Abstract
In patients with chronic myeloid leukemia (CML), kinase domain mutations account for imatinib resistance in the majority of cases. Mutations cause either a direct steric hindrance to drug binding or a conformational change that favors kinase activation, which therefore precludes imatinib binding. We have previously characterized the dual Src-Abl kinase inhibitor AP23464 and found it to effectively suppress the growth of cells expressing native and essentially all imatinib-resistant variants of BCR-ABL, with the notable exception of the gatekeeper T315I mutant (
Refinement of small-molecule kinase inhibitors by the integration of sequential screening of panels of mutants coupled with structural analysis is a powerful drug discovery paradigm that is applicable to an increasing number of targeted therapeutic agents.
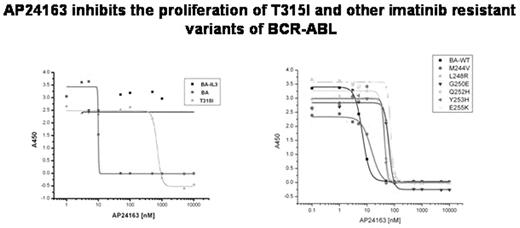
INHIBITION OF PROLIFERATION OF BAF3 CELLS EXPRESSING BCR-ABL AND ITS VARIANTS (IC50 in nM)
| . | IMATINIB . | AP23464 . | AP23846 . | AP24163 . |
|---|---|---|---|---|
| WT | 600 | 14 | 500 | 7 |
| T315I | >20000 | >1000 | 500 | 480 |
| L248R | >20000 | 92 | ND | 64 |
| G250E | 5000 | 25 | ND | 63 |
| Q252H | 3000 | 40 | ND | 42 |
| Y253H | 18000 | 32 | ND | 44 |
| E255K | 12000 | 74 | ND | 24 |
| BAF3+IL3 | >20000 | >1000 | 500 | >10000 |
| . | IMATINIB . | AP23464 . | AP23846 . | AP24163 . |
|---|---|---|---|---|
| WT | 600 | 14 | 500 | 7 |
| T315I | >20000 | >1000 | 500 | 480 |
| L248R | >20000 | 92 | ND | 64 |
| G250E | 5000 | 25 | ND | 63 |
| Q252H | 3000 | 40 | ND | 42 |
| Y253H | 18000 | 32 | ND | 44 |
| E255K | 12000 | 74 | ND | 24 |
| BAF3+IL3 | >20000 | >1000 | 500 | >10000 |
Figure
Disclosure: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal