To gain insight into the poorly understood pathophysiology of the myelodysplastic syndromes (MDSs), we have determined the gene expression profiles of the CD34+ cells of 55 patients with MDS by using a comprehensive array platform. These profiles showed many similarities to reported interferon-γ-induced gene expression in normal CD34+ cells; indeed the 2 most up-regulated genes, IFIT1 and IFITM1, are interferon-stimulated genes (ISGs). Alterations in the expression of ISGs may play a role in the hematologic features of MDS, such as peripheral blood cytopenias. Up-regulation of IFIT1 is a potential diagnostic marker for MDS. We determined whether distinct gene expression profiles were associated with specific FAB and cytogenetic groups. CD34+ cells from patients with refractory anemia with ringed sideroblasts (RARS) showed a particular gene expression profile characterized by up-regulation of mitochondrial-related genes and, in particular, of those of heme synthesis (eg, ALAS2). CD34+ cells from patients with the del(5q) had a distinct gene expression profile, characterized by down-regulation of genes assigned to 5q, and up-regulation of the histone HIST1 gene cluster at chromosome 6p21 and of genes related to the actin cytoskeleton. This study provides important and new insights into the pathophysiology of MDS. (Blood. 2006;108:337-345)
Introduction
The myelodysplastic syndromes (MDSs) are a heterogeneous group of hematopoietic malignancies, characterized by blood cytopenias, ineffective hematopoiesis, and a hypercellular bone marrow.1,2 Dysplasia of at least one lineage (myeloid, erythroid, or megakaryocyte-platelet) is a characteristic feature of MDSs. Typically the bone marrow is hypercellular which contrasts to the cytopenias observed in the peripheral blood.1,2 The MDSs are preleukemic conditions with transformation into acute myeloid leukemia (AML) occurring in approximately 30% to 40% of cases.1,2 The classification of MDS is based on morphologic anomalies according to the French-American-British (FAB) Cooperative Study Group.3 This group defined 5 subtypes based on the percentage of immature blasts in the bone marrow, the presence of ringed sideroblasts, and the degree of monocytosis. The 5 groups are refractory anemia (RA), RA with ringed sideroblasts (RARS), RA with excess blasts (RAEB), RAEB in transformation (RAEB-T), and chronic myelomonocytic leukemia (CMML).3 More recently this classification has been modified by the World Health Organization (WHO).4 For example, RAEB has been subdivided into 2 categories, RAEBI and RAEBII, based on the percentage of bone marrow blasts.4 While the WHO classification system has some clear advantages, the FAB classification remains widely used in clinical practice.
Patients with RA and RARS have anemia, often transfusion dependent, and have a relatively low risk of progression to acute myeloid leukemia.1,5 Patients with RA and RARS have ineffective erythropoiesis, primarily because of a high rate of erythroid apoptosis.6,7 RARS is a subtype of MDS in which excess iron accumulates in the mitochondria of the erythroid precursors.1,8 Patients with RAEB and those with RAEB-T generally have a poor prognosis, with a median survival ranging from 5 to 12 months.9 MDS represents an excellent model of leukemic development with a progressive increase of blastic bone marrow involvement, but little is known about the genetic events that lead to this evolution.2,10 The disease course and prognosis are highly variable in MDS.1,2 It would be of great value for the management of patients with MDS if a set of molecular markers could be identified enabling better prediction of disease progression and prognosis. With the exception of lenalidomide, which is effective in early MDS,11 there are few effective drug treatments for this disorder, and most patients receive supportive care only (blood transfusions). Therefore, the identification of new targets in MDS for drug treatment is a priority.
Karyotypic abnormalities are detected by conventional analysis in about 50% of patients with primary MDS and 80% of patients with secondary MDS.12 The characteristic MDS abnormalities involve loss of genetic material, whereas balanced translocations are rare. The most common cytogenetic abnormalities in MDS include the del(5q), trisomy 8, and del(7q)/-7.12 Few specific gene abnormalities have been implicated in the development or progression of MDS.10,13 The MDSs remain poorly understood: the pathophysiology is uncertain and classification is based on morphology.
Progress has been made in the areas of classification, outcome prediction, pathway delineation, and treatment target identification of various hematologic malignancies through the application of gene expression profiling.14-19 We have studied the transcriptome of the CD34+ cells of a large group of patients with MDS to gain insight into the pathophysiology of MDS, to discover molecular markers for diagnosis and prognosis, and to determine whether distinct gene expression profiles could be identified for specific groups of MDSs as defined by either the FAB classification or karyotype.
Materials and methods
Sample collection and cell separation
Fifty-five patients with MDS and 11 healthy control subjects were included in the study. Classification of patients with MDS was according to the FAB criteria,3 and the patients were selected solely on the basis of having MDS. The MDS patient samples were obtained from several sources: Oxford and Bournemouth (United Kingdom), Duisburg (Germany), and Pavia (Italy). At the time of investigation, 18 patients had RA, 19 had RARS, and 18 had RAEB. The RAEB group was further subdivided into RAEBI (9 patients) and RAEBII (9 patients) according to the WHO classification4 for a selected part of analysis. The International Prognostic Scoring System (IPSS)5 score could be calculated for 54 of the 55 patients with MDS: 18 patients were low risk, 26 patients were intermediate-1 (Int-1), 7 patients were intermediate-2 (Int-2), and 3 patients were high risk. Twenty of 55 patients with MDS had a del(5q). At the time of investigation, only 1 of the 55 patients with MDS was receiving treatment (thalidomide). The median disease duration was 12 months; 12 patients were newly diagnosed. The study was approved by the ethics committees (Oxford C00.196, Bournemouth 9991/03/E, Duisburg 2283/03, Pavia 26264/2002), and informed consent was obtained. Bone marrow samples were obtained, and CD34+ cells isolated from patients with MDS and from healthy control subjects. Mononuclear cells were purified by Histopaque (Sigma-Aldrich, Gillingham, United Kingdom) density gradient centrifugation and labeled with CD34 MicroBeads, and CD34+ cells were isolated using MACS magnetic cell separation columns (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's recommendations. The median number of CD34+ cells isolated from MDS samples was 4 × 105. CD34+ cell purity was evaluated with fluorescence-activated cell sorting (FACS) and was at least 90%.
RNA extraction
Total RNA was extracted using TRIZOL (Invitrogen, Paisley, United Kingdom) following the protocol supplied by the manufacturer. The median RNA yield from MDS samples was 260 ng. An aliquot of the RNA samples was conserved for evaluation of quality using Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA).
Labeling and hybridization
For each sample, 50 ng total RNA was amplified and labeled with the 2-Cycle cDNA Synthesis and the 2-Cycle Target Labeling and Control Reagent packages (Affymetrix, Santa Clara, CA) following the manufacturer's recommendations. Biotin-labeled fragmented cRNA (15 μg) was hybridized to GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix), covering more than 47 000 transcripts representing 39 000 human genes. Hybridization occurred at 45°C for 16 hours in a Hybridization Oven 640 (Affymetrix). Chips were then washed and stained in a Fluidics Station 450 (Affymetrix) and scanned with a GeneChip Scanner 3000 (Affymetrix).
Data analysis
Cell intensity calculation and scaling was performed using GeneChip Operating Software (GCOS) and data analysis using GeneSpring 7.2 (Agilent Technologies). Quality control was performed within the GCOS software after scaling the signal intensities of all arrays to a target of 100. The scale factors for all samples were comparable and less than 3.0. Background levels, percentage of present calls, intensities of spike hybridization controls were within the acceptable range for all samples. The 3′/5′ GAPDH ratio was less than 3.0 for all samples. Affymetrix CEL files were uploaded and preprocessed using Robust MultiChip Analysis (RMA).20 The raw signal of each gene for each patients with MDS was divided by the median raw signal of the gene in the healthy control subjects to obtain an expression ratio. Genes showing small variability between conditions were excluded prior to data analysis. Statistical analysis was performed on log-transformed data using Welch approximate t test or ANOVA and Benjamini-Hochberg21 multiple testing correction to control false discovery rate. SAM (significance analysis for microarrays)22 was also used to identify differentially expressed genes. Hierarchical clustering was performed using standard correlation. Biologic themes within lists of genes were identified using the Gene Ontology tool within the NetAffx analysis center resource available at www.affymetrix.com. The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE4679.
Real-time quantitative PCR
Real-time quantitative polymerase chain reaction (PCR)23,24 was used to validate expression data for selected genes. The expression level of the beta-2-microglobulin (B2M) gene was used to normalize for differences in input cDNA. Predeveloped TaqMan Assays were used (Assays-onDemand; Applied Biosystems, Foster City, CA). Each sample was performed in triplicate, and a reverse-transcriptase negative control was also tested to exclude any contaminating DNA amplification. The expression ratio was calculated as 2n, where n is the CT value difference for each patient (selected gene minus B2M) normalized by the average CT difference of the samples from healthy control subjects (ΔΔCT method).25
Results
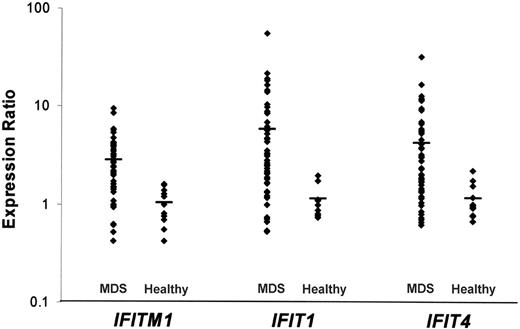
The CD34+ cells obtained from 55 patients with MDS were compared with the CD34+ cells of healthy individuals using gene expression profiling. We identified several genes that were up- or down-regulated in the CD34+ cells of most patients with MDS. Twenty-one genes were up-regulated with a ratio greater than 2.0 in at least 28 patients with MDS (Table 1). IFITM1 and IFIT1, 2 interferon-stimulated genes (ISGs), were up-regulated in 37 of 55 patients with MDS (Table 1; Figure 1). DLK1 was up-regulated in 33 of 55 patients with MDS (9 RA, 14 RARS, and 10 RAEB). The most represented gene category for commonly up-regulated genes was “signal transduction” (4 genes; eg, IFITM1, RAB27B, RGS18, and PTHR2).
The 10 most up-regulated and the 10 most down-regulated genes in CD34+ cells of patients with MDS
Affymetrix ID . | Common name . | Map . | No. of patients; n = 55 . | RA; n = 18 . | RARS; n = 19 . | RAEB; n = 18 . |
|---|---|---|---|---|---|---|
| Most up-regulated | ||||||
| 203153_at | IFIT1 | 10q25-q26 | 37 | 13 | 15 | 9 |
| 214022_s_at | IFITM1 | 11p15.5 | 37 | 13 | 11 | 13 |
| 219777_at | hIAN2 | 7q36.1 | 35 | 12 | 10 | 13 |
| 209560_s_at | DLK1 | 14q32 | 33 | 9 | 14 | 10 |
| 228708_at | RAB27B | 18q21.2 | 32 | 10 | 11 | 11 |
| 206772_at | PTHR2 | 2q33 | 31 | 8 | 12 | 11 |
| 202382_s_at | GNPDA1 | 5q21 | 30 | 6 | 14 | 10 |
| 1554242_a_at | COCH | 14q12-q13 | 30 | 8 | 16 | 6 |
| 221081_s_at | FLJ22457 | 1p13.2 | 30 | 10 | 14 | 6 |
| 206698_at | XK | xp21.1 | 30 | 10 | 14 | 6 |
| Most down-regulated | ||||||
| 230493_at | TMEM46 | 13q12,13 | 45 | 16 | 13 | 16 |
| 241679_at | AKAP12 | 6q24-q25 | 44 | 13 | 13 | 18 |
| 231935_at | ARPP-21 | 3p22.3 | 44 | 13 | 13 | 18 |
| 221969_at | PAX5 | 9p13 | 43 | 14 | 12 | 17 |
| 227846_at | GPR | 15q14 | 43 | 14 | 13 | 16 |
| 210450_at | LOC90925 | 14q32.33 | 43 | 13 | 12 | 18 |
| 208078_s_at | TCF8 | 10p11.2 | 43 | 16 | 14 | 13 |
| 220068_at | VPREB3 | 22q11.23 | 43 | 13 | 12 | 18 |
| 232204_at | EBF | 5q34 | 42 | 13 | 12 | 17 |
| 203434_s_at | MME | 3q25.1-q25.2 | 42 | 12 | 12 | 18 |
Affymetrix ID . | Common name . | Map . | No. of patients; n = 55 . | RA; n = 18 . | RARS; n = 19 . | RAEB; n = 18 . |
|---|---|---|---|---|---|---|
| Most up-regulated | ||||||
| 203153_at | IFIT1 | 10q25-q26 | 37 | 13 | 15 | 9 |
| 214022_s_at | IFITM1 | 11p15.5 | 37 | 13 | 11 | 13 |
| 219777_at | hIAN2 | 7q36.1 | 35 | 12 | 10 | 13 |
| 209560_s_at | DLK1 | 14q32 | 33 | 9 | 14 | 10 |
| 228708_at | RAB27B | 18q21.2 | 32 | 10 | 11 | 11 |
| 206772_at | PTHR2 | 2q33 | 31 | 8 | 12 | 11 |
| 202382_s_at | GNPDA1 | 5q21 | 30 | 6 | 14 | 10 |
| 1554242_a_at | COCH | 14q12-q13 | 30 | 8 | 16 | 6 |
| 221081_s_at | FLJ22457 | 1p13.2 | 30 | 10 | 14 | 6 |
| 206698_at | XK | xp21.1 | 30 | 10 | 14 | 6 |
| Most down-regulated | ||||||
| 230493_at | TMEM46 | 13q12,13 | 45 | 16 | 13 | 16 |
| 241679_at | AKAP12 | 6q24-q25 | 44 | 13 | 13 | 18 |
| 231935_at | ARPP-21 | 3p22.3 | 44 | 13 | 13 | 18 |
| 221969_at | PAX5 | 9p13 | 43 | 14 | 12 | 17 |
| 227846_at | GPR | 15q14 | 43 | 14 | 13 | 16 |
| 210450_at | LOC90925 | 14q32.33 | 43 | 13 | 12 | 18 |
| 208078_s_at | TCF8 | 10p11.2 | 43 | 16 | 14 | 13 |
| 220068_at | VPREB3 | 22q11.23 | 43 | 13 | 12 | 18 |
| 232204_at | EBF | 5q34 | 42 | 13 | 12 | 17 |
| 203434_s_at | MME | 3q25.1-q25.2 | 42 | 12 | 12 | 18 |
The genes are ranked by number of patients showing up-regulation (ratio > 2.0) or down-regulation (ratio < 0.5). The breakdown according to FAB subtype of patients showing up- or down-regulation is also indicated.
We found that several of the genes commonly up-regulated in the patients with MDS included in our study were ISGs. Twenty-two ISGs previously described in the literature were confirmed by Zeng et al26(Tab 2) as increased by interferon-γ (IFN-γ) in normal CD34+ cells. Fifteen of these 22 ISGs were up-regulated in the CD34+ cells of a significant number of the patients with MDS in our study. In particular, the genes IFITM1, IFIT1, and IFIT4 were elevated by greater than 2-fold in most of our patients with MDS (Figure 1).
One hundred nine genes were down-regulated with a ratio less than 0.5 in at least 28 patients with MDS, including Gravin/AKAP12, ARPP-21, ELA2, CD24, MME, and EBF (Table 1). The most represented gene categories for commonly down-regulated genes were signal transduction, (26 genes; eg, Gravin/AKAP12, LEF1) immune response (16 genes; eg, CD24, PAX5), and regulation of transcription (26 genes; eg, EBF, FOSB).
We sought to identify diagnostic markers for patients with MDS with RA and a normal karyotype. One up-regulated gene (IFIT1) and 3 down-regulated genes (TMEM46, TCF8, and NR4A2) were identified in 6 of 7 such patients with MDS.
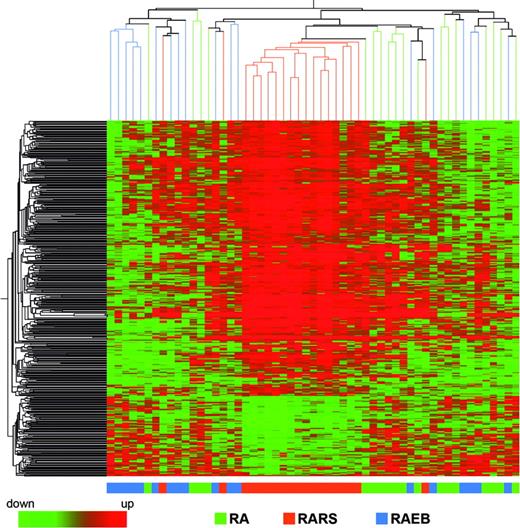
We investigated differences in gene expression that could distinguish patients with MDS according to their FAB subtype classification. We applied ANOVA using P value cutoff of .01 and Benjamini-Hochberg multiple testing correction, and 457 significantly different probe sets were identified. A similar result was obtained with SAM analysis. Hierarchical clustering performed using these 457 probe sets showed that patients with MDS with RARS constitute a homogeneous group, whereas patients with MDS with RA and RAEB show more overlap (Figure 2). Leave one out cross-validation classified correctly 62% of the samples, with the percentage increasing to 79% when considering only patients with MDS with RARS.
Gene expression levels for the interferon-stimulated genes IFITM1, IFIT1, and IFIT4. Each point represents 1 patient with MDS or a healthy individual, and the bar represents the mean value.
Gene expression levels for the interferon-stimulated genes IFITM1, IFIT1, and IFIT4. Each point represents 1 patient with MDS or a healthy individual, and the bar represents the mean value.
Hierarchical clustering of 457 differentially expressed genes in the patients with MDS with RA, RARS, and RAEB. Each row represents a single Affymetrix (or Affy) probe set. Each column represents a separate patient CD34+ sample.
Hierarchical clustering of 457 differentially expressed genes in the patients with MDS with RA, RARS, and RAEB. Each row represents a single Affymetrix (or Affy) probe set. Each column represents a separate patient CD34+ sample.
The most significant gene ontology categories represented in the biological process branch were heme biosynthesis (7 probe sets; eg, ALAS2, ALAD), regulation of transcription (33 probe sets; eg, GATA1, KLF1), and protein biosynthesis (25 probe sets; eg, EIF2B3, KIAA0664). We observed increased gene expression of many enzymes in the heme biosynthesis pathway; 5 genes involved in this pathway (ALAS2, ALAD, HMBS, UROD, FECH) were differentially expressed with higher average expression levels in patients with MDS with RARS (Table 2). Other genes expressed at higher levels in RARS include GATA-1, CA2, and EPO-R (Table 2). The most significant gene ontology category represented in the cellular component branch was mitochondrion which included 44 probe sets (eg, CGI-69, TRAP1, TIMM10, and several mitochondrial ribosomal proteins) (Table 2). Forty-three of these 44 probe sets had higher average expression levels in patients with MDS with RARS than in patients with MDS with RA or RAEB.
Genes differentially expressed in RARS
. | RA . | RAEB . | RARS . |
|---|---|---|---|
| Heme pathway genes | |||
| FECH | 1.35 (0.69-3.89) | 1.07 (0.45-2.02) | 2.20 (0.79-5.78) |
| ALAS2 | 2.00 (0.65-20.44) | 1.60 (0.60-29.17) | 12.77 (1.16-104.4) |
| ALAD | 1.12 (0.74-2.84) | 0.97 (0.66-2.19) | 1.93 (0.88-4.90) |
| HMBS | 1.10 (0.60-2.43) | 0.94 (0.52-2.02) | 2.41 (0.89-8.39) |
| UROD | 1.03 (0.43-2.24) | 0.91 (0.30-2.22) | 1.73 (0.87-2.83) |
| Other erythroid genes | |||
| GATA1 | 1.22 (0.79-2.38) | 0.90 (0.52-2.21) | 1.91 (0.80-3.36) |
| CA2 | 1.15 (0.17-8.97) | 0.88 (0.12-2.63) | 1.90 (0.28-10.36) |
| EPO-R | 1.40 (0.70-3.36) | 1.02 (0.41-2.70) | 2.08 (1.19-4.66) |
| Mitochondrial genes | |||
| CGI-69 | 1.31 (0.75-3.09) | 1.49 (0.77-4.71) | 3.03 (1.23-9.33) |
| TRAP1 | 0.92 (0.42-1.59) | 0.93 (0.43-2.58) | 1.62 (0.79-2.75) |
| TIMM10 | 0.96 (0.59-1.90) | 0.93 (0.39-2.24) | 1.64 (0.65-2.79) |
. | RA . | RAEB . | RARS . |
|---|---|---|---|
| Heme pathway genes | |||
| FECH | 1.35 (0.69-3.89) | 1.07 (0.45-2.02) | 2.20 (0.79-5.78) |
| ALAS2 | 2.00 (0.65-20.44) | 1.60 (0.60-29.17) | 12.77 (1.16-104.4) |
| ALAD | 1.12 (0.74-2.84) | 0.97 (0.66-2.19) | 1.93 (0.88-4.90) |
| HMBS | 1.10 (0.60-2.43) | 0.94 (0.52-2.02) | 2.41 (0.89-8.39) |
| UROD | 1.03 (0.43-2.24) | 0.91 (0.30-2.22) | 1.73 (0.87-2.83) |
| Other erythroid genes | |||
| GATA1 | 1.22 (0.79-2.38) | 0.90 (0.52-2.21) | 1.91 (0.80-3.36) |
| CA2 | 1.15 (0.17-8.97) | 0.88 (0.12-2.63) | 1.90 (0.28-10.36) |
| EPO-R | 1.40 (0.70-3.36) | 1.02 (0.41-2.70) | 2.08 (1.19-4.66) |
| Mitochondrial genes | |||
| CGI-69 | 1.31 (0.75-3.09) | 1.49 (0.77-4.71) | 3.03 (1.23-9.33) |
| TRAP1 | 0.92 (0.42-1.59) | 0.93 (0.43-2.58) | 1.62 (0.79-2.75) |
| TIMM10 | 0.96 (0.59-1.90) | 0.93 (0.39-2.24) | 1.64 (0.65-2.79) |
The average ratio in each group is given with the range in parentheses.
To identify genes differentially expressed between early and advanced MDS, a comparison was made between the 18 patients with RA and the 9 patients with MDS with RAEBII. We applied a t test using a P value cutoff of .05 and Benjamini-Hochberg multiple testing correction, and 762 significantly different probe sets were identified. Hierarchical clustering performed using these 762 probe sets could group together patients with MDS with RAEBII (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). Leave one out cross-validation classified correctly 78% of the samples. The most significant gene ontology categories represented were cell-matrix adhesion (18 probe sets; eg CD44, TNXB) and positive regulation of signal transduction (12 probe sets; eg, HIPK2, NDFIP2). The CASP3 gene was one of the genes with the lowest P value (.012) with higher expression levels in patients with RA and decreasing in patients with RAEBII. Other genes showing higher expression levels in patients with RA, decreasing in patients with RAEBII, include EMS1 (P = .021), PRKAR2B (P = .016), and LEF1 (P = .02). Genes with lower expression levels in patients with RA and increasing in patients with RAEBII include FLT3 (P = .021) and DHRS8 (P = .018). Two hundred ninety-one probe sets were significantly different (P < .01, Benjamini-Hochberg multiple testing correction) between low-risk (IPSS score, low or Int-1) and high-risk (IPSS score, Int-2 or high) patients with MDS. Most of these 291 probe sets, including the 20 most significant ones, overlap with the 762 probe sets that could distinguish patients with MDS with RA from patients with MDS with RAEBII.
We determined whether a distinct gene expression profile was associated with specific cytogenetic groups. We investigated differences in gene expression between the 20 patients with MDS with a del(5q), 25 patients with MDS with a normal karyotype, and 10 patients with MDS with other karyotypes. We applied ANOVA using a P value cutoff of .01 and Benjamini-Hochberg multiple testing correction, and 889 significantly different probe sets were identified (Table 3), 266 of which map to chromosome 5q. A similar result was obtained with SAM analysis. Hierarchical clustering performed using these 889 probe sets could group separately the patients with MDS with a del(5q), whereas patients with MDS with normal karyotype and patients with MDS with other karyotypes could not be separated (Figure 3). Leave one out cross-validation classified correctly 95% of the patients with MDS with a del(5q). The most significant gene ontology categories represented for the 889 probe sets were translational initiation (12 probe sets; eg, EIF3S4, EIF3S7) and nucleosome assembly (21 probe sets; eg, HIST1 gene family).
The most significant genes expressed at higher or lower levels in MDS del(5q)
Affymetrix ID . | Common . | P . | MDS del5q . | MDS normal . | MDS other . | Map . |
|---|---|---|---|---|---|---|
| Genes expressed at higher levels in MDS del(5q) | ||||||
| 213513_x_at | ARPC2 | < .001 | 1.26 (1.05-1.62) | 0.96 (0.72-1.16) | 1.00 (0.84-1.18) | 2q36.1 |
| 208496_x_at | HIST1H3G | < .001 | 1.42 (0.91-2.35) | 0.87 (0.56-1.31) | 0.87 (0.70-1.04) | 6p21.3 |
| 208180_s_at | HIST1H4H | < .001 | 9.03 (0.91-71.12) | 1.13 (0.54-3.22) | 0.92 (0.63-1.54) | 6p21.3 |
| 201938_at | CDK2AP1 | < .001 | 1.60 (1.17-2.38) | 1.02 (0.39-1.43) | 1.03 (0.56-1.34) | 12q24.31 |
| 214469_at | HIST1H2AE | < .001 | 6.38 (1.09-46.71) | 1.11 (0.57-3.10) | 0.90 (0.41-1.62) | 6p22.2-p21.1 |
| 202746_at | ITM2A | < .001 | 1.97 (1.22-3.92) | 1.16 (0.70-2.33) | 1.21 (0.67-2.07) | Xq13.3-Xq21.2 |
| 221676_s_at | CORO1C | < .001 | 1.86 (1.08-3.37) | 0.93 (0.35-1.65) | 0.93 (0.67-1.33) | 12q24.1 |
| 201312_s_at | SH3BGRL | < .001 | 1.28 (0.87-1.75) | 0.96 (0.77-1.22) | 0.97 (0.88-1.10) | xq13.3 |
| 214472_at | HIST1H3D | < .001 | 4.14 (1.60-25.44) | 1.15 (0.54-6.90) | 1.16 (0.50-5.23) | 6p21.3 |
| 235479_at | CPEB2 | < .001 | 1.39 (0.71-3.18) | 0.91 (0.58-1.33) | 0.71 (0.55-0.89) | 4p15.33 |
| 206110_at | HIST1H3H | < .001 | 11.43 (2.19-120.60) | 1.16 (0.60-2.75) | 0.92 (0.49-2.05) | 6p22-p21.3 |
| 238199_x_at | MYBPC3 | < .001 | 1.28 (0.65-1.90) | 0.89 (0.59-1.19) | 0.80 (0.64-0.93) | 11p11.2 |
| 215779_s_at | HIST1H2BC | < .001 | 3.64 (1.15-17.89) | 1.19 (0.75-2.71) | 0.97 (0.65-1.72) | 6p21.3 |
| 208579_x_at | HIST1H2BM | < .001 | 5.46 (1.23-25.16) | 1.35 (0.77-3.26) | 1.45 (0.34-5.23) | 6p22-p21.3 |
| 223144_s_at | C6orf166 | < .001 | 1.49 (0.73-5.00) | 0.92 (0.35-1.91) | 0.77 (0.60-0.99) | 6q15 |
| 212082_s_at | MYL6 | < .001 | 1.28 (0.90-1.97) | 0.94 (0.70-1.14) | 0.92 (0.79-1.08) | 12q13.13 |
| 238585_at | FLJ11753 | < .001 | 1.18 (0.79-1.61) | 0.84 (0.59-1.13) | 0.92 (0.71-1.20) | 2q22.3-q23.1 |
| 202821_s_at | LPP | .001 | 1.20 (0.76-1.86) | 0.98 (0.73-1.48) | 0.84 (0.74-0.98) | 3q27-q28 |
| 221613_s_at | AWP1 | .001 | 1.26 (0.82-1.80) | 0.95 (0.70-1.21) | 0.85 (0.63-0.99) | 15q24.3 |
| 201238_s_at | CAPZA2 | .001 | 1.79 (1.22-3.08) | 1.19 (0.77-1.67) | 1.13 (0.75-1.63) | 7q31.2-q31.3 |
| Genes, localized to 5q, expressed at lower levels in MDS del(5q) | ||||||
| 203538_at | CAMLG | < .001 | 0.53 (0.36-0.85) | 1.13 (0.84-1.52) | 1.15 (0.58-1.78) | 5q23 |
| 203024_s_at | C5orf15 | < .001 | 0.53 (0.36-0.77) | 1.02 (0.81-1.35) | 1.01 (0.77-1.32) | 5q31.1 |
| 220495_s_at | C5orf14 | < .001 | 0.45 (0.28-0.95) | 1.10 (0.56-1.60) | 1.14 (0.85-1.73) | 5q31.2 |
| 224068_x_at | RBM22 | < .001 | 0.54 (0.29-1.08) | 1.21 (0.79-1.82) | 1.41 (1.08-1.90) | 5q33.1 |
| 202227_s_at | BRD8 | < .001 | 0.63 (0.40-1.04) | 1.25 (0.80-1.66) | 1.19 (0.79-2.04) | 5q31 |
| 206562_s_at | CSNK1A1 | < .001 | 0.60 (0.36-0.93) | 1.06 (0.65-1.38) | 1.09 (0.76-1.29) | 5q32 |
| 201977_s_at | KIAA0141 | < .001 | 0.71 (0.46-1.18) | 1.23 (0.66-1.89) | 1.45 (1.17-2.01) | 5q31.3 |
| 225698_at | TIGA1 | < .001 | 0.43 (0.19-1.04) | 1.23 (0.81-2.24) | 1.37 (0.66-1.80) | 5q21-q22 |
| 232181_at | LOC153346 | < .001 | 0.57 (0.28-1.08) | 1.23 (0.63-2.03) | 1.31 (1.05-1.97) | 5q33.1 |
| 202042_at | HARS | < .001 | 0.52 (0.28-0.85) | 1.16 (0.76-1.50) | 1.21 (0.65-1.76) | 5q31.3 |
| Genes, not localized to 5q, expressed at lower levels in MDS del(5q) | ||||||
| 223157_at | C4orf14 | < .001 | 0.64 (0.25-1.07) | 1.44 (0.88-2.86) | 1.59 (0.98-2.84) | 4q12 |
| 1556967_at | ZDHHC14 | < .001 | 0.84 (0.73-1.01) | 1.17 (0.87-1.61) | 0.95 (0.81-1.30) | 6q25.3 |
| 207170_s_at | HCCR1 | < .001 | 0.86 (0.52-1.35) | 1.63 (0.89-2.74) | 1.62 (1.02-2.32) | 12q13.13 |
| 228557_at | FLJ35936 | < .001 | 0.63 (0.30-1.50) | 1.47 (0.81-3.62) | 1.50 (0.90-2.30) | 18p11.31-p11.23 |
| 202080_s_at | OIP106 | < .001 | 0.75 (0.44-1.21) | 1.17 (0.79-1.82) | 1.23 (1.02-1.39) | 3p25.3-p24.1 |
| 206019_at | RBM19 | < .001 | 0.79 (0.58-1.21) | 1.22 (0.80-1.89) | 1.13 (0.90-1.68) | 12q24.21 |
| 201871_s_at | LOC51035 | < .001 | 0.70 (0.43-1.17) | 1.25 (0.73-2.03) | 1.41 (0.88-2.07) | 11q12.3 |
| 205321_at | EIF2S3 | < .001 | 0.78 (0.47-1.29) | 1.32 (0.71-2.30) | 1.63 (1.03-2.88) | Xp22.2-p22.1 |
| 201682_at | PMPCB | < .001 | 0.71 (0.47-0.98) | 1.02 (0.81-1.21) | 0.96 (0.62-1.19) | 7q22-q32 |
| 212307_s_at | OGT | < .001 | 0.49 (0.24-0.83) | 1.04 (0.58-2.51) | 0.94 (0.62-1.97) | Xq13 |
Affymetrix ID . | Common . | P . | MDS del5q . | MDS normal . | MDS other . | Map . |
|---|---|---|---|---|---|---|
| Genes expressed at higher levels in MDS del(5q) | ||||||
| 213513_x_at | ARPC2 | < .001 | 1.26 (1.05-1.62) | 0.96 (0.72-1.16) | 1.00 (0.84-1.18) | 2q36.1 |
| 208496_x_at | HIST1H3G | < .001 | 1.42 (0.91-2.35) | 0.87 (0.56-1.31) | 0.87 (0.70-1.04) | 6p21.3 |
| 208180_s_at | HIST1H4H | < .001 | 9.03 (0.91-71.12) | 1.13 (0.54-3.22) | 0.92 (0.63-1.54) | 6p21.3 |
| 201938_at | CDK2AP1 | < .001 | 1.60 (1.17-2.38) | 1.02 (0.39-1.43) | 1.03 (0.56-1.34) | 12q24.31 |
| 214469_at | HIST1H2AE | < .001 | 6.38 (1.09-46.71) | 1.11 (0.57-3.10) | 0.90 (0.41-1.62) | 6p22.2-p21.1 |
| 202746_at | ITM2A | < .001 | 1.97 (1.22-3.92) | 1.16 (0.70-2.33) | 1.21 (0.67-2.07) | Xq13.3-Xq21.2 |
| 221676_s_at | CORO1C | < .001 | 1.86 (1.08-3.37) | 0.93 (0.35-1.65) | 0.93 (0.67-1.33) | 12q24.1 |
| 201312_s_at | SH3BGRL | < .001 | 1.28 (0.87-1.75) | 0.96 (0.77-1.22) | 0.97 (0.88-1.10) | xq13.3 |
| 214472_at | HIST1H3D | < .001 | 4.14 (1.60-25.44) | 1.15 (0.54-6.90) | 1.16 (0.50-5.23) | 6p21.3 |
| 235479_at | CPEB2 | < .001 | 1.39 (0.71-3.18) | 0.91 (0.58-1.33) | 0.71 (0.55-0.89) | 4p15.33 |
| 206110_at | HIST1H3H | < .001 | 11.43 (2.19-120.60) | 1.16 (0.60-2.75) | 0.92 (0.49-2.05) | 6p22-p21.3 |
| 238199_x_at | MYBPC3 | < .001 | 1.28 (0.65-1.90) | 0.89 (0.59-1.19) | 0.80 (0.64-0.93) | 11p11.2 |
| 215779_s_at | HIST1H2BC | < .001 | 3.64 (1.15-17.89) | 1.19 (0.75-2.71) | 0.97 (0.65-1.72) | 6p21.3 |
| 208579_x_at | HIST1H2BM | < .001 | 5.46 (1.23-25.16) | 1.35 (0.77-3.26) | 1.45 (0.34-5.23) | 6p22-p21.3 |
| 223144_s_at | C6orf166 | < .001 | 1.49 (0.73-5.00) | 0.92 (0.35-1.91) | 0.77 (0.60-0.99) | 6q15 |
| 212082_s_at | MYL6 | < .001 | 1.28 (0.90-1.97) | 0.94 (0.70-1.14) | 0.92 (0.79-1.08) | 12q13.13 |
| 238585_at | FLJ11753 | < .001 | 1.18 (0.79-1.61) | 0.84 (0.59-1.13) | 0.92 (0.71-1.20) | 2q22.3-q23.1 |
| 202821_s_at | LPP | .001 | 1.20 (0.76-1.86) | 0.98 (0.73-1.48) | 0.84 (0.74-0.98) | 3q27-q28 |
| 221613_s_at | AWP1 | .001 | 1.26 (0.82-1.80) | 0.95 (0.70-1.21) | 0.85 (0.63-0.99) | 15q24.3 |
| 201238_s_at | CAPZA2 | .001 | 1.79 (1.22-3.08) | 1.19 (0.77-1.67) | 1.13 (0.75-1.63) | 7q31.2-q31.3 |
| Genes, localized to 5q, expressed at lower levels in MDS del(5q) | ||||||
| 203538_at | CAMLG | < .001 | 0.53 (0.36-0.85) | 1.13 (0.84-1.52) | 1.15 (0.58-1.78) | 5q23 |
| 203024_s_at | C5orf15 | < .001 | 0.53 (0.36-0.77) | 1.02 (0.81-1.35) | 1.01 (0.77-1.32) | 5q31.1 |
| 220495_s_at | C5orf14 | < .001 | 0.45 (0.28-0.95) | 1.10 (0.56-1.60) | 1.14 (0.85-1.73) | 5q31.2 |
| 224068_x_at | RBM22 | < .001 | 0.54 (0.29-1.08) | 1.21 (0.79-1.82) | 1.41 (1.08-1.90) | 5q33.1 |
| 202227_s_at | BRD8 | < .001 | 0.63 (0.40-1.04) | 1.25 (0.80-1.66) | 1.19 (0.79-2.04) | 5q31 |
| 206562_s_at | CSNK1A1 | < .001 | 0.60 (0.36-0.93) | 1.06 (0.65-1.38) | 1.09 (0.76-1.29) | 5q32 |
| 201977_s_at | KIAA0141 | < .001 | 0.71 (0.46-1.18) | 1.23 (0.66-1.89) | 1.45 (1.17-2.01) | 5q31.3 |
| 225698_at | TIGA1 | < .001 | 0.43 (0.19-1.04) | 1.23 (0.81-2.24) | 1.37 (0.66-1.80) | 5q21-q22 |
| 232181_at | LOC153346 | < .001 | 0.57 (0.28-1.08) | 1.23 (0.63-2.03) | 1.31 (1.05-1.97) | 5q33.1 |
| 202042_at | HARS | < .001 | 0.52 (0.28-0.85) | 1.16 (0.76-1.50) | 1.21 (0.65-1.76) | 5q31.3 |
| Genes, not localized to 5q, expressed at lower levels in MDS del(5q) | ||||||
| 223157_at | C4orf14 | < .001 | 0.64 (0.25-1.07) | 1.44 (0.88-2.86) | 1.59 (0.98-2.84) | 4q12 |
| 1556967_at | ZDHHC14 | < .001 | 0.84 (0.73-1.01) | 1.17 (0.87-1.61) | 0.95 (0.81-1.30) | 6q25.3 |
| 207170_s_at | HCCR1 | < .001 | 0.86 (0.52-1.35) | 1.63 (0.89-2.74) | 1.62 (1.02-2.32) | 12q13.13 |
| 228557_at | FLJ35936 | < .001 | 0.63 (0.30-1.50) | 1.47 (0.81-3.62) | 1.50 (0.90-2.30) | 18p11.31-p11.23 |
| 202080_s_at | OIP106 | < .001 | 0.75 (0.44-1.21) | 1.17 (0.79-1.82) | 1.23 (1.02-1.39) | 3p25.3-p24.1 |
| 206019_at | RBM19 | < .001 | 0.79 (0.58-1.21) | 1.22 (0.80-1.89) | 1.13 (0.90-1.68) | 12q24.21 |
| 201871_s_at | LOC51035 | < .001 | 0.70 (0.43-1.17) | 1.25 (0.73-2.03) | 1.41 (0.88-2.07) | 11q12.3 |
| 205321_at | EIF2S3 | < .001 | 0.78 (0.47-1.29) | 1.32 (0.71-2.30) | 1.63 (1.03-2.88) | Xp22.2-p22.1 |
| 201682_at | PMPCB | < .001 | 0.71 (0.47-0.98) | 1.02 (0.81-1.21) | 0.96 (0.62-1.19) | 7q22-q32 |
| 212307_s_at | OGT | < .001 | 0.49 (0.24-0.83) | 1.04 (0.58-2.51) | 0.94 (0.62-1.97) | Xq13 |
The average ratio and range is shown for each gene in the MDS del(5q), MDS with a normal karyotype, and MDS with other karyotype groups.
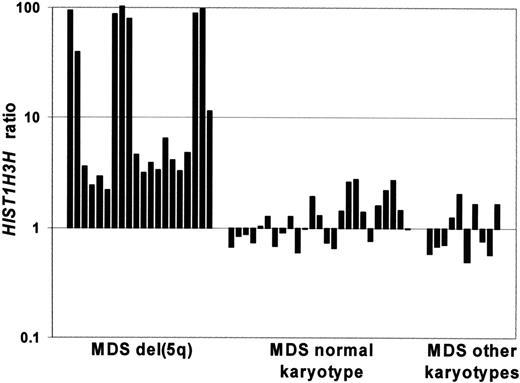
We found that many histone genes within the HIST1 gene cluster on chromosome 6p21 were expressed at higher levels in the patients with MDS with a del(5q) (Table 3; Figure 4). We investigated the expression levels of all probe sets (total number 65) for the members of this gene family and noted that average expression levels for 62 of 65 HIST1 probe sets were higher in the del(5q) group (range, 0.86-11.43) than the MDS with normal karyotype group (range, 0.75-1.35) and the MDS with other karyotype group (range, 0.61-1.45).
Several of the genes most significantly up-regulated in patients with a del(5q) encode actin-binding proteins or myosin-related proteins, including ARPC2, CORO1C, MYL6, CAPZA2, and WASPIP (Table 3). We also observed increased expression of megakaryocyte/platelet-associated genes, including PF4V1, PPBP, THBS1, GP1BA, and CD61, in several patients with MDS with a del(5q) compared with all other patients with MDS. One of the most elevated genes was PF4V1 with a greater than 2-fold increase in the expression levels in 15 patients with del(5q) and greater than 100-fold increase in 5 of these cases.
We plotted the ratios of 2 of the most discriminating genes (HIST1H3H and C5orf15) for the del(5q) in a scatterplot to determine how well just 2 genes could separate the del(5q) group from all other MDSs. Figure 5A shows that there is no overlap. Similarly, we sought to determine whether 2 actin-related genes (ARPC2 and WASPIP) plotted in the same way could separate the del(5q) group, and Figure 5B shows there is little overlap.
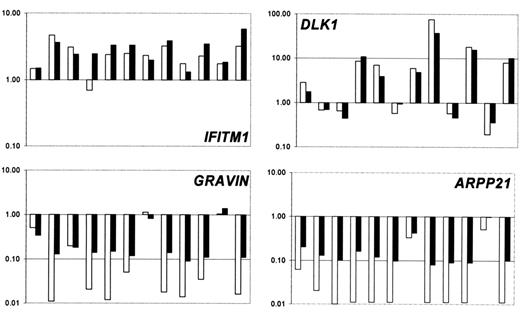
The gene expression results for several genes were validated using the TaqMan 5′ nuclease fluorogenic quantitative PCR assay. Expression levels obtained with Affymetrix chips were compared with the expression levels determined by using real-time quantitative PCR. We chose to investigate IFITM1, DLK1, Gravin/AKAP12, and ARPP21 (Figure 6), and PTHR2, RAB27B, ALAS2, HIST1H2AC, MME. The concordance between the 2 assays for all genes validated was high, indicating a good level of agreement between the 2 assays for the identification of deregulated genes.
Discussion
We have used the Affymetrix GeneChip U133 Plus 2.0 platform, on which most human genes are represented, to determine the transcriptome of the CD34+ cells of a large group of patients with MDS as compared with CD34+ cells of healthy control subjects. In particular, we sought to identify genes commonly deregulated in MDS and to determine whether distinct gene expression profiles could be identified for specific FAB subtypes or cytogenetic groups.
The 2 most up-regulated genes across the MDS spectrum were the interferon-stimulated genes, IFITM1 and IFIT1. This finding highlights the complex role that cytokines are thought to have in the regulation of hematopoiesis generally and, more particularly, in the bone marrow failure syndromes such as aplastic anemia, paroxysmal nocturnal hemoglobinuria, and MDS.27,28 IFN-γ,an inhibitory cytokine, can negatively affect hematopoietic stem cell self-renewal,29 and overexpression of IFN-γ has been demonstrated in the bone marrow of patients with MDS.30 Interestingly, we have found that the expression profiles of the CD34+ cells of the patients with MDS show similarities to IFN-γ-induced gene expression in CD34+ cells from healthy individuals.26 Of the 22 interferon-stimulated genes (ISGs) in the CD34+ cells of healthy individuals reported in the paper by Zeng et al,26(Tab 2) 15 were up-regulated in a significant number of the patients included in our study. In particular, the genes IFITM1, IFIT1, and IFIT4 were elevated by greater than 2-fold in most patients, with many patients showing expression levels greater than 10-fold. The interferoninduced transmembrane protein encoded by IFITM1 plays a role in the antiproliferative activity of interferons.31 IFITM1 associates with other proteins at the cell surface, forming a complex which relays growth inhibitory and cell adhesion signals.32 IFITM1 was positively correlated to IFITM2 (0.93, standard correlation) and IFITM3 (0.91, standard correlation). TNFSF10 (TRAIL), another ISG,26 was also up-regulated in the CD34+ cells of approximately 50% of the patients in our study.
The paradox between the hypercellular bone marrow and the cytopenias of the MDS is well recognized.1,2 It is often considered that increased apoptosis is the underlying cause of the ineffective hematopoiesis and peripheral blood cytopenia found in this disorder, although the exact cause-effect relation between these 2 features is not clear.33,34 We suggest that the up-regulation of ISG may be responsible for some of the hematologic characteristics of MDS, including cytopenias. Because IFN-γ is an important stimulator of apoptosis in erythroid progenitor cells,35 it may also play a role in the anemia that characterizes MDS.
Hierarchical clustering of 889 differentially expressed genes in patients with MDS with a del(5q), patients with MDS with normal karyotype, and patients with MDS with other karyotypes. Each row represents a single Affy probe set. Each column represents a separate patient CD34+ sample.
Hierarchical clustering of 889 differentially expressed genes in patients with MDS with a del(5q), patients with MDS with normal karyotype, and patients with MDS with other karyotypes. Each row represents a single Affy probe set. Each column represents a separate patient CD34+ sample.
The DLK1 gene, encoding a transmembrane protein belonging to the epidermal growth factor-like superfamily,36 was another commonly up-regulated gene in the patients with MDS included in our study. The up-regulation of DLK1 has been previously reported in patients with MDS in 2 studies.37,38 DLK1 was up-regulated by greater than 2-fold in 60% of patients with MDS included in our study. However, all FAB subtypes were represented, and DLK1 up-regulation was not characteristic of either early38 or advanced37 MDS, as previously reported.
Many genes were down-regulated in the CD34+ cells of most patients, including Gravin/AKAP12, ARPP-21, ELA2, and MME, some of which show absent expression in some patients with MDS. Gravin/AKAP12, a putative tumor suppressor gene, previously shown to be down-regulated in MDS and other myeloid malignancies,39 was down-regulated in the CD34+ cells of 44 of 55 patients with MDS studied and is one of the most commonly downregulated genes in MDS.
Bar graph showing the expression levels of HIST1H3H in patients with MDS. Expression levels of HIST1H3H, one of the differentially expressed genes of the HIST1 family, in the MDS del(5q), MDS with normal karyotype and MDS with other karyotype groups. Each bar represents the expression ratio in 1 patient.
Bar graph showing the expression levels of HIST1H3H in patients with MDS. Expression levels of HIST1H3H, one of the differentially expressed genes of the HIST1 family, in the MDS del(5q), MDS with normal karyotype and MDS with other karyotype groups. Each bar represents the expression ratio in 1 patient.
Scatterplots of genes which discriminate patients with MDS with del(5q) from other patients with MDS. Scatterplot of the ratios for the genes HIST1H3H and C5orf15 (A), and ARPC2 and WASPIP (B), for patients with MDS with del(5q) as compared with the other patients with MDS.
Scatterplots of genes which discriminate patients with MDS with del(5q) from other patients with MDS. Scatterplot of the ratios for the genes HIST1H3H and C5orf15 (A), and ARPC2 and WASPIP (B), for patients with MDS with del(5q) as compared with the other patients with MDS.
Reduced expression of genes involved in B-lymphocyte development in RA with a normal karyotype was recently reported.40 We similarly observed lower expression levels in RA of the 9 B-lymphocyte development genes previously shown to be downregulated by Sternberg et al.40 Moreover, with the exception of DNTT and BACH1, we observed lower expression levels of these genes in patients with MDS with RAEB than in patients with MDS with RA or RARS, presumably reflecting the progressive myeloid nature of this disease.
Confirmation of gene expression data. Comparison of the expression ratios obtained from real-time quantitative PCR (□) and Affymetrix experiments (▪) for selected genes in 12 patients with MDS from which enough material was left for real-time quantitative PCR.
Confirmation of gene expression data. Comparison of the expression ratios obtained from real-time quantitative PCR (□) and Affymetrix experiments (▪) for selected genes in 12 patients with MDS from which enough material was left for real-time quantitative PCR.
One of the goals of our gene expression profiling studies in MDS is to identify diagnostic markers for MDS and, in particular, for patients with RA and a normal karyotype, which is the group with the most diagnostic uncertainty. Among the genes commonly up- and down-regulated in patients with MDS, one gene was up-regulated (IFIT1) and 3 were down-regulated (TMEM46, TCF8, and NR4A2) in 6 of the 7 patients with MDS with RA and a normal karyotype. Further studies are needed to investigate the value of these markers for diagnosis.
We investigated differences in gene expression that could distinguish patients with MDS according to their FAB subtype classification. The patients were divided into 3 groups, RA, RARS, and RAEB, and hierarchical clustering was performed by using the 457 probe sets identified as significantly different. Patients with MDS with RARS were grouped into a cluster, with only 3 of 19 patients with RARS being placed outside this cluster. Patients with RA and RAEB showed considerable overlap. The demonstration that RARS has a particular gene expression profile suggests a common underlying pathophysiologic basis to the disease.
The abnormal mitochondrial iron metabolism that characterizes RARS is poorly understood. This has led to the suggestion that RARS is a disease resulting from a specific alteration in a gene(s) involved in mitochondrial function, iron distribution, or both.8,41 Interestingly, the most significant gene ontology category represented in the cellular component branch was mitochondrion which included 44 probe sets. Forty-three of these 44 probe sets had higher average expression levels in patients with MDS with RARS than RA or RAEB, including, for example, mitochondrial carrier CGI-69, mitochondrial matrix protein TRAP1, and mitochondrial membrane chaperone TIMM10. It is possible that the mitochondrialrelated genes shown to be differentially expressed in RARS by this study may represent candidate genes for this disorder.
We also observed up-regulation of several genes in the heme biosynthesis pathway in RARS; 5 genes involved in this pathway (ALAS2, ALAD, HMBS, UROD, FECH) were differentially expressed with higher average expression levels in patients with RARS. ALAS2 (aminolevulinate, delta-, synthase 2), the first enzyme of heme synthesis, showed the highest expression levels with marked up-regulation (an average increase of greater than 12-fold, with some cases showing greater than 100-fold increase) in patients with RARS compared with healthy control subjects. The other enzymes in the heme pathway (ALAD, HMBS, UROD, FECH) showed an average increase of approximately 2-fold in RARS. The increase in gene expression of ALAS2 may relate to the increased erythropoiesis observed in RARS or to a more specific deregulation of the gene.
We sought to identify genes differentially expressed between early and advanced MDS. The 9 patients with RAEBII4 could be distinguished from the 18 patients with RA according to their gene expression profiles. The most significant genes identified with higher expression levels in patients with RA than RAEBII include CASP3, PRKAR2B, LEF1, and EMS1. It is well recognized that there is a higher bone marrow apoptotic index in the bone marrow of patients with low-risk MDS compared with high-risk MDS and MDS-AML.42,43 Interestingly CASP3, the key effector caspase, was shown to have higher expression levels in RA than in RAEBII in our study. Enhanced CASP3 activation has been described in the mononuclear bone marrow cells of patients with RA and RARS.6 LEF1, a transcriptional regulator in the TGF-β and Wnt signaling cascade,44 is expressed at high levels in carcinomas,45 and EMS1, encoding an actin-associated protein involved in cytoskeleton organization, is located within a chromosomal region frequently amplified in cancer.46 The most significant genes identified with lower expression levels in patients with RA than RAEBII include FLT3 and DHRS8. Abnormalities involving FLT3, such as internal tandem duplications (FLT3-ITD), are common in AML and have prognostic value. Several groups have demonstrated an association between elevated FLT3 mRNA expression levels and poor clinical outcome in AML, both in patients with and without FLT3-ITD.15,47 Our demonstration that an increase in FLT3 expression levels is associated with disease progression in MDS is of particular interest, other genes differentially expressed between early and advanced MDS may have value as prognostic markers or markers of disease progression.
In AML the global gene expression patterns are strongly linked to karyotype as observed, for example, in t(8;21), t(15;17), inv(16), and 11q23 rearrangement.48,49 In this study we present the results of 2 karyotypic groups, namely del(5q) and normal karyotype MDS, and compare them with the remaining patients with MDS. We have shown that patients with MDS with the del(5q), the most frequently reported karyotypic abnormality in MDS,12,50 have a distinct gene expression profile. Hierarchical clustering grouped patients with a del(5q) into a distinct cluster, separate from both patients with a normal karyotype and those with other karyotypic abnormalities. The normal karyotype group did not have a distinct gene expression signature and considerable overlap with the residual patients (possessing a variety of karyotypes) was observed. Approximately 40% of the significant probe sets showing lower expression levels in patients with a del(5q) map to chromosome 5q, suggesting that the loss of 1 allele in these patients has a gene dosage effect. A similar observation has recently been made in patients with AML with the del(5q).51 The CTNNA1 gene, mapping to 5q31, was among the chromosome 5 probe sets expressed at lower levels in patients with a del(5q). Low or absent expression of CTNNA1 has been recently reported in a proportion of patients with MDS/AML with a del(5q).52 In our study none of the patients with del(5q) showed absent expression of CTNNA1, with most showing expression levels consistent with loss of 1 allele.
We found that many histone genes within the HIST1 gene cluster on chromosome 6p21 were expressed at significantly higher levels in the patients with MDS with a del(5q). The average expression levels for 62 of 65 HIST1 probe sets were higher in the del(5q) group than the MDS with normal karyotype and the MDS with other karyotypes groups. Indeed a number of patients with the del(5q) showed greater than 100-fold up-regulation of certain HIST1 genes, for example HIST1H3H. The overexpression of HIST1 genes in patients with MDS with a del(5q) may affect chromatin remodeling and expression of other genes.
Several of the genes most significantly up-regulated in patients with a del(5q), including ARPC2, CORO1C, and CAPZA2, encode actin-binding proteins or myosin-related proteins. ARPC2, for example, is one of the subunits of the Arp2/3 protein complex that controls actin polymerization in cells.53 Disruption of the actin cytoskeleton, and concomitant deregulation of related signal transduction pathways, is known to play a role in tumorigenesis.54 The CD34+ cells of patients with MDS with the del(5q) clearly have a particular abnormality in the expression of genes involved in the regulation or maintenance of the actin cytoskeleton. We also observed increased expression of megakaryocyte/platelet associated genes, including PF4V1, PPBP, and CD61, in several patients with MDS with a del(5q). Interestingly, the 5q-syndrome is associated with abnormal megakaryocyte morphology.55,56
We have identified many genes that are differentially expressed in patients with MDS with a del(5q). We sought to identify a minimum number of genes that could distinguish patients with MDS with a del(5q) from all other patients with MDS. We plotted the ratios for 2 of the most significant genes in a scatterplot to see whether they alone could separate these groups. The scatterplot obtained using the genes HIST1H3H and C5orf15 showed that indeed the patients with a del(5q) could be separated from all other MDS cases with no overlap. Similarly, we plotted the ratios for 2 actin-related genes, ARPC2 and WASPIP, and the scatterplot obtained showed a good separation between patients with a del(5q) and all other MDS cases. This illustrates the diagnostic potential for microarray gene expression in MDS, and clearly there is much scope for development here.
Finally, it has to be recognized that MDS bone marrows have both clonal and nonclonal CD34+ cells and that gene expression profiles may reflect this complexity. However, the percentage of cytogenetically abnormal cells was high (≥ 80%) in those cases where this information was available. In addition, circumstantial evidence that nonclonal CD34+ cells were not predominant is our demonstration of the marked down-regulation of certain genes, such as Gravin/AKAP12 and ARPP21, in most cases (clearly a high proportion of nonclonal CD34+ cells would mask these findings).
This is the first study to determine gene expression profiles in a large group of patients with MDS using an array platform containing most human genes. We have found that the upregulation of ISGs is a major feature of MDS hematopoietic stem cells, and we suggest that this may be important in the pathophysiology of MDS. We have shown that characteristic gene expression profiles could be identified for patients with MDS with RARS and patients with MDS with the del(5q). We have also identified several molecular markers that may be of value in diagnosis and prognosis in MDS.
Prepublished online as Blood First Edition Paper, March 9, 2006; DOI 10.1182/blood-2005-12-4769.
Supported by the Leukaemia Research Fund of the United Kingdom and in part by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan (M.C.); Fondazione Cariplo, Milan (M.C.); and IRCCS Policlinico San Matteo, Pavia, Italy (M.C.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal