling conditions: melt at 95°C for 20 seconds, anneal at 60°C for 45 seconds, and elongate at 68°C for 90 seconds. First, single-stranded copies of the viral 3′ long terminal repeat (LTR)–host junction region were made by 100 cycles of linear amplification using a biotinylated LTR primer (biotin-5′AAGCCTCAATAAAGCTTGCC). The product was bound to streptavidin-coated magnetic M280 Dynabeads (Dynal Biotech, Oslo, Norway). Subsequent reactions and washes (3 × 200 μL after every reaction) took place on this matrix. Matrix-bound DNA was resuspended in 20 μL of a random-priming reaction containing 500 nM dNTP, 100 ng/μL random hexamers, 2 to 5 U Klenow enzyme (New England Biolabs, Beverly, MA), and manufacturer-supplied buffer at 1 ×, and incubated at 37°C for 60 minutes. Products were divided into 3 equal arms and digested with Tsp509I, HaeIII, or RsaI (New England BioLabs). Matrix-bound DNA was ligated to a double-stranded anchor primer (Fast Link Ligase; Epicenter Technologies, Madison, WI). For Tsp509I, component primers GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG and AATTCCTAACTGCTGTGCCACTGAATTCAGATC were used, yielding a Tsp509I-compatible end. For HaeIII and RsaI, a blunt-ended anchor primer was used, with CCTAACTGCTGTGCCACTGAATTCAGATC as the second component. The double-stranded junction fragments, containing host DNA flanked by LTR and anchor-primer (ANCH) sequences, was eluted from the matrix in 5 μL 0.1 N NaOH, and amplified by 2 rounds of nested PCR. A portion (2 μL) of the eluate was amplified in a 50-μL PCR reaction with LTR primer 1; TGAGTGCTTCAAGTAGTGTGTGC and ANCH primer 1; and GACCCGGGAGATCTGAATTC. This reaction was diluted 100-fold in water and a 2-μL template was amplified by 30 cycles of nested PCR, using LTR primer 2; ACTCTGGTAACTAGAGATCC and ANCH primer 2; and GATCTGAATTCAGTGGCACAG. The LAM-PCR products were separated by electrophoresis on Novex 4% to 20% polyacrylamide, TBE gels (Invitrogen, Carlsbad, CA), stained with ethidium bromide, and visualized by UV light fluorescence.
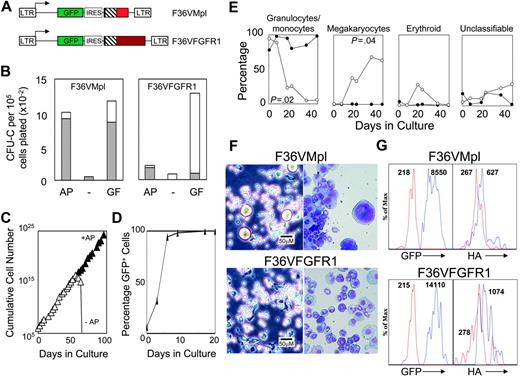
F36VFGFR1 supports the expansion of transduced mouse bone marrow cells in culture. (A) Schematic depiction of F36VMpl and F36VFGFR1 retrovirus vectors. LTR indicates MSCV long terminal repeat; GFP, green fluorescent protein; IRES, encephalomyocarditis virus internal ribosomal entry site; ▪, myristylation domain; ▧, F36V-modified FKBP12; and red and maroon boxes, cytoplasmic domains of mouse Mpl and rat FGFR1. At the carboxyterminal end of both F36VMpl and F36VFGFR1isan epitope tag from the HA protein of influenza31 (not shown). (B) Colony assays of marrow cells transduced with the F36VMpl or F36VFGFR1 vectors performed in the presence (GF) or absence (–) of a combination of growth factors, or in the presence of 100 nM AP20187 without growth factors (AP). Total colonies (bars) and GFP+ colonies (▦) are indicated. (C) Logarithmic expansion of F36VFGFR1-transduced marrow cells over time in serum-containing cultures plus AP20187 (100 nM) without added cytokines. Cell growth ceased following CID withdrawal (–AP, open symbols). We have previously reported a similar exponential expansion of mouse marrow cells in response to CID-activated derivatives of Mpl,10-14 but not CID-activated derivatives of Flt3 or G-CSF receptor.12 (D) Rise in the percentage of GFP+-expressing F36VFGFR1-transduced marrow cells over time in culture with CID. (E) Differences in cell types emerging from cultures of F36VFGFR1-transduced (•) versus F36VMpl-transduced (○) marrow cells. Similar results were obtained in 7 independent experiments using F36VFGFR1, and in previously published reports using F36VMpl.10 (F) Phase-contrast microscopy (left panels) and Wright-Giemsa staining (right panels) (× 200) of F36VMpl (top) or F36VFGFR1 (bottom) transduced lin–c-kit+, sca-1+ cells after 6 days (phase contrast) or 8 days (Wright-Giemsa) of culture in AP20187. Phase-contrast images were visualized using a Leica DMIL inverted microscope (Leica, Wetzlar, Germany) equipped with a C PLAN L 20×/0.30 numerical aperture (NA) PH1 objective. Wright-Giemsa stains were visualized using a Leica DMLB microscope equipped with an HC PL FLUOTAR 20×/0.50 NA objective. Images were captured using a Leica DFC480 digital camera and Leica Image Management software, and were processed using Adobe Photoshop software version 8.0 (Adobe Systems, San Jose, CA). (G) Flow histograms of lin–c-kit+, sca-1+ cells from mice that received transplants 4 months earlier with F36VMpl-(top) or F36VFGFR1 (bottom)–transduced marrow cells. Expression levels of GFP and the HA-tagged fusions overlap significantly, but average 1.6-fold higher for F36VFGFR1. Values indicate mean fluorescent intensity (MFI). Red lines in the left panels indicate GFP– cells. Blue lines in the left panels, GFP+ cells; red lines in the right panels, cells stained with isotype control antibody; and the blue lines in the right panels, cells stained with HA antibody.
F36VFGFR1 supports the expansion of transduced mouse bone marrow cells in culture. (A) Schematic depiction of F36VMpl and F36VFGFR1 retrovirus vectors. LTR indicates MSCV long terminal repeat; GFP, green fluorescent protein; IRES, encephalomyocarditis virus internal ribosomal entry site; ▪, myristylation domain; ▧, F36V-modified FKBP12; and red and maroon boxes, cytoplasmic domains of mouse Mpl and rat FGFR1. At the carboxyterminal end of both F36VMpl and F36VFGFR1isan epitope tag from the HA protein of influenza31 (not shown). (B) Colony assays of marrow cells transduced with the F36VMpl or F36VFGFR1 vectors performed in the presence (GF) or absence (–) of a combination of growth factors, or in the presence of 100 nM AP20187 without growth factors (AP). Total colonies (bars) and GFP+ colonies (▦) are indicated. (C) Logarithmic expansion of F36VFGFR1-transduced marrow cells over time in serum-containing cultures plus AP20187 (100 nM) without added cytokines. Cell growth ceased following CID withdrawal (–AP, open symbols). We have previously reported a similar exponential expansion of mouse marrow cells in response to CID-activated derivatives of Mpl,10-14 but not CID-activated derivatives of Flt3 or G-CSF receptor.12 (D) Rise in the percentage of GFP+-expressing F36VFGFR1-transduced marrow cells over time in culture with CID. (E) Differences in cell types emerging from cultures of F36VFGFR1-transduced (•) versus F36VMpl-transduced (○) marrow cells. Similar results were obtained in 7 independent experiments using F36VFGFR1, and in previously published reports using F36VMpl.10 (F) Phase-contrast microscopy (left panels) and Wright-Giemsa staining (right panels) (× 200) of F36VMpl (top) or F36VFGFR1 (bottom) transduced lin–c-kit+, sca-1+ cells after 6 days (phase contrast) or 8 days (Wright-Giemsa) of culture in AP20187. Phase-contrast images were visualized using a Leica DMIL inverted microscope (Leica, Wetzlar, Germany) equipped with a C PLAN L 20×/0.30 numerical aperture (NA) PH1 objective. Wright-Giemsa stains were visualized using a Leica DMLB microscope equipped with an HC PL FLUOTAR 20×/0.50 NA objective. Images were captured using a Leica DFC480 digital camera and Leica Image Management software, and were processed using Adobe Photoshop software version 8.0 (Adobe Systems, San Jose, CA). (G) Flow histograms of lin–c-kit+, sca-1+ cells from mice that received transplants 4 months earlier with F36VMpl-(top) or F36VFGFR1 (bottom)–transduced marrow cells. Expression levels of GFP and the HA-tagged fusions overlap significantly, but average 1.6-fold higher for F36VFGFR1. Values indicate mean fluorescent intensity (MFI). Red lines in the left panels indicate GFP– cells. Blue lines in the left panels, GFP+ cells; red lines in the right panels, cells stained with isotype control antibody; and the blue lines in the right panels, cells stained with HA antibody.
Inhibitor assays
Mouse marrow cells transduced with F36VMpl or F36VFGFR1 and cultured for at least 1 month were plated in individual wells of a 6-well dish at a density of 1 × 106 cells/mL in IMDM, 10% FCS, and 100 nM AP20187 in the absence or presence of a range of concentrations of U73122, SU6656, Ly294002 U0126, AG490, or Jak Inhibitor 1 (all from Calbiochem, La Jolla, CA).29,30 After 3 to 4 days in culture, cell numbers were determined using a hemocytometer and trypan blue exclusion.
Results
F36VFGFR1 is a proliferation switch for hematopoietic cells
Activation of both F36VMpl and F36VFGFR1 (Figure 1A) using the CID AP20187 triggered the proliferation of virtually all transduced marrow progenitors as determined by expression of the GFP marker in colony-forming cells (Figure 1B). The composition of colonies generated in response to F36VMpl and F36VFGFR1 was similar (data not shown). In suspension, F36VFGFR1-transduced marrow cells grew exponentially for more than 200 days in serum containing medium supplemented with AP20187 but no other growth factor (Figure 1C; data not shown). All AP20187-expanded cells expressed GFP (Figure 1D), and all died promptly following CID withdrawal (Figure 1C). This logarithmic pattern of growth is very similar to our previous observations using F36VMpl,10-14 and does not occur with CID-activated derivatives of Flt3 or granulocyte colony-stimulating factor (G-CSF) receptor.12 Limiting dilution showed that the frequency of cells capable of sustaining CID-dependent growth for at least 30 days was, for both F36VMpl and F36VFGFR1, about 0.5% (data not shown). However, the cell types generated by F36VFGFR-1 signaling differed markedly from suspension cultures expanded by F36VMpl. Granulocytes and macrophages disappeared rapidly from cultures of F36VMpl-transduced cells, replaced by megakaryocytes (as reported previously10 ), whereas granulocytes and macrophages remained at high levels in suspension cultures of F36VFGFR1-transduced cells (Figure 1E). These differences in frequencies of granulocytes/macrophages and megakaryocytes were statistically significant (P = .02 and P = .04, respectively), and were highly reproducible.
Similar differences between F36VMpl and F36VFGFR1 rapidly developed in cultures initiated using purified c-kit+, sca1+, lin– cells. Megakaryocytes were readily identified in cultures of F36VMpl-transduced cells by day 6 of culture, whereas granulocytes and macrophages were more abundant in the culture of F36VFGFR1-transduced cells (Figure 1F). These findings demonstrate the differential regulation of primitive mouse hematopoietic cells using these 2 growth factor receptors.
To compare F36VMpl and F36VFGFR1 expression in primitive hematopoietic cells, we examined the fluorescence intensity of GFP expression in lin–c-kit+sca1+ cells isolated from mice that received transplants of either F36VMpl or F36VFGFR1-transduced marrow cells 4 months earlier. While considerable overlap was observed between the 2 populations, the mean fluorescence intensity of GFP expression was 1.6-fold higher in cells containing F36VFGFR1 relative to cells containing F36VMpl. A direct comparison of the fusion proteins, using an antibody directed against the HA epitope tag (Figure 1 legend), yielded very similar results (Figure 1G).
F36VFGFR1-induced proliferation requires MAP kinase signaling, whereas F36VMpl-induced proliferation does not
To identify which signaling pathways might underlie the different hematologic responses to F36VFGFR1 versus F36VMpl, a variety of inhibitors were tested (Figure 2A). Antagonists of phospholipase Cγ (U71322), Src kinase (SU6656), and phosphoinositol 3′ kinase (Ly294002) inhibited proliferation induced by both receptors, with somewhat higher concentrations being required for F36VMpl inhibition. Virtually identical patterns of inhibition by the Jak2 inhibitor AG490, and the pan-Jak inhibitor (Jak1), indicated a strong dependency of F36VFGFR1 on Jak-Stat signaling, a finding confirmed by the demonstration that F36VFGFR1 could not sustain the ex vivo expansion of marrow cells derived from STAT5a/bΔNΔN mice25 (Figure 2B). Different responses were observed using U0126, an inhibitor of mitogen-activated protein (MAP) kinase kinase, which potently inhibited F36VFGFR1-mediated proliferation while leaving F36VMpl-induced proliferation unaffected. F36VFGFR1's specific dependency on MAP kinase signaling for proliferation was not reflected in more intense Erk1/2 phosphorylation; in fact, Erk1/2 phosphorylation was less intense following F36VFGFR1 activation compared with F36VMpl (Figure 2C).
The MAP kinase kinase inhibitor, U0126, distinguishes F36VMpl- and F36VFGFR1-induced cell proliferation. (A) F36VMpl-transduced (•) and F36VFGFR1-transduced (▴) mouse marrow cells were cultured in suspension in the presence of AP20187 (100 nM) and a range of concentrations of the following inhibitors: U71322 (phospholipase Cγ), SU6656 (Src kinases), Ly294002 (phosphatidylinositol-3 kinase), AG490 (Jak2), Jak inhibitor-1 (all Jak family members), and U0126 (MAP kinase kinase). Each inhibitor was evaluated at least twice with very similar results. (B) F36VFGFR1 was unable to sustain the ex vivo expansion of marrow cells from Stat5a/bΔNΔN mice (▵), whereas sustained growth was observed using marrow cells from C57Bl6/J mice (▴). No expansion of marrow cells from wild-type or Stat5a/bΔNΔN mice occurred in the absence of CID (data not shown). Initial growth of Stat5a/b knockout mouse marrow cells in response to F36VFGFR1 signaling was confirmed in a second independent experiment. (C) Intracellular flow cytometry shows only faint Erk1/2 phosphorylation following CID stimulation (solid lines) of both F36VFGFR1-transduced (left) and F36VMpl-transduced (right) marrow cells. Dashed line indicates before stimulation.
The MAP kinase kinase inhibitor, U0126, distinguishes F36VMpl- and F36VFGFR1-induced cell proliferation. (A) F36VMpl-transduced (•) and F36VFGFR1-transduced (▴) mouse marrow cells were cultured in suspension in the presence of AP20187 (100 nM) and a range of concentrations of the following inhibitors: U71322 (phospholipase Cγ), SU6656 (Src kinases), Ly294002 (phosphatidylinositol-3 kinase), AG490 (Jak2), Jak inhibitor-1 (all Jak family members), and U0126 (MAP kinase kinase). Each inhibitor was evaluated at least twice with very similar results. (B) F36VFGFR1 was unable to sustain the ex vivo expansion of marrow cells from Stat5a/bΔNΔN mice (▵), whereas sustained growth was observed using marrow cells from C57Bl6/J mice (▴). No expansion of marrow cells from wild-type or Stat5a/bΔNΔN mice occurred in the absence of CID (data not shown). Initial growth of Stat5a/b knockout mouse marrow cells in response to F36VFGFR1 signaling was confirmed in a second independent experiment. (C) Intracellular flow cytometry shows only faint Erk1/2 phosphorylation following CID stimulation (solid lines) of both F36VFGFR1-transduced (left) and F36VMpl-transduced (right) marrow cells. Dashed line indicates before stimulation.
Repopulation by F36VFGFR1-expanded cells
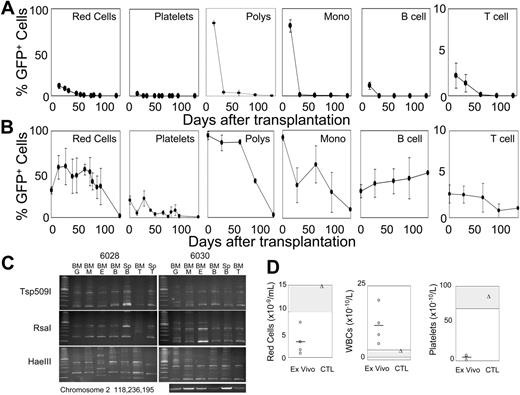
Our previous results have shown that F36VMpl cannot support the expansion of HSCs.10-14 In contrast, 8 × 106 cells, taken from cultures initiated with 2.5 × 106 F36VFGFR1-transduced marrow cells, and expanded 11 million–fold over 28 days in AP20187, reconstituted all hematopoietic lineages in each of 10 lethally irradiated mice (Figure 3A-B). In 5 mice given cotransplants of 1 × 105 fresh bone marrow cells (Figure 3A), ex vivo–expanded cells contributed to all hematopoietic lineages at day 15 after transplantation, and GFP+ granulocytes and monocytes surpassed 80%. However, by day 36 the progeny of cultured cells had virtually disappeared. One plausible explanation for these findings is that F36VFGFR1 supports the expansion of HSCs that are rapidly exhausted or out-competed by the fresh marrow cells. An equally plausible explanation is that F36VFGFR1 supports the expansion of various types of committed progenitors that can in aggregate generate all hematopoietic lineages, without expanding HSCs. To distinguish between these possibilities we examined 5 mice that received transplants of ex vivo–expanded cells only, without coinjection of fresh marrow cells. All mice in this group survived, and approximately 90% of granulocytes/monocytes, 59% of red cells, 51% of B cells, 22% of platelets, and 3% of T cells were of donor origin (Figure 3B). GFP-expressing Kupfer cells and lymphocytes repopulated the livers and lymph nodes, respectively, of mice that received transplants (data not shown). Sorted GFP+ granulocytes, macrophages, erythroid cells, B cells, and T cells from 2 mice that received transplants (no. 6028 and no. 6030) analyzed by LAM-PCR with 3 different restriction enzymes showed similar provirus insertion patterns (Figure 3C), Sequencing of LAM-PCR products from the RsaI arm of mouse 6030 revealed a common provirus insertion in B cells and granulocytes in chromosome 7 (position 30126217; data not shown) and a second insertion in chromosome 2 (position 118236195) that was present in all lineages by direct PCR using primers spanning the insert junction (Figure 3C, bottom panel). These findings confirm that F36VFGFR1 had induced the ex vivo expansion of 1 or more hematopoietic stem cells. However, beyond day 60 after transplantation, GFP+ red cells, platelets, granulocytes, and macrophages fell, dropping to very low levels by 4 months after transplantation (Figure 3B). Pancytopenia at 3 months after transplantation (Figure 3D; white cell counts were spuriously elevated due to an abundance of nucleated and macrocytic red cells) was consistent with a loss of hematopoietic reconstitution by the ex vivo–expanded cells, with incomplete compensation by residual host hematopoietic cells (data not shown). These findings are consistent with the interpretation that F36VFGFR1 signaling expanded HSCs with short-term repopulating ability (stHSCs). The magnitude and persistence of repopulation by F36VFGFR1-expanded cells varied considerably between different experiments, and consistently fell when cells were beyond 28 days in culture (data not shown).
F36VFGFR1 can expand stHSCs. (A-B) Average percentages of GFP+ red cells, platelets, granulocytes (polys), monocytes (monos), B cells, and T cells in 10 mice that were given transplants of 8 × 106 F36VFGFR1-transduced cells obtained from cultures initiated with 2.5 × 106 transduced marrow cells, and expanded 11 × 106-fold over 28 days in AP20187. (A) Results from 5 mice that were given cotransplants of 1 × 105 fresh bone marrow cells. (B) Results from 5 mice that were given transplants of F36VFGFR1-expanded cells alone. Error bars depict standard deviations. (C) Three-arm LAM-PCR of sorted granulocytes (G), monocytes (M), erythroid cells (E), B cells (B), and T cells (T) from the marrow (BM) and spleens (Sp) of 2 mice (numbers 6028 and 6030) from panel B. Sequencing of LAM-PCR products from mouse 6030 revealed a provirus insertion in chromosome 2 at position 118236195. DNAs from the indicated cell isolates were subjected to 38 cycles of amplification using an LTR primer and a host genomic primer. (D) Individual (symbols) and average (line) red cell, white cell, and platelet counts of mice depicted in panel B, 3 months after transplantation (ex vivo; circles) and a normal control mouse (CTL; triangles). A second identical experiment showed lower levels of donor-origin cells for only 65 days after transplantation. The repopulating ability of F36VFGFR1-transduced cells fell dramatically when cultured for longer than 28 days (data not shown).
F36VFGFR1 can expand stHSCs. (A-B) Average percentages of GFP+ red cells, platelets, granulocytes (polys), monocytes (monos), B cells, and T cells in 10 mice that were given transplants of 8 × 106 F36VFGFR1-transduced cells obtained from cultures initiated with 2.5 × 106 transduced marrow cells, and expanded 11 × 106-fold over 28 days in AP20187. (A) Results from 5 mice that were given cotransplants of 1 × 105 fresh bone marrow cells. (B) Results from 5 mice that were given transplants of F36VFGFR1-expanded cells alone. Error bars depict standard deviations. (C) Three-arm LAM-PCR of sorted granulocytes (G), monocytes (M), erythroid cells (E), B cells (B), and T cells (T) from the marrow (BM) and spleens (Sp) of 2 mice (numbers 6028 and 6030) from panel B. Sequencing of LAM-PCR products from mouse 6030 revealed a provirus insertion in chromosome 2 at position 118236195. DNAs from the indicated cell isolates were subjected to 38 cycles of amplification using an LTR primer and a host genomic primer. (D) Individual (symbols) and average (line) red cell, white cell, and platelet counts of mice depicted in panel B, 3 months after transplantation (ex vivo; circles) and a normal control mouse (CTL; triangles). A second identical experiment showed lower levels of donor-origin cells for only 65 days after transplantation. The repopulating ability of F36VFGFR1-transduced cells fell dramatically when cultured for longer than 28 days (data not shown).
In contrast to F36VFGFR1, 8 × 106 cultured marrow cells similarly expanded for 28 days in response to F36VMpl signaling were unable to rescue lethally irradiated mice (data not shown). Nevertheless, at a very early timepoint after transplantation (day 15), a significant proportion of circulating cells were GFP+ in the recipients of F36VMpl-expanded cells, albeit at levels generally lower than in recipients of F36VFGFR1-expanded cells (Table 1). T cells were a notable exception, which, although evanescent, were generated at much higher frequencies in the mice given transplants of F36VMpl-expanded cells (P < .001). Despite the narrow window of time in which F36VFGFR1-expanded cells possessed repopulating ability, and the variability in the magnitude of repopulation, F36VFGFR1 consistently outperformed F36VMpl in these assays.
Percentage of GFP+ cells in mice that were given transplants of marrow cells expanded in response to F36VFGFR1 or F36VMpl signaling
. | . | . | F36VFGFR1 with helper . | . | F36VMpl with helper . | . | ||
|---|---|---|---|---|---|---|---|---|
| Cell type . | F36VFGFR1 without helper, 15 d after BMT . | F36VMpl without helper, 15 d after BMT . | 15 d after BMT . | 36 d after BMT . | 15 d after BMT . | 36 d after BMT . | ||
| RBCs | 31.4 (4.3) | 11.1 (5.2) | 11.2 (2.8) | 5.7 (1.2) | 1.8 (0.3) | 0.9 (0.1) | ||
| Platelets | 20.1 (4.7) | 20.1 (10.7) | 2.5 (0.3) | 0.1 (0.04) | 0.37 (0.1) | 0.02 (0.02) | ||
| Granulocytes | 94.8 (1.9) | 88.4 (10.9) | 83.3 (3.1) | 4.3 (2.4) | 15.5 (5.0) | 0.1 (0.09) | ||
| B cells | 30.5 (7.6) | 14.5 (14.8) | 11.7 (3.8) | 0.2 (0.1) | 0.6 (0.2) | 0.09 (0.02) | ||
| T cells | 2.7 (1.05) | 65.6 (9.9) | 2.3 (1.4) | 1.4 (1.0) | 43.5 (12.5) | 0.08 (0.04) | ||
| Monocytes | 91.4 (3.1) | 69.2 (26.6) | 80.3 (7.1) | 1.1 (0.6) | 2.3 (0.5) | 0.1 (0.09) | ||
. | . | . | F36VFGFR1 with helper . | . | F36VMpl with helper . | . | ||
|---|---|---|---|---|---|---|---|---|
| Cell type . | F36VFGFR1 without helper, 15 d after BMT . | F36VMpl without helper, 15 d after BMT . | 15 d after BMT . | 36 d after BMT . | 15 d after BMT . | 36 d after BMT . | ||
| RBCs | 31.4 (4.3) | 11.1 (5.2) | 11.2 (2.8) | 5.7 (1.2) | 1.8 (0.3) | 0.9 (0.1) | ||
| Platelets | 20.1 (4.7) | 20.1 (10.7) | 2.5 (0.3) | 0.1 (0.04) | 0.37 (0.1) | 0.02 (0.02) | ||
| Granulocytes | 94.8 (1.9) | 88.4 (10.9) | 83.3 (3.1) | 4.3 (2.4) | 15.5 (5.0) | 0.1 (0.09) | ||
| B cells | 30.5 (7.6) | 14.5 (14.8) | 11.7 (3.8) | 0.2 (0.1) | 0.6 (0.2) | 0.09 (0.02) | ||
| T cells | 2.7 (1.05) | 65.6 (9.9) | 2.3 (1.4) | 1.4 (1.0) | 43.5 (12.5) | 0.08 (0.04) | ||
| Monocytes | 91.4 (3.1) | 69.2 (26.6) | 80.3 (7.1) | 1.1 (0.6) | 2.3 (0.5) | 0.1 (0.09) | ||
After 28 days of ex vivo culture in the presence of AP20187, 8 × 106 cells containing either F36VFGFR1 or F36VMpl were transplanted into lethally irradiated mice, which were in turn evaluated on days 15 and 36 after transplantation. Without helper indicates 8 × 106 ex vivo—expanded cells transplanted alone, without fresh marrow cells; with helper, ex vivo—expanded cells transplanted in combination with 1 × 105 fresh marrow cells. All values are mean percentages of GFP+ cells (SD).
F36VFGFR1 signaling allows long-term repopulating HSCs to survive growth factor deprivation
Our studies of ex vivo expansion demonstrated that F36VFGFR1 signaling was unable to expand HSCs with long-term repopulating ability (ltHSCs). Among the potential reasons for a lack of effect on ltHSCs is that they may be impervious to F36VFGFR1 signaling; alternatively, AP20187 may not efficiently enter, or may be efficiently extruded from ltHSCs. Relevant to the latter possibility is that ABC transporters, capable of extruding a wide range of pharmacologic agents, are known to be highly expressed on ltHSCs.32 We therefore tested whether AP20187 exposure allowed for the survival of F36VFGFR1-transduced ltHSCs under conditions of growth factor deprivation. Marrow cells from 5FU-treated mice were transduced with the F36VFGFR1 vector, then plated in 6-well dishes using 5 × 105 cells/well in serum-containing media without added cytokines, with or without AP20187 (100 nM). After 1 or 5 days of culture, the entire contents of each well were collected and transplanted into lethally irradiated congenic recipients at a ratio of 1 well per recipient. Mice reconstituted with CID-exposed cells were compared with recipients of cells cultured for the same period of time without CID, and with mice that received transplants of 5 × 105 cells from the same transduction that were not cultured but injected immediately following gene transfer (Figure 4A).
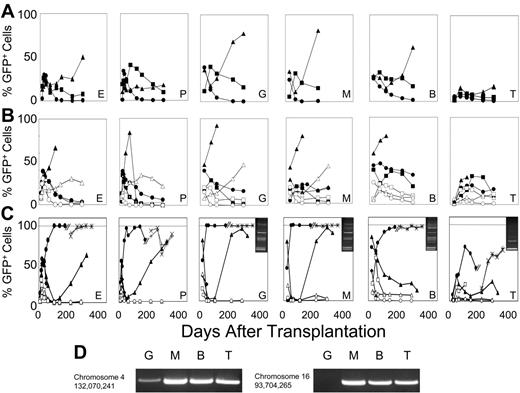
F36VFGFR1 supports the survival of long-term repopulating HSCs during growth factor deprivation. Marrow cells (5 × 105) transduced with the F36VFGFR1 vector were transplanted into lethally irradiated recipients either immediately following transduction (A) or after 1 (B) or 5 days (C) of culture in the presence (solid symbols) or absence (open symbols) of 100 nM AP20187. Mice that were given transplants of cultured cells received all the progeny generated in cultures initiated with 5 × 105 transduced cells at day 0. Each line depicts results from a single mouse. One mouse (6281) that was given transplants of cells cultured for 5 days in AP20187 was killed at day 174 after transplantation and 5 × 106 marrow cells from this mouse were transplanted into 2 lethally irradiated secondary recipients (symbols × and +). E indicates erythroid; P, platelets, G, granulocytes; M, monocytes; B, B cells; and T, T cells. (C; inserts) LAM-PCR using RsaI confirms common provirus insertion patterns in the granulocytes (G), monocytes (M), B cells (B), and T cells (T) of mouse 6281. MW indicates DNA ladder. Similar results were obtained using 2 other restriction enzymes, Tsp509 I and HaeIII. Sequencing of LAM-PCR products from mouse 6281 (panel D) revealed provirus insertion sites at the indicated positions in chromosomes 4 and 16, and their presence in the indicated lineages was confirmed by PCR using using an LTR primer, a host genomic primer, and 38 cycles of amplification. A second independent experiment performed in serum-free conditions showed a similar trend at 64 days after transplantation.
F36VFGFR1 supports the survival of long-term repopulating HSCs during growth factor deprivation. Marrow cells (5 × 105) transduced with the F36VFGFR1 vector were transplanted into lethally irradiated recipients either immediately following transduction (A) or after 1 (B) or 5 days (C) of culture in the presence (solid symbols) or absence (open symbols) of 100 nM AP20187. Mice that were given transplants of cultured cells received all the progeny generated in cultures initiated with 5 × 105 transduced cells at day 0. Each line depicts results from a single mouse. One mouse (6281) that was given transplants of cells cultured for 5 days in AP20187 was killed at day 174 after transplantation and 5 × 106 marrow cells from this mouse were transplanted into 2 lethally irradiated secondary recipients (symbols × and +). E indicates erythroid; P, platelets, G, granulocytes; M, monocytes; B, B cells; and T, T cells. (C; inserts) LAM-PCR using RsaI confirms common provirus insertion patterns in the granulocytes (G), monocytes (M), B cells (B), and T cells (T) of mouse 6281. MW indicates DNA ladder. Similar results were obtained using 2 other restriction enzymes, Tsp509 I and HaeIII. Sequencing of LAM-PCR products from mouse 6281 (panel D) revealed provirus insertion sites at the indicated positions in chromosomes 4 and 16, and their presence in the indicated lineages was confirmed by PCR using using an LTR primer, a host genomic primer, and 38 cycles of amplification. A second independent experiment performed in serum-free conditions showed a similar trend at 64 days after transplantation.
Mice reconstituted with cells cultured for only 1 day in CID did not differ markedly from the recipients of cells cultured for the same period without CID (Figure 4B). In contrast, 5 days of culture produced a striking and sustained effect (Figure 4C). The percentage of GFP+ cells in 2 mice surviving 174 days after transplantation averaged 63%, 67%, 94%, 59%, and 28% for red cells, platelets, granulocytes, B cells, and T cells, respectively, compared with average values of 0.075%, 0.025%, 0%, 4%, and 1.4% in 2 recipients of cultured, non-CID–exposed cells. Sorted granulocytes, monocytes, and B cells from 1 of the recipients of CID-exposed marrow cells contained multiple shared provirus insertions, whereas T cells harbored 2 major insertions, 1 of which was present in the other lineages (Figure 4C inserts). Shared provirus insertion sites were confirmed by sequencing of LAM-PCR products, and by PCR using primers spanning the insert junctions (Figure 4D). Two secondary transplant recipients from this animal averaged more than 95% GFP+ cells in all lineages at 4 months after secondary transplantation (Figure 4C), confirming that 5 days of ex vivo CID exposure resulted in selection of ltHSCs.
F36VFGFR1's ability to support the survival of ltHSCs in culture contrasts with that of F36VMpl, which we have previously found to be unable to support HSC survival during growth factor deprivation10 (and data not shown).
Receptor-specified in vivo selection
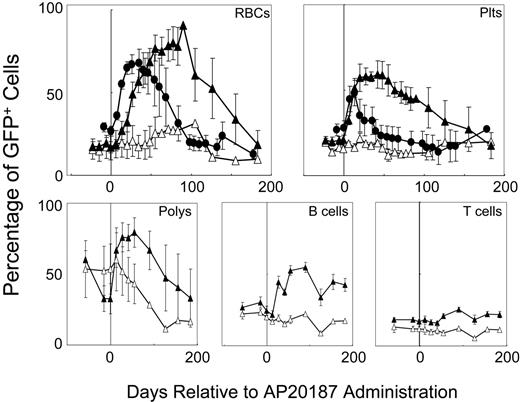
To determine whether the different hematopoietic responses to F36VFGFR1 and F36VMpl signaling in vitro are reflected in vivo, we compared the responses to a 14-day course of AP20187 administration in 5 mice that received transplants of F36VFGFR1-transduced marrow cells with the responses of 5 recipients of F36VMpl-transduced marrow cells (Figure 5). Mice harboring F36VMpl responded to CID administration with a 2.5-fold increase in GFP+ red cells (from 26.6% to a peak of 67.8%), and a 1.7-fold rise in GFP+ platelets (27.7% to 48.2%). The duration of these responses from the initiation of CID treatment was 91 days and 40 days, respectively. Much more pronounced responses were obtained in recipients of F36VFGFR1-transduced marrow cells. GFP+ red cells rose 5.3-fold (from an baseline of 17.1% to a peak of 90.6%) and platelets rose 2.8-fold (from 20.8% to 59.2%). Also in contrast to F36VMpl, which has modest effects on murine leukocytes,10 GFP+ granulocytes in the F36VFGFR1 group rose 2.4-fold (from 32.7% to 79.5%), B cells rose 2.3-fold (from 24.3% to 54.7%), and T cells rose 1.49-fold (from 17% to 25.4%). Even more striking than the differences in magnitude of the F36VFGFR1 response (versus F36VMpl) was the duration of the F36VFGFR1 response. Area under the curve (AUC) measurements following CID treatment were 2-fold greater for F36VFGFR1 relative to F36VMpl for GFP+ red cells (P = .12) and 9.8-fold higher for GFP+ platelets (P = .04). Increases in GFP+ red cells, platelets, granulocytes, B cells, and T cells were sustained for a minimum of 6 months in the F36VFGFR1 group following the initiation of CID treatment. The prolonged duration of the response to F36VFGFR1 signaling is consistent with a CID-induced expansion of primitive hematopoietic cells, possibly stHSCs, in vivo. The eventual decline in GFP+ cells, combined with the lack of persistently high levels of GFP+ cells in secondary transplant recipients (data not shown), is consistent with F36VFGFR1's inability to expand ltHSCs. While the F36VMpl response was quantitatively inferior to that of F36VFGFR1, its onset was more rapid than that of F36VFGFR1 (Figure 5).
Receptor-modulated in vivo selection. Percentages of GFP+ red cells, platelets (plts), granulocytes (polys), B cells, and T cells in 5 mice that were given transplants of F36VFGFR1-transduced marrow cells (▴) and GFP+ red cells and platelets in 4 mice that were given transplants of F36VMpl-transduced marrow cells (•) prior to and following a 14-day course of AP20187. Vertical line depicts the start of CID treatment; ▵ depicts mean values of 4 control mice that were given transplants of F36VFGFR1-transduced marrow cells but not treated with CID. Error bars depict SEM.
Receptor-modulated in vivo selection. Percentages of GFP+ red cells, platelets (plts), granulocytes (polys), B cells, and T cells in 5 mice that were given transplants of F36VFGFR1-transduced marrow cells (▴) and GFP+ red cells and platelets in 4 mice that were given transplants of F36VMpl-transduced marrow cells (•) prior to and following a 14-day course of AP20187. Vertical line depicts the start of CID treatment; ▵ depicts mean values of 4 control mice that were given transplants of F36VFGFR1-transduced marrow cells but not treated with CID. Error bars depict SEM.
F36VMpl accelerates hematologic recovery following myeloablative irradiation
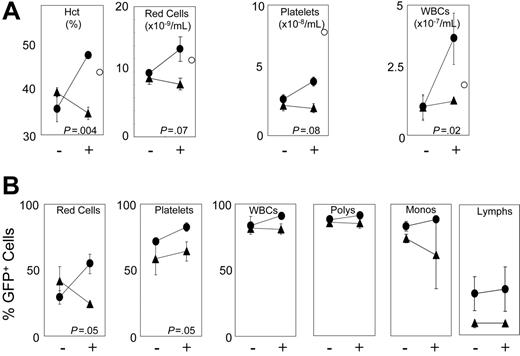
To examine whether receptors can increase absolute blood counts in a clinically relevant setting, we tested marrow cells transduced with either F36VFGFR1 or F36VMpl for their ability to accelerate hematologic recovery following myeloablative conditioning. Mice that received transplants of F36VFGFR1-transduced or F36VMpl-transduced marrow cells were treated, immediately following transplantation, with either a 3-day course of AP20187 or vehicle alone. Complete blood counts on day 6 after transplantation showed no difference between groups, and an extremely low frequency of GFP+ cells (< 2%) in all lineages (data not shown). However, by day 12 after transplantation, CID-treated mice that received transplants of F36VMpl-transduced marrow cells had significantly higher hematocrits (P = .004) and a trend toward higher platelet counts (P = .08) relative to non-CID–treated controls (Figure 6A). Leukocyte counts were spuriously elevated in CID-treated mice that received transplants of F36VMpl-transduced marrow cells due to the incorrect assignment of reticulocytes by the particle counter (data not shown). Responses in CID-treated recipients of F36VMpl-transduced marrow cells were also reflected in increased percentages of GFP+ red cells, platelets, and (to a modest degree) leukocyte subsets (Figure 6B). In contrast, no response to CID treatment was observed in recipients of F36VFGFR1-transduced marrow cells day 12 after transplantation, despite levels of GFP+ cells approximating those observed in the non-CID–treated F36VMpl controls (Figure 6).
F36VMpl accelerates hematologic recovery following myeloablative irradiation. Twelve mice exposed to myeloablative irradiation (1050 cGy) were given transplants of either 7.6 × 105 F36VMpl-transduced (circles) or 6.6 × 105 F36VFGFR1-transduced (triangles) marrow cells, with gene transfer rates into progenitors of 65.6% and 46.6%, respectively. Beginning immediately following the infusion of transduced marrow cells, half of the mice in each group were treated with daily injections of AP20187 (10 mg/kg) for 3 doses (+). – indicates no CID. Complete blood counts were obtained 12 days after transplantation. (A) Complete blood counts. White cells were spuriously elevated in CID-treated mice that were given transplants of F36VMpl-transduced marrow cells due to the incorrect assignment of reticulocytes by the particle counter. ○ indicates measurements obtained from a normal control mouse. (B) Percentages of GFP+ red cells, platelets, WBCs, polymorphonuclear cells (polys), monocytes (monos), and lymphocytes (lymphs). Values reflect means of 3 mice. Error bars indicate SDs. All P values reflect differences between CID-versus non-CID–treated mice that were given transplants of F36VMpl-transduced marrow cells.
F36VMpl accelerates hematologic recovery following myeloablative irradiation. Twelve mice exposed to myeloablative irradiation (1050 cGy) were given transplants of either 7.6 × 105 F36VMpl-transduced (circles) or 6.6 × 105 F36VFGFR1-transduced (triangles) marrow cells, with gene transfer rates into progenitors of 65.6% and 46.6%, respectively. Beginning immediately following the infusion of transduced marrow cells, half of the mice in each group were treated with daily injections of AP20187 (10 mg/kg) for 3 doses (+). – indicates no CID. Complete blood counts were obtained 12 days after transplantation. (A) Complete blood counts. White cells were spuriously elevated in CID-treated mice that were given transplants of F36VMpl-transduced marrow cells due to the incorrect assignment of reticulocytes by the particle counter. ○ indicates measurements obtained from a normal control mouse. (B) Percentages of GFP+ red cells, platelets, WBCs, polymorphonuclear cells (polys), monocytes (monos), and lymphocytes (lymphs). Values reflect means of 3 mice. Error bars indicate SDs. All P values reflect differences between CID-versus non-CID–treated mice that were given transplants of F36VMpl-transduced marrow cells.
Discussion
Beginning more than 15 years ago, erythropoietin, G-CSF, and granulocyte-macrophage CSF (GM-CSF), in rapid succession, transformed many areas of medical practice. Since then, relatively few growth factors with novel biological activity have entered the clinic, and none has gained widespread use. The paucity of new factors in the clinic comes not from a slowdown in their discovery, but from difficulties in delivery and a lack of specificity, reducing efficacy and promoting side effects. FGFs illustrate the problem. A number of different FGFs (23) bind 7 different receptor isoforms that are variably expressed in all tissues. Orchestrating this complexity requires that FGFs be presented in temporally and spatially distinct contexts.18,19 Furthermore, full FGFR activation requires the added presence of heparin,33-35 and some FGFs appear to act intracellularly.36 Unsurprisingly, this physiologic elegance has been difficult to exploit therapeutically.37-40
A focus on receptors and other signaling molecules,17,41 rather than ligands, might address many of these issues. In contrast to the promiscuity of ligands that can activate multiple receptors, or receptors expressed by multiple tissues, CIDs can activate specific receptors in specific tissues. Both AP20187 and a closely related CID currently under development for clinical use, AP1903, were specifically designed to bind the F36V-modified FK506-binding protein (FKBP) variant, and both exhibit a dramatically reduced affinity for endogenous FKBP.42 In a phase 1 trial in healthy human volunteers, the side effects of AP1903 were negligible, restricted to a single episode of facial flushing.43 Beyond hematopoiesis, applications of CID-regulated cell proliferation might extend to many tissues,3 including myoblasts26 and pancreatic beta cells.34 Applying CIDs within the hematopoietic system might open the door to novel biological activities unattainable using growth factors, and need not be constrained to receptors expressed endogenously. However, realizing this potential depends on the ability to use different signaling molecules to achieve different outcomes.
Results presented here demonstrate that hematopoiesis can be differentially regulated by 2 different growth factor receptors. HSCs recognize the F36VFGFR1 and F36VMpl signals differently, since only F36VFGFR1 allowed for the survival of ltHSCs and variable expansion of stHSCs. Distinct patterns of differentiation were also apparent. Starting either with unfractionated or purified (c-kit+sca-1+lin–) marrow cells, morphologic differences between F36VMpl and F36VFGFR1 were evident by day 6 of culture, with the former inducing the emergence of megakaryocytes and the latter, granulocytes and monocytes. These rapidly apparent differences in mature cell types are consistent with the possibility that F36VMpl or F36VFGFR1 might be influencing lineage commitment decisions in HSCs; however, they are equally well explained by differences among hematopoietic lineages in their susceptibility to F36VFGFR1- or F36VMpl-induced proliferation and differentiation. Further insight into the basis for these differences, and their relationship to relative levels of transgene expression (Figure 1G) will come from a direct prospective comparison between F36VMpl and F36VFGFR1 in highly purified ltHSCs, stHSCs, and various subsets of lineage-committed progenitors.2
F36VFGFR1 joins F36VMpl10-16 and a conditional derivative of Jak2 (V′VJak2)17,41 as the third CID-regulated “proliferation switch” with in vivo hematopoietic activity identified to date. With the caveat that direct comparisons of these signaling proteins are limited, and that the hematologic responses may depend on the duration of CID treatment, these signaling proteins appear to possess a progressively additive range of activities (Table 2), ranking from V′VJak2 to F36VMpl to F36VFGFR1. Each regulates the CID-dependent in vivo expansion of transduced red cells, suggesting that red cell expansion may represent a basal response. In addition to being a more potent expander of red cells than V′VJak2,17 F36VMpl has the added ability to expand platelets in vivo (albeit modestly), and to promote the sustained growth of transduced mouse bone marrow cells in culture, without added growth factors.10 By comparison, F36VFGFR1 can regulate an even broader repertoire of hematopoietic cell types (stHSCs and ltHSCs in vitro; all hematopoietic lineages in vivo), and surpasses F36VMpl in the magnitude and persistence of the hematopoietic response in vivo. Thus, the activities of each signaling domain fall within a series of concentric spheres, with the in vivo expansion of red cells at the center, and effects on granulocytes/monocytes and HSCs at the periphery. As new proliferation switches are developed, it will be of interest to determine whether their activities fall outside this spectrum by expanding myelomonocytic cells or HSCs, without also expanding red cells.
Hematologic responses to 3 different conditional signaling proteins
Response . | F36VFGFR1 . | F36VMpl . | V′ VJak2 . |
|---|---|---|---|
| In vitro | |||
| Sustained expansion in culture | + | + | - |
| Expansion of stHSCs | + | - | ND |
| Survival of ltHSCs | + | - | ND |
| In vivo | |||
| Red cells | +++ | ++ | + |
| Platelets | +++ | + | - |
| Granulocytes/monocytes | +++ | +/- | - |
Response . | F36VFGFR1 . | F36VMpl . | V′ VJak2 . |
|---|---|---|---|
| In vitro | |||
| Sustained expansion in culture | + | + | - |
| Expansion of stHSCs | + | - | ND |
| Survival of ltHSCs | + | - | ND |
| In vivo | |||
| Red cells | +++ | ++ | + |
| Platelets | +++ | + | - |
| Granulocytes/monocytes | +++ | +/- | - |
+ indicates present; -, absent; ND, not done; +++, strongest effect; ++, strong effect; +/-, variable and modest effect.
Although F36VFGFR1 outperformed F36VMpl during steady-state hematopoiesis, only F36VMpl accelerated recovery from anemia following myeloablative conditioning. F36VMpl's ability to mitigate anemia following a myelosuppressive insult might be applied to the treatment of cancer-related anemia, where potential off-target effects of erythropoietin were associated with an increased risk of death due to tumor progression in 2 phase 3 clinical trials.44,45 This application is just 1 example of how a CID-regulated receptor may hold a significant theoretic advantage over conventional growth factors. The delay between the initiation of F36VFGFR1 signaling and the subsequent hematologic response may be too long to allow for its use in this particular application. However, in the context of a linked therapeutic gene, F36VFGFR1's ability to expand primitive hematopoietic cells may be advantageous for gene therapy of Fanconi anemia, or its effect on myelomonocytic cells may be useful for gene therapy of diseases such as mucopolysaccharidoses, leukocyte adhesion deficiency, or chronic granulomatous disease.
The different hematopoietic responses to F36VFGFR1 versus F36VMpl are ultimately based in differences in signal transduction. In this regard, cells expressing F36VFGFR1 or F36VMpl were strikingly similar in their susceptibilities to most signaling inhibitors. Notably different responses occurred with the Mek1/2 antagonist U0126, which significantly inhibited F36VFGFR1 proliferation at concentrations permissive for F36VMpl-induced expansion. It will be of interest to determine whether Mek1/2 signaling underlies F36VFGFR1's distinctive effects on HSCs. A dependency of F36VFGFR1 on Jak-Stat signaling was confirmed by its failure to support the sustained ex vivo expansion of marrow cells from Stat5a/bΔNΔN mice, consistent with a requirement for Stat5 activation in transformation mediated by an oncogenic FGFR1 fusion.46 The ability of marrow cells from Stat5a/bΔNΔN mice to expand for up to 8 weeks in response to F36VFGFR1 signaling, but no longer (Figure 2B), suggests that cells capable of only transient proliferation are Stat5 independent, whereas cells capable of sustaining the culture beyond 8 weeks are Stat5 dependent. These findings may be consistent with the previously demonstrated ability of hematopoietic progenitor cells from Stat5a/bΔNΔN mice to respond to a variety of cytokines,25 and with the inability of marrow cells from Stat5a/bΔNΔN mice to radioprotect quaternary transplant recipients.47
In summary, our results demonstrate that HSCs can distinguish signals delivered by 2 different receptors, and that CID-based methods for regulation can elicit a diversity of hematologic responses, both in vitro and in vivo. Further studies using F36VFGFR1 may provide the opportunity to establish causality between specific signaling pathways and desired HSC behaviors.
Authorship
One of the authors (C.A.B.) is named as an inventor on a patent application owned by the University of Washington that is related to the work described in this study.
Prepublished online as Blood First Edition Paper, August 10, 2006; DOI 10.1182/blood-2006-01-012278.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank James Ihle and Evan Parganas for Stat5a/bΔNΔN mice, and Nancy Lin and Virginia Broudy for technical advice.
This work was supported by National Institutes of Health grants DK52997, DK61844, DK074522, and HL53750.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal