Cell-type-specific transcription of mouse high-affinity IgE receptor (FcϵRI) β-chain is positively regulated by the transcription factor GATA-1. Although GATA-1 is expressed in erythroid cells, megakaryocytes, and mast cells, the expression of mouse FcϵRI β-chain is restricted to mast cells. In the present study, we characterized the role of GATA-associated cofactor FOG-1 in the regulation of the FcϵRI β-chain promoter. The expression levels of FOG-1, GATA-1, and β-chain in each hematopoietic cell line were analyzed by reverse transcriptase-polymerase chain reaction (RT-PCR) and Western blotting. FOG-1 expression was higher in the β-chain-negative hematopoietic progenitor cell line Ba/F3 than in the β-chain-positive mast cell line PT18. By contrast, GATA-1 expression was similar when comparing the 2 cell lines. A transient reporter assay demonstrated that the β-chain promoter functioned in PT18 but not in Ba/F3 and that the transcription activity of the β-chain promoter in PT18 was markedly suppressed by overexpression of FOG-1. Although the activity of the β-chain promoter, which was upregulated by coexpression of GATA-1, was significantly suppressed by coexpression of FOG-1 in the simian kidney CV-1 cells (β-chain-, GATA-1-, and FOG-1-), the transactivation of the β-chain promoter by the GATA-1 mutant V205G, which cannot bind FOG-1, was not affected by coexpression of FOG-1. Further, overexpression of FOG-1 in PT18 resulted in decreases in cell surface expression of FcϵRI and β-chain transcription. Finally, suppression of FOG-1 expression using an siRNA approach resulted in increased β-chain promoter activity in Ba/F3. These results suggest that FOG-1 expression level regulates the GATA-1-dependent FcϵRI β-chain promoter. (Blood. 2006;108:262-269)
Introduction
The high-affinity IgE receptor FcϵRI is composed of an α-chain, a β-chain, and a γ-chain. Allergen-IgE antibody complex-induced cross-linking of FcϵRI results in activation of mast cells, which subsequently secrete various chemical mediators that induce the symptoms of an allergic response.
In humans, FcϵRI is expressed as a tetramer (αβγ2) on mast cells and basophils and as a trimer (αγ2) on Langerhans cells, monocytes, and dendritic cells.1-5 Thus, while β-chain may facilitate cell-surface expression of FcϵRI,6 it is not necessarily required for cell-surface expression of human FcϵRI. By contrast, in mice, FcϵRI is expressed as a tetramer (αβγ2) only on mast cells and basophils, and the β-chain is necessary for cell-surface expression of mouse FcϵRI7 and acts as an amplifier for FcϵRI signaling by increasing phosphorylation of the γ-chain8 and by enhancing signaling protein recruitment.9 Therefore, characterization of the mechanisms of mouse β-chain expression is critical for the understanding of mast-cell- and basophil-specific transcriptional regulatory systems.
We previously reported that the transcription factor GATA-1 positively regulated cell-type-specific β-chain expression via 4 GATA motifs in the promoter.10 GATA-1 mediates the maturation of various cell lineages, including erythroid cells, megakaryocytes, eosinophils, basophils, and mast cells.11-13 However, the expression of mouse β-chain is limited to mast cells but not observed in other GATA-1-positive cell lineages. Thus, other factors may regulate cell-type-specific transcription of the β-chain.
A zinc finger cofactor, FOG-1, interacts with GATA-1 and can either enhance or repress GATA-1-dependent gene expression.14-17 FOG-1 is abundantly expressed in erythroid cells and in megakaryocytes, where it regulates growth and differentiation. Erythroid and megakaryocyte lineage development is arrested at proerythroblast stage in Gata-1-/- or Fog-1-/- mice,18,19 and FOG-1 mutants that lack GATA-binding activity result in abnormal differentiation of megakaryotic cells.19,20 Recent studies have implicated abnormalities in GATA-1 and FOG-1 in various human diseases, including thrombocytopenia and idiopathic myelofibrosis (IM).21-23 Thus, the goal of the present study was to characterize the role of GATA-associated cofactor FOG-1 in the regulation of the FcϵRI β-chain promoter.
Materials and methods
Cell culture
The mouse mast cell line PT18 was cultured in RPMI 1640 (Sigma-Aldrich, St Louis, MO) containing 10% fetal bovine serum (FBS; Sigma-Aldrich). The mouse hematopoietic progenitor cell line Ba/F3 was purchased from ATCC (Manassas, VA) and maintained in RPMI 1640 containing 10% FBS and 10% pokeweed mitogen-stimulated spleen-condition medium (PWM-SCM).24 The simian kidney cell line CV-1 was purchased from RIKEN Cell Bank (Tsukuba, Japan) and maintained in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich) with 10% FBS.
Plasmids
A series of reporter plasmids carrying the luciferase gene under the control of various lengths of wild-type or mutant β-chain promoters were generated in a previous study.10 In brief, β-2357/pGL3-Basic, β-69/pGL3-Basic, and βM1234/pGL3-Basic, including β-chain 5′-flanking region -2357/+103, -69/+103, and -69/+103 lacking 4 GATA-1 binding sites by nucleotide replacement, respectively, were used for a reporter assay. The plasmid GATA-1/pCR3.1 (pCR-GATA-1), generated in a previous study,25 was used to produce GATA-1. Plasmid GATA-1 V205G/pCR3.1 (pCR-GATA-1 V205G) was used to express a GATA-1 mutant that is unable to bind FOG-1.26 This plasmid was generated by nucleotide replacement from GTG (Val) to GGG (Gly) at the 205th residue of wild-type GATA-1 using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). FOG-1 open reading frame cDNA was obtained from Ba/F3 mRNA by reverse transcriptase-polymerase chain reaction (RT-PCR) and subcloned into pCR3.1 expression plasmid (Invitrogen Life Technologies, Leek, The Netherlands). Three FOG-1 mutant expression plasmids (pCR-ΔN-FOG-1,27 pCR-ΔCtBP-FOG-1,28 and pCR-ΔN/ΔCtBP-FOG-1 [double mutant]) were constructed by using a QuikChange site-directed mutagenesis kit (Stratagene). The 5′-flanking region of the mouse αIIb gene (accession number AF169829), -324/+32, was obtained using a Genome Walker kit (BD Clontech, Palo Alto, CA) according to the manufacturer's instructions and was introduced upstream of the luciferase gene in a reporter plasmid (pGL3-Basic; Promega, Madison, WI) to generate the final reporter plasmid, αIIb/pGL3-Basic.
RT-PCR
Reverse transcription to detect the mRNA for FOG-1, GATA-1, and GAPDH was performed using SuperScript II reverse transcriptase (Invitrogen Life Technology). Total RNA was isolated from each cell line by RNA STAT-60 (Tel-Test, Friendswood, TX) and used as a template for RT. PCR was performed using an Advantage2 polymerase mix (BD Clontech). The following oligonucleotides were used as primers: mouse FOG-1, 5′-ATGTCCAGGAGGAAACAGAGC-3′ and 5′-GAGCTCCCTGCACCCTGCTGTAGC-3′; mouse GATA-1, 5′-ATGGATTTTCCTGGTCTA GGGGC-3′ and 5′-TCAAGAACTGAGTGGGGCGATCACG-3′. A mouse GAPDH primer set was purchased from BD Clontech. The thermal cycling was conducted for 25 to 30 cycles at the following conditions: 95°C for 30 seconds, 60°C for 1 minute, and 68°C for 30 seconds.
Luciferase assay
Ba/F3 or PT18 cells (5 × 106) were transfected with 5 μg reporter plasmid, 3 μg expression plasmid (for coexpression analysis), and 10 ng pRL-null (Promega) by electroporation as described previously.10 CV-1 cells (1 × 106) were transfected with 500 ng reporter plasmid, 100 ng expression plasmid, and 0.5 ng pRL-null by FuGENE 6 (Roche Diagnostics, Indianapolis, IN) in accordance with the manufacturer's instruction. The pRL-null plasmid was used as an internal control of transfection efficiency.
After 20 to 24 hours of culture, cells were harvested and a Dual-luciferase assay kit (Promega) and Micro Lumat Plus (Berthold, Bad Wildbad, Germany) were used to assess luciferase activity. Firefly luciferase activity derived from reporter plasmid was normalized to Renilla luciferase activity derived from pRL-null.
Introduction of the FOG-1 expression plasmid into mast cells
PT18 cells were transfected with pCR-FOG-1 or pCR3.1 by electroporation using a Bio-Rad Gene Pulsar II (Bio-Rad, Hercules, CA) set at 300 V and 950 μF. Cells were then cultured in growth medium containing G418 (1 mg/mL) to obtain transfectants. After 2 weeks of culture, transfected cells were analyzed by flow cytometry, Western blotting, and quantitative real-time PCR.
Flow cytometry
For detection of cell-surface expression of FcϵRI, cells (1 × 106) were incubated with 1 μg of FITC-conjugated mouse IgE anti-DNP monoclonal antibody (mAb; clone SPE-7; Sigma-Aldrich) in the presence of 1 μg of 2.4G2 (BD PharMingen, San Diego, CA) for 1 hour at 4°C in 50 μL of PBS containing 2% FBS. Cells were washed with PBS and analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Diego, CA).
Western blot analysis
Cell lysates (from 1 × 106 cells) were subjected to electrophoresis on a 7.5% SDS polyacrylamide gel. The transferred membrane was soaked with an enhanced chemiluminescence (ECL) plus Western blotting detection reagent (Amersham Pharmacia Biotech, Uppsala, Sweden), and chemiluminescence was detected by LAS-1000 plus (Fuji Film, Tokyo, Japan). Anti-GATA-1 Ab (clone N6, rat monoclonal IgG2a), anti-FOG-1 Ab (clone A-20, goat polyclonal IgG), and anti-YY1 Ab (clone H-10, mouse monoclonal IgG1) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Peroxidase-conjugated anti-goat IgG donkey Ab (Santa Cruz Biotechnology), anti-rat IgG rabbit Ab (Zymed, San Francisco, CA), and anti-mouse IgG rabbit Ab (Zymed) were used as the secondary Abs.
Quantitative PCR
Quantitative PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and a 7500 Real-Time PCR System with Assays-on-Demand gene expression products of the mouse target genes, including the FcϵRI α-chain (Mm00 438 867_m1), the β-chain (Mm00 442 780_m1), FOG-1 (Mm00 494 336_m1), and an endogenous control (GAPDH; Mm99 999 915_g1). The expression levels of the α-chain, the β-chain, and FOG-1 were expressed relative to those of GAPDH by calculation of cycle threshold (Ct) values in amplification plots with 7500 SDS software (Applied Biosystems), as recommended by the supplier. The following equations were used: the mRNA expression level (%) = 2 (Ct FOG-1 transfectant) - (Ct mock transfectant) × 100; Ct FOG-1 transfectant = (Ct value of α-chain, β-chain, or FOG-1 of FOG transfectant) - (Ct value of GAPDH of FOG transfectant); Ct mock transfectant = (Ct value of α-chain, β-chain, or FOG-1 of mock transfectant) - (Ct value of GAPDH of mock transfectant).
The siRNA-mediated inhibition of FOG-1 expression
The FOG-1 siRNA sequence was selected using BLOCK-iT RNAi designer (Invitrogen). Approximately 2.5 or 5 μL of 20 μM double-stranded FOG-1 Stealth siRNA (NM_009 569_stealth_1926; Invitrogen), 1 μg of β-69/pGL3-Basic, and 1 ng of pRL-null were introduced into 2 × 106 of Ba/F3 cells with Nucleofector II (Amaxa GmbH, Koeln, Germany) set at program T-16 using a Cell Line Nucleofector Kit V (Amaxa) according to the manufacturer's instruction. Another siRNA with a shuffled sequence of FOG-1 Stealth siRNA was constructed and used as nonsilencing control. After 20 or 44 hours of culture, FOG-1 mRNA, FOG-1 protein, and β-chain promoter activity were analyzed in transfected Ba/F3 cells by quantitative real-time PCR, Western blot analysis, and luciferase assay, respectively. FOG-1 double-stranded siRNA was prepared by annealing of the sense strand (5′-CCGGGACACUCUUUCUGCCACAAUA) and the antisense strand (5′-UAUUGUGGCAGAAAGAGUGUCCCGG). Nonsilencing control siRNA was annealed with sense strand (5′-CCGCACAUUCUGUCUCACCAGGAUA) and antisense strand (5′-UAUCCUGGUGAGACAGAAUGUGCGG).
ChIP assay
Lin- cells were prepared from mouse bone marrow cells by the BD IMag separation technique according to the manufacturer's instruction. In brief, bone marrow cells from BALB/c mice (Japan SLC, Hamamatsu, Japan) were treated with biotinylated mAbs to mouse CD3e, CD11b, CD45R/B220, Ly-6G, and Ly-6C, and TER-119/erythroid cells, in a Mouse Lineage Panel (BD Biosciences Pharmingen), as the first step. The cell suspension treated with biotinylated mAbs was then reacted with a BD IMag Streptavidin Particles Plus-DM (BD Biosciences) and was applied to a BD IMagnet (BD Biosciences). The purity of the isolated Lin- cells, which were collected as negative fraction using this BD magnetic separation system, was confirmed to be over 95% by flow cytometry. Mouse bone marrow-derived mast cells (BMMCs) were generated from Lin- cells by 4- to 5-week culture in RPMI supplemented with 10% FBS, 100 μM 2-ME, 10 μM MEM nonessential amino acids solution (Sigma-Aldrich), antibiotics, 10 ng/mL mouse recombinant IL-3 (rIL-3; PeproTech, Rocky Hill, NJ), and 10 ng/mL mouse SCF (PeproTech). After 4- to 5-week culture, over 90% of cells were identifiable as mast cells by toluidine blue staining and as c-kit+/FcϵRI+ by flow cytometric analysis. FOG-1 binding to β-chain promoter region in Lin- cells and BMMCs at different developing stages was analyzed by chromatin immunoprecipitation (ChIP) assay. ChIP assay was performed using a ChIP Assay Kit (Upstate, Lake Placid, NY) according to the manufacturer's instruction with a modification for use of protein G instead of protein A. Anti-FOG-1 Ab, anti-GATA-1 Ab, the same as Abs for Western blotting analysis, goat IgG (Sigma), and rat IgG (Zymed) were used in ChIP assay. PCRs were performed with the same condition as that in RT-PCR.
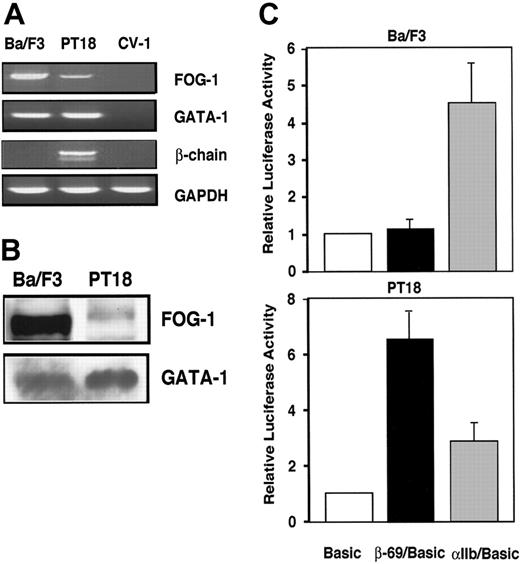
The β-chain, GATA-1, and FOG-1 mRNA levels in Ba/F3 and PT18. (A) RT-PCR was performed to measure FcϵRI β-chain, FOG-1, GATA-1, and GAPDH mRNA levels, as visualized by ethidium bromide staining of agarose gels. (B) Western blotting analyses of FOG-1 and GATA-1. Whole-cell lysates (1 × 106 cells per lane) were applied into each well and were analyzed with anti-FOG-1 or anti-GATA-1 Ab as the primary Abs followed by peroxidase-conjugated anti-goat IgG donkey Ab or peroxidase-conjugated anti-rat IgG rabbit Ab as the secondary Abs, respectively. (C) Transcription activities of the β-chain and αIIb promoters. Five micrograms of each reporter plasmid was introduced into Ba/F3 or PT18. The relative luciferase activity driven by β-69/pGL3-Basic (β-69; ▪) or αIIb/pGL3-Basic (αIIb; ▦)is represented as the ratio to the activity driven by pGL3-Basic (Basic; □). Each experiment was conducted in duplicate for each sample, and the results are expressed as mean ± SD for more than 3 independent experiments in Figures 1, 2, 3, and 5.
The β-chain, GATA-1, and FOG-1 mRNA levels in Ba/F3 and PT18. (A) RT-PCR was performed to measure FcϵRI β-chain, FOG-1, GATA-1, and GAPDH mRNA levels, as visualized by ethidium bromide staining of agarose gels. (B) Western blotting analyses of FOG-1 and GATA-1. Whole-cell lysates (1 × 106 cells per lane) were applied into each well and were analyzed with anti-FOG-1 or anti-GATA-1 Ab as the primary Abs followed by peroxidase-conjugated anti-goat IgG donkey Ab or peroxidase-conjugated anti-rat IgG rabbit Ab as the secondary Abs, respectively. (C) Transcription activities of the β-chain and αIIb promoters. Five micrograms of each reporter plasmid was introduced into Ba/F3 or PT18. The relative luciferase activity driven by β-69/pGL3-Basic (β-69; ▪) or αIIb/pGL3-Basic (αIIb; ▦)is represented as the ratio to the activity driven by pGL3-Basic (Basic; □). Each experiment was conducted in duplicate for each sample, and the results are expressed as mean ± SD for more than 3 independent experiments in Figures 1, 2, 3, and 5.
Primers used in this assay to amplify target genes are as follows: the β-chain gene (-197/+128), mBeta-197F (5′-GATGCTTCAGTATTAGGGTCC-3′), and mBeta+128R (5′-CTGCTCCTATTTTCTGTGTCC-3′); GAPDH,29 GAPDH-F (5′-ACCACAGTCCATGCCATCAC-3′), and GAPDH-R (5′-TCCACCACCCTGTTGCTGTA-3′).
Amounts of FOG-1 and GATA-1 bound to target DNA were also quantified using 7500 Real-Time PCR System. Ratio of a specific DNA fragment in each immunoprecipitate to that fragment in the DNA before immunoprecipitation (input DNA) was calculated from each Ct value as described previously.30 Quantitative ChIP was carried out with the β-chain promoter region (-73/-6) along with the region (+6980/+7086) of the β-chain gene that has no GATA sequence as a cis control.
Primers and TaqMan probe sequences used for this analysis are as follows: for the promoter region, mBeta-pro-73F (5′-ACAGCAAGAGAAAGGAGTCACTGAT-3′), mBeta-pro-6R (5′-CATGCGGAACCTACTTGTCAGA-3′), and TaqMan probe-1 (5′-FAM-CAATCAGCCTGGAGACT-MGB-3′); for the cis control region, mBeta+6980F (5′-GAAGGTAGTCAATGGGAATGACAA-3′), mBeta+7086R (5′-TGTGCTGAGATTCTAGGCAAACA-3′), and TaqMan probe-2 (5′-FAM-AAGTTGGTAGAGATTGGCA-MGB-3′).
Results
Suppression of FcϵRI β-chain promoter activity by FOG-1
To elucidate the involvement of GATA-associated cofactor FOG-1 in the expression of β-chain, β-chain expression and FOG-1 expression were characterized in 2 hematopoietic cell lines: the β-chain+ cell line PT18 and the β-chain- cell line Ba/F3. The β-chain mRNA was detected in PT18 but not in Ba/F3. By contrast, FOG-1 mRNA levels were significantly higher in Ba/F3 than in PT18 (Figure 1A). Western blotting also demonstrated higher FOG-1 protein expression in Ba/F3 than in PT18 (Figure 1B). However, GATA-1 mRNA level in both cell lines was similar and GATA-1 protein level in PT18 was somewhat higher than that of Ba/F3 (Figure 1A-B). From these data we hypothesize that FOG-1 may act as a negative regulator of the GATA-1-dependent β-chain promoter.
Next, a reporter assay was used to evaluate the transcriptional activity of the GATA-1-dependent promoter. The luciferase activity derived from PT18 cells transfected with β-69/pGL3-Basic was significantly higher than that of a promoterless reporter plasmid, pGL3-Basic, whereas the promoter activity of β-chain was similar to that of pGL3-Basic in Ba/F3 (Figure 1C). These data are consistent with the notion of mast-cell-specific transcription of β-chain in PT18 (Figure 1A) and suggest that the β-chain promoter, which functions in mast cells, was repressed in Ba/F3 cells, despite the fact that GATA-1 levels were similar in PT18 and Ba/F3.
The αIIb gene encodes the α integrin chain of the platelet fibrinogen receptor αIIb/B3, and previous studies have demonstrated that the αIIb gene promoter was synergistically transactivated by GATA-1 and FOG-1.29 Experiments in the present study demonstrated significant αIIb promoter activity in Ba/F3 cells but not in PT18 (Figure 1C). These data suggest that FOG-1 can either enhance or repress expression of different GATA-1-dependent promoters.
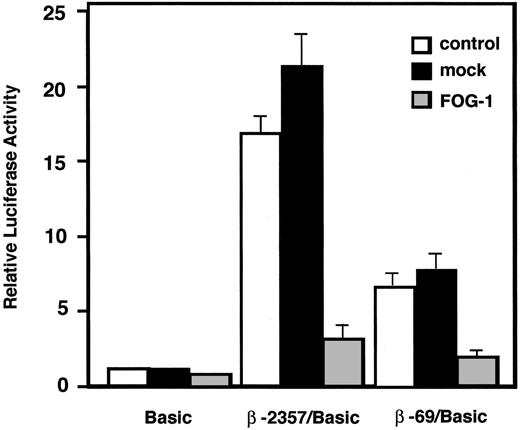
To confirm that expression of FOG-1 reduces β-chain promoter activity, coexpression analysis was performed in PT18. Coexpression of FOG-1 resulted in a marked reduction of luciferase activities driven by -2357/+103 and -69/+103 β-chain promoters, which have been identified to be the full-length promoter and the minimum promoter required for binding and regulation by GATA-1, respectively,10 whereas cotransfection of mock vector had no apparent effect on either β-chain promoter (Figure 2). These results suggest that increased FOG-1 expression suppresses the promoter activity of the β-chain specifically in mast cells and that the suppressive effect of FOG-1 is mediated through the -69/+103 region of the β-chain promoter.
FOG-1/GATA-1 interaction is required for downregulation of the β-chain promoter
FOG-1 augments or inhibits GATA-1-dependent gene expression via the formation of regulatory complexes. To investigate the role of GATA-1/FOG-1 interactions in the suppression of the β-chain promoter, studies were performed with the GATA-1 mutant V205G, in which the Val at the 205th residue, which is essential for FOG-1 binding, was replaced with Gly.26 The GATA-1 V205G retains DNA binding activity but has a defect in FOG-1 association activity. CV-1 cells, which do not express GATA-1 or FOG-1, were transfected with plasmids encoding the cDNAs for wild-type or mutant GATA-1 along with the reporter plasmid and FOG-1 expression plasmid (Figure 3B). Coexpression of wild-type GATA-1 or GATA-1 V205G resulted in marked upregulation of β-chain promoter activity (Figure 3A), whereas coexpression of FOG-1 with wild-type GATA-1 resulted in a dose-dependent attenuation of the GATA-1-induced increase in β-chain promoter activity (Figure 3A,C) without reducing GATA-1 gene expression (Figure 3C bottom). By contrast, coexpression of FOG-1 produced a modest attenuation of the GATA-1 V205G-induced increase in β-chain promoter activity.
Transcriptional activity of β-chain promoter was downregulated by FOG-1 expression in mast cells. Five micrograms of each reporter plasmid was introduced into PT18 cells with or without 3 μg of expression plasmid. The ratio of luciferase activity of each construct in the absence of expression plasmid to that of pGL3-Basic was represented as relative luciferase activity. □ indicates without expression plasmid; ▪, with mock expression plasmid; and ▦, with FOG-1 expression plasmid.
Transcriptional activity of β-chain promoter was downregulated by FOG-1 expression in mast cells. Five micrograms of each reporter plasmid was introduced into PT18 cells with or without 3 μg of expression plasmid. The ratio of luciferase activity of each construct in the absence of expression plasmid to that of pGL3-Basic was represented as relative luciferase activity. □ indicates without expression plasmid; ▪, with mock expression plasmid; and ▦, with FOG-1 expression plasmid.
Direct interaction between FOG-1 and GATA-1 is required for suppression of the β-chain promoter. (A) CV-1 cells were transfected with 500 ng each of reporter plasmids (pGL3-Basic; β-69/pGL3-Basic; or β M1234/pGL3-Basic [β-chain mutant promoter lacking 4 GATA motifs by nucleotide replacement]) and 100 ng each of expression plasmids (pCR-GATA-1; pCR-GATA-1 V205G [GATA-1 mutant lacking FOG-1 binding activity]; and/or pCR-FOG-1). Total amount of expression plasmids was adjusted to 200 ng by addition of the empty plasmid pCR3.1. The ratio of luciferase activity of each expression plasmid in the presence of reporter plasmid to that of the empty expression plasmid was represented as fold activation. (B) Transcriptional level of endogenously produced GATA-1 and FOG-1 in CV-1 cells. RT-PCR was performed with total RNA prepared from each transfectant. The expression level of GATA-1 V205G was similar to that of wild-type GATA-1 (data not shown). (C) Dose dependency of the FOG-1 effect on β-chain promoter activity. CV-1 cells were transfected with 100 ng of reporter plasmid β-69/pGL3-Basic with or without 100 ng of pCR-3.1 (empty vector), pCR-GATA-1, or various amounts of pCR-FOG-1 (10, 30, 100, or 300 ng). The ratio of luciferase activity of each construct in the absence of expression plasmid to that of pGL3-Basic was represented as relative luciferase activity. Protein levels of exogenously expressed GATA-1 and FOG-1 and control YY1 are analyzed by Western blotting (bottom).
Direct interaction between FOG-1 and GATA-1 is required for suppression of the β-chain promoter. (A) CV-1 cells were transfected with 500 ng each of reporter plasmids (pGL3-Basic; β-69/pGL3-Basic; or β M1234/pGL3-Basic [β-chain mutant promoter lacking 4 GATA motifs by nucleotide replacement]) and 100 ng each of expression plasmids (pCR-GATA-1; pCR-GATA-1 V205G [GATA-1 mutant lacking FOG-1 binding activity]; and/or pCR-FOG-1). Total amount of expression plasmids was adjusted to 200 ng by addition of the empty plasmid pCR3.1. The ratio of luciferase activity of each expression plasmid in the presence of reporter plasmid to that of the empty expression plasmid was represented as fold activation. (B) Transcriptional level of endogenously produced GATA-1 and FOG-1 in CV-1 cells. RT-PCR was performed with total RNA prepared from each transfectant. The expression level of GATA-1 V205G was similar to that of wild-type GATA-1 (data not shown). (C) Dose dependency of the FOG-1 effect on β-chain promoter activity. CV-1 cells were transfected with 100 ng of reporter plasmid β-69/pGL3-Basic with or without 100 ng of pCR-3.1 (empty vector), pCR-GATA-1, or various amounts of pCR-FOG-1 (10, 30, 100, or 300 ng). The ratio of luciferase activity of each construct in the absence of expression plasmid to that of pGL3-Basic was represented as relative luciferase activity. Protein levels of exogenously expressed GATA-1 and FOG-1 and control YY1 are analyzed by Western blotting (bottom).
Four GATA-1 binding sites in -55/-26 of β-chain promoter are critically required for GATA-1-dependent transactivation.10 When using the βM1234/pGL3-Basic reporter plasmid, which carries the mutant β-chain promoter that lacks all 4 of the GATA-1 binding sites, activation of β-chain promoter by GATA-1 and repression of GATA-1-dependent upregulation by FOG-1 are reduced compared with the case of the wild-type promoter β-69/Basic (Figure 3A). Slight activation by GATA-1, which was decreased by coexpression of FOG-1, was observed in the cells cotransfected with the promoterless plasmid (Basic) or βM1234/Basic. Cryptic GATA motifs present in the reporter vector31 may be responsible for the moderate activity. As for βM1234/pGL3-Basic, 3 GATA-like motifs in the 5′-untranslated region may also be attributed to the response to GATA-1. In any case, from these observations we conclude that FOG-1-mediated downregulation of the β-chain promoter is primarily mediated by GATA-1 binding to the promoter via the GATA-1 binding sites in β-chain promoter.
Overexpression of FOG-1 induces downregulation of β-chain transcription and surface FcϵRI in mast cells
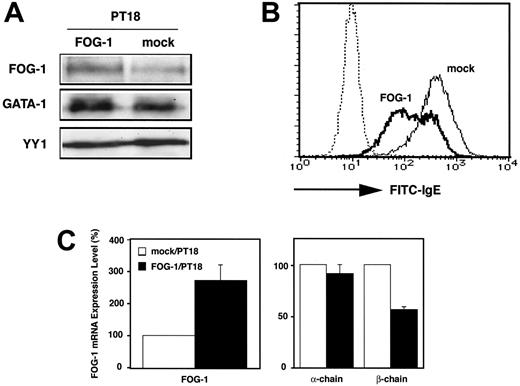
To further investigate the effect of FOG-1 overexpression on FcϵRI expression, PT18 cells were transfected with the pCR-FOG-1 using electroporation. PT18 transfectants were selected after culture in growth medium containing G418. After 2 weeks of culture, Western blotting demonstrated a higher amount of FOG-1 expression in PT18 cells transfected with pCR-FOG-1 than in PT18 cells undergoing mock transfection (Figure 4A). In contrast, protein levels of GATA-1 and of the ubiquitously expressed irrelevant transcription factor YY-1 were similar when comparing both transfectants. These data suggest that FOG-1 overexpression has no effect on the expression of endogenous GATA-1 protein in PT18 cells. Cell-surface FcϵRI on the transfectants was stained with FITC-conjugated IgE and analyzed by fluorescence-activated cell sorter (FACS), which revealed decreased cell-surface FcϵRI expression in PT18 cells that were transfected with FOG-1 expression plasmid (Figure 4B).
FcϵRI is composed of 3 subunits (α-, β-, and γ-chain), and the transcription of α-chain gene is regulated by GATA-1.25,32 To determine whether decreases in β-chain expression mediate the decrease in cell-surface FcϵRI expression in response to FOG-1 overexpression, quantitative real-time PCR analysis was performed to measure mRNA levels of the α- and β-chains in PT18 cells overexpressing FOG-1. The α-chain and β-chain transcripts were not detected in Ba/F3, whereas FOG-1 transcripts were approximately 120-fold higher in Ba/F3 than in PT18 mock transfectant (data not shown). FOG-1 mRNA increased 2.7-fold in FOG-1 transfectants (Figure 4C), which correlated with the increase in FOG-1 protein levels in the transfectants (Figure 4A). The amount of β-chain mRNA in FOG-1-overexpressing cells was 57% of that of mock transfectant, whereas the amount of α-chain mRNA was not significantly affected by FOG-1 overexpression (Figure 4C). These results indicate that FOG-1 overexpression reduced surface expression of FcϵRI on PT18, which correlated with the reduction in β-chain transcription.
Overexpression of FOG-1 induced the downregulation of β-chain transcription and surface expression of FcϵRI in PT18 cells. (A) Western blot analysis of transfected cells. PT18 cells were transfected with pCR-FOG-1 or pCR3.1 by electroporation and cultured in the presence of G418 for longer than 2 weeks. Lysates (1 × 106 cells per lane) were analyzed using anti-FOG-1, anti-GATA-1, or anti-YY1 Abs. (B) Surface expression of FcϵRI on transfectants. PT18 cells transfected with pCR-FOG-1 (FOG-1, bold line) or empty vector (mock, solid line) were stained with FITC-conjugated mouse IgE antibody. FOG-1 transfectants without antibody are shown by the dotted line. (C) Quantitative real-time RT-PCR analysis of transfected cells. The mRNA levels of FOG-1, α-chain, and β-chain in FOG-overproducing cells were represented as the ratio to that of mock transfectants. □ indicates PT18 cells transfected with pCR3.1; and ▪, PT18 cells transfected with pCR-FOG-1. Data are represented as the average ± SD of triplicate samples.
Overexpression of FOG-1 induced the downregulation of β-chain transcription and surface expression of FcϵRI in PT18 cells. (A) Western blot analysis of transfected cells. PT18 cells were transfected with pCR-FOG-1 or pCR3.1 by electroporation and cultured in the presence of G418 for longer than 2 weeks. Lysates (1 × 106 cells per lane) were analyzed using anti-FOG-1, anti-GATA-1, or anti-YY1 Abs. (B) Surface expression of FcϵRI on transfectants. PT18 cells transfected with pCR-FOG-1 (FOG-1, bold line) or empty vector (mock, solid line) were stained with FITC-conjugated mouse IgE antibody. FOG-1 transfectants without antibody are shown by the dotted line. (C) Quantitative real-time RT-PCR analysis of transfected cells. The mRNA levels of FOG-1, α-chain, and β-chain in FOG-overproducing cells were represented as the ratio to that of mock transfectants. □ indicates PT18 cells transfected with pCR3.1; and ▪, PT18 cells transfected with pCR-FOG-1. Data are represented as the average ± SD of triplicate samples.
Inhibition of FOG-1 expression by siRNA resulted in upregulation of β-chain promoter activity. (A) Quantification of FOG-1 mRNA in siRNA transfectants using real-time RT-PCR. Ba/F3 cells (2 × 106) were transfected with 5 μL of 20 μM FOG-1 siRNA or shuffled control siRNA. After 20-hour culture, total RNA was extracted from each transfectant, and the amount of FOG-1 and GAPDH mRNAs was analyzed by ABI7500. FOG-1 mRNA levels are represented as the ratio to that of control (without siRNA). (B) Western blot analysis of transfectants. After 44 hours of culture, lysates (5 × 105 cells per lane) were analyzed using anti-FOG-1 or anti-YY1 Abs. (C) Transcription activity of the β-chain promoter. One microgram of reporter plasmid was introduced into Ba/F3 cells with or without 2.5 or 5 μL of 20 μM siRNA or shuffled control siRNA. The ratio of luciferase activity of each transfectant to that of control transfectant (without siRNA) was represented as fold activation.
Inhibition of FOG-1 expression by siRNA resulted in upregulation of β-chain promoter activity. (A) Quantification of FOG-1 mRNA in siRNA transfectants using real-time RT-PCR. Ba/F3 cells (2 × 106) were transfected with 5 μL of 20 μM FOG-1 siRNA or shuffled control siRNA. After 20-hour culture, total RNA was extracted from each transfectant, and the amount of FOG-1 and GAPDH mRNAs was analyzed by ABI7500. FOG-1 mRNA levels are represented as the ratio to that of control (without siRNA). (B) Western blot analysis of transfectants. After 44 hours of culture, lysates (5 × 105 cells per lane) were analyzed using anti-FOG-1 or anti-YY1 Abs. (C) Transcription activity of the β-chain promoter. One microgram of reporter plasmid was introduced into Ba/F3 cells with or without 2.5 or 5 μL of 20 μM siRNA or shuffled control siRNA. The ratio of luciferase activity of each transfectant to that of control transfectant (without siRNA) was represented as fold activation.
The β-chain promoter activity increases with decreasing FOG-1 expression
To confirm the effect of FOG-1 on β-chain expression, FOG-1 siRNA was introduced into Ba/F3 cells by electroporation, and the FOG-1 mRNA and protein levels in each transfectant were determined using quantitative real-time PCR and Western blot analysis, respectively. FOG-1 siRNA reduced FOG-1 mRNA levels by 70%, whereas scrambled siRNA had no effect on FOG-1 mRNA levels (Figure 5A). Western blot analysis demonstrated that FOG-1 protein levels were decreased in cells treated with FOG-1 siRNA when compared with control cells or control siRNA transfectants (Figure 5B). Cotransfection of Ba/F3 with 2.5 μL or 5 μL of 20 μM FOG-1 siRNA and β-69/pGL3-Basic reporter plasmid revealed that the luciferase activity was significantly higher in FOG-1 siRNA transfectant than in control siRNA transfectants or in those cells not treated with siRNA (Figure 5C). These results suggest that FOG-1 suppresses β-chain promoter activity and modulates cell-type-specific β-chain expression.
Binding of FOG-1 and GATA-1 to β-chain promoter in BMMCs at different developing stages
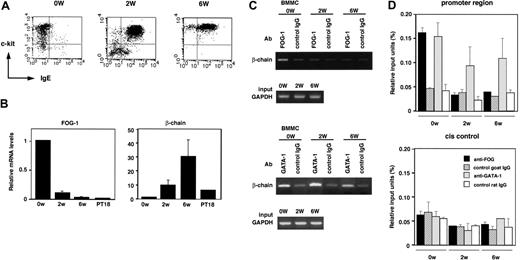
GATA-1 expression level varies depending on the stage of mast-cell development, with a most abundant GATA-1 expression in matured mast cells.33,34 To evaluate the relevance of FOG-1 in repressing β-chain expression, expression levels of FcϵRI and related molecules in different developing stages of mast cells were analyzed. BMMCs were generated from Lin- cells, which were purified from mouse bone marrow cells, by 4- to 5-week culture in the presence of IL-3 and SCF. Flow cytometric analysis demonstrated that expression of FcϵRI was observed in most cells after 2-week culture (approximately 85% was positive) as well as 6-week culture (approximately 90% was positive), whereas freshly purified Lin- cells are c-kit+/FcϵRI- (Figure 6A). The β-chain transcription level analyzed by real-time PCR was significantly higher in 2- and 6-week cultured BMMCs than that of Lin- cells (Figure 6B). In contrast, FOG-1 transcription level in Lin--derived cells was markedly reduced to the level in mast cells as the culture proceeded (Figure 6B). These results indicate that β-chain promoter is already transactivated in immature BMMCs (cultured 2 weeks) as well as matured BMMCs (cultured 6 weeks), accompanied by reduced expression of FOG-1, resulting in the cell-surface expression of FcϵRI. To examine whether FOG-1 was bound to β-chain promoter in vivo, ChIP assay was performed (Figure 6C-D). In Lin- cells, a significantly higher amount of chromatin containing β-chain promoter region (-197/+128) was immunoprecipitated by anti-FOG-1 Ab compared with control IgG. In contrast, apparent difference was not observed between anti-FOG-1 Ab and control IgG when 2- and 6-week cultured BMMCs were analyzed. When anti-GATA-1 Ab was used, a significant amount of β-chain promoter region was immunoprecipitated irrespective of the culture stages. On the other hand, both anti-FOG-1 and anti-GATA-1 Abs did not immunoprecipitate cis control region of β-chain gene (about 7 kb downstream of β-chain promoter) specifically. These results suggest that both FOG-1 and GATA-1 are recruited to β-chain promoter in FcϵRI- hematopoietic stem cells and a proportion of the promoter bound to FOG-1 is apparently reduced as the cells differentiate, due to the decrease in FOG-1 expression. In developing and developed BMMCs expressing FcϵRI, GATA-1 but not FOG-1 is bound to β-chain promoter, whereas GATA-1 occupies β-chain promoter constantly throughout the process of the mast-cell development from hematopoietic stem cells. This is the first study comparing FOG-1 and GATA-1 occupancy on β-chain promoter in different developing stages of BMMCs from Lin- cells.
Structure-function mapping of FOG-1 relating to CtBP and MeCP1/NuRD
C-terminal binding protein (CtBP) is a corepressor that can bind to FOG-1.15,28 In addition, recent studies have reported that FOG-1 inhibits expression of early erythroid genes via promotion of recruitment of MeCP1/NuRD complex to GATA-1.27,35 However, the presence and/or formation of the repressive CtBP or MeCP1/NuRD complex on the mouse β-chain promoter have not yet been demonstrated. To evaluate the involvement of this complex on FOG-1-mediated inhibition, reporter assay was performed using the expression plasmids of 3 FOG-1 mutants: ΔN-FOG-1 lacking NuRD-binding region, ΔCtBP-FOG-1 lacking CtBP-binding site, and ΔN/ΔCtBP-FOG-1 carrying both mutations (Figure 7A). As shown in Figure 7B, coexpression of ΔN-FOG-1 and ΔCtBP-FOG-1 inhibited β-chain promoter activity in PT18 as well as wild-type FOG-1, suggesting that FOG-1-mediated inhibition of the β-chain promoter may occur independent of recruitment of MeCP/NuRD to GATA-1 and the CtBP binding. The repressive effect of ΔN/ΔCtBP was almost the same as that of wild-type. Regardless, these results demonstrate that the FOG-1-dependent repression of β-chain promoter is mainly mediated by other unidentified factor(s).
Binding of FOG-1 and GATA-1 to β-chain promoter in BMMCs at different developing stages. (A) Expression profile of c-kit and FcϵRI analyzed by flow cytometry. 0W indicates freshly prepared Lin- cells; 2W, immature BMMCs developed from Lin- cells after 2-week culture; and 6W, mature BMMCs developed from Lin- cells after 6-week culture. (B) Transcription levels of FOG-1 and β-chain during BMMC development. The mRNA levels of FOG-1 and β-chain in different developing stages of BMMCs and PT18 were measured by quantitative PCR and are represented as the ratio to that of Lin- (0W). Data are represented as the average ± SD of triplicate samples. (C) FOG-1 and GATA-1 in vivo binding to β-chain promoter. Binding between β-chain promoter (-197/+128) and FOG-1 or GATA-1 was analyzed by ChIP assay using anti-FOG-1, anti-GATA-1, or each isotype control Ab. The GAPDH gene served as control. (D) Quantitative analysis of FOG-1 and GATA-1 binding to β-chain gene by ChIP assay using real-time PCR. ▪ indicates anti-FOG-1 Ab; ▨, goat IgG (control of anti-FOG-1 Ab); ▦, anti-GATA-1 Ab; □, rat IgG (control of anti-GATA-1 Ab). The results are expressed as mean ± SD for 2 PCRs with duplicate samples on each 2 independent ChIPs. Relative input units are calculated from Ct values as described in “ChIP assay.”
Binding of FOG-1 and GATA-1 to β-chain promoter in BMMCs at different developing stages. (A) Expression profile of c-kit and FcϵRI analyzed by flow cytometry. 0W indicates freshly prepared Lin- cells; 2W, immature BMMCs developed from Lin- cells after 2-week culture; and 6W, mature BMMCs developed from Lin- cells after 6-week culture. (B) Transcription levels of FOG-1 and β-chain during BMMC development. The mRNA levels of FOG-1 and β-chain in different developing stages of BMMCs and PT18 were measured by quantitative PCR and are represented as the ratio to that of Lin- (0W). Data are represented as the average ± SD of triplicate samples. (C) FOG-1 and GATA-1 in vivo binding to β-chain promoter. Binding between β-chain promoter (-197/+128) and FOG-1 or GATA-1 was analyzed by ChIP assay using anti-FOG-1, anti-GATA-1, or each isotype control Ab. The GAPDH gene served as control. (D) Quantitative analysis of FOG-1 and GATA-1 binding to β-chain gene by ChIP assay using real-time PCR. ▪ indicates anti-FOG-1 Ab; ▨, goat IgG (control of anti-FOG-1 Ab); ▦, anti-GATA-1 Ab; □, rat IgG (control of anti-GATA-1 Ab). The results are expressed as mean ± SD for 2 PCRs with duplicate samples on each 2 independent ChIPs. Relative input units are calculated from Ct values as described in “ChIP assay.”
Effect of FOG-1 mutants lacking NuRD or CtBP binding on β-chain promoter. (A) Structure of FOG-1 mutants: ΔN-FOG-1, lacking NuRD-binding N-terminus27 ; ΔCtBP-FOG-1, with replacement of 2 amino acids essential for CtBP binding28 ; ΔN/ΔCtBP-FOG-1, carrying both mutations of ΔN and ΔCtBP. (B) Coexpression analysis using FOG-1 mutants. PT18 cells were transfected with 5 μg of reporter plasmid β-69/pGL3-Basic with or without 3 or 10 μg of pCR-3.1 (mock), pCR-FOG-1 (wild-type), pCR-ΔN-FOG-1 (ΔN), pCR-ΔCtBP-FOG-1 (ΔCtBP), or pCR-ΔN/ΔCtBP-FOG-1 (ΔN/ΔCtBP). The ratio of each luciferase activity to that without coexpression plasmid was represented as relative luciferase activity. Data represent the average ± SD of triplicate samples.
Effect of FOG-1 mutants lacking NuRD or CtBP binding on β-chain promoter. (A) Structure of FOG-1 mutants: ΔN-FOG-1, lacking NuRD-binding N-terminus27 ; ΔCtBP-FOG-1, with replacement of 2 amino acids essential for CtBP binding28 ; ΔN/ΔCtBP-FOG-1, carrying both mutations of ΔN and ΔCtBP. (B) Coexpression analysis using FOG-1 mutants. PT18 cells were transfected with 5 μg of reporter plasmid β-69/pGL3-Basic with or without 3 or 10 μg of pCR-3.1 (mock), pCR-FOG-1 (wild-type), pCR-ΔN-FOG-1 (ΔN), pCR-ΔCtBP-FOG-1 (ΔCtBP), or pCR-ΔN/ΔCtBP-FOG-1 (ΔN/ΔCtBP). The ratio of each luciferase activity to that without coexpression plasmid was represented as relative luciferase activity. Data represent the average ± SD of triplicate samples.
Discussion
Cell-type-specific transcription of mouse FcϵRI β-chain gene (Ms4a2) is regulated by the transcription factor GATA-1.10 The present study demonstrated that the GATA-associated cofactor FOG-1 acted as a negative regulator of FcϵRI β-chain gene expression. When the expression level of GATA-1 and FOG-1 was compared between the β-chain-positive mast cell line PT18 and the β-chain-negative hematopoietic progenitor cell line Ba/F3, FOG-1 expression was markedly higher in Ba/F3 than in PT18 (Figure 1). Further, β-chain promoter activity was down-regulated by FOG-1 expression in PT18 (Figure 2), and FOG-1 expression suppressed GATA-1-mediated transactivation of the β-chain promoter through interaction with GATA-1 in a dose-dependent manner in CV-1 cells (β-chain-/FOG-1-/GATA-1-; Figure 3). FOG-1 overexpression also inhibited transcription of the β-chain and subsequently reduced the surface expression of FcϵRI in PT18 cells (Figure 4). In addition, reduction of Ba/F3 FOG-1 expression using FOG-1 siRNA resulted in a significant increase in β-chain promoter activity (Figure 5). FOG-1 and GATA-1 are recruited to β-chain promoter in FcϵRI- hematopoietic stem cells but only GATA-1 among the 2 transcription factors binds to β-chain promoter in FcϵRI+ primary mast cells (Figure 6). From these results, we conclude that FOG-1 suppresses GATA-1-dependent β-chain promoter activity through interaction with GATA-1, which subsequently determines mast-cell-specific FcϵRI expression. Coexpression analysis using 3 FOG-1 mutants lacking the binding ability to either and both of NuRD and CtBP suggested that FOG-1 suppresses β-chain promoter in a manner independent of these 2 molecules (Figure 7), although further detailed analysis using various FOG-1 mutants should be performed to clarify the structure-function relationship of FOG-1.
In the present study, overexpression of FOG-1 suppressed β-chain transcription in mast cells. By contrast, transcription of the α-chain was not affected by FOG-1 overexpression, despite the fact that the α-chain promoter is also transactivated by GATA-1.25,32,36 In addition, Ba/F3 exhibited significant activity of GATA-1-dependent αIIb promoter despite the fact that Ba/F3 expresses high levels of endogenous FOG-1 that inhibit β-chain promoter function. Therefore, the suppressive effect of FOG-1 on GATA-1-dependent promoter appears to be β-chain specific. However, the mechanism by which FOG-1 can augment or inhibit GATA-1 activity in different promoter contexts remains unclear and requires further study.
GATA-1 and FOG-1 are essential for regulation of a tissue-specific chromatin loop, which functions to place a long-range enhancer element in close proximity to the β-globin gene major promoter.37 In fact, the GATA-1- and FOG-1-mediated change in chromatin conformation may occur within the β-chain gene. Further study to test this hypothesis would be of benefit.
Considering that FcϵRI β-chain is involved in IgE-dependent allergic reaction, there is a possibility that mice and humans with GATA-1 mutations lacking FOG-1-binding ability show hypersensitivity to allergens and IgE. However, disorder of mast-cell development or function has not been reported in the previous in vivo studies of mice overexpressing V205G mutant GATA-1 instead of wild-type GATA-138 and several human individuals with missense mutations in the N-terminal GATA-1 zinc finger,21,22,39-41 which display an abrogated interaction between FOG-1 and GATA-1. One possible reason why abnormal function of mast cells was not found in previous in vivo studies is that function of mast cells is observed when IgE-dependent allergic disease is caused by immunization with allergens, whereas thrombocytopenia and anemia were observed at birth and thereafter. Allergic model study with V205G mice might clarify the involvement of FOG-1 in FcϵRI β-chain expression in vivo. Transcriptional regulatory mechanism of the β-chain is different between human and mouse. In brief, Oct-1 and MZF-1 but not GATA1 have been identified as transcription factors regulating human β-chain gene expression, although mast-cell-specific activation of mouse β-chain promoter is critically dependent on GATA-1.10,42,43 Furthermore, human FcϵRI can be expressed on the cell surface as a functional trimer form, αγ2, even in the absence of β-chain, whereas β-chain is critically required for expression of FcϵRI in mice. Therefore, lack of interaction between FOG-1 and GATA1 may not induce severe phenotypic change of human mast cells. Regardless, detailed analysis of mast cells or basophils isolated from V205G mice and human individuals carrying GATA-1 mutations is required to evaluate the in vivo relevance of the repression of GATA-1-dependent FcϵRI β-chain expression by FOG-1.
GATA-1 transactivates megakaryocyte-specific promoters GPIX and GPIbα in a cooperative manner with Fli-144 and mast-cell-specific promoter FcϵRI α-chain with PU.1.32 We have found an Ets motif close to GATA-1 binding sites in the mouse β-chain promoter as an Fli-1- and PU.1-bindable sequence. However, introduction of nucleotide mutation in this Ets motif had no effect on β-chain promoter activity (data not shown), suggesting that binding of Ets-family transcription factors to an Ets motif close to 4 GATA motifs may not be critical to regulate GATA-1-dependent β-chain promoter activity. Although we cannot rule out the possibilities that (i) other transcription factors regulate β-chain promoter synergistically with GATA-1 via elements far from GATA motifs and (ii) FOG-1 also indirectly regulates the β-chain gene expression through controlling the function of the unidentified factors, necessity of GATA-1 binding for suppressive effect of FOG-1 and binding of FOG-1 to β-chain promoter in vivo observed in this study are evidences indicating FOG-1 directly regulates GATA-1-dependent β-chain promoter activity. When compared between Ba/F3 and PT18, GATA-1 protein level is higher in PT18 cells (Figure 1B). This fact suggests an additional possibility that some other factors may downregulate the translational expression of GATA-1 in Ba/F3 cells and subsequently block the expression of β-chain. Regardless, further studies are required to reveal fully the regulatory mechanism of cell-type-specific β-chain expression depending on GATA-1.
Several recent studies have suggested that mast-cell-specific gene expression is regulated by cooperation between GATA-1/2 and the myeloid- and lymphoid-specific transcription factor PU.1.25,32,45 The present study suggests that mast-cell-specific gene expression is also regulated by the inhibitory effect of FOG-1 on GATA-1-mediated transcription. In fact, this is the first report to demonstrate the involvement of FOG-1 in mast-cell-specific gene regulation. GATA-1 or GATA-2 can transcriptionally regulate other mast-cell-specific molecules in addition to FcϵRI α- and β-chains, including carboxypeptidase A46 and chymase.47 In addition, when FOG-1 was overexpressed in mouse bone marrow cells developing toward mast cells, c-kit expression was markedly reduced and the number of cells was much decreased compared with mock transfectant in our preliminary experiments (data not shown). These observations may suggest a possibility that overexpressed FOG-1 inhibits development and function of mast cells globally as observed in eosinophils.48 These observations indicate that the mast cell is an excellent model to study the role of transcription factors and cofactors involved in GATA-1/2-mediated gene regulation. Further study will be required to identify other mast-cell-specific target genes that are regulated by GATA proteins and FOG-1 in order to characterize the specific mechanisms by which FOG-1 regulates gene expression.
Prepublished online as Blood First Edition Paper, March 7, 2006; DOI 10.1182/blood-2005-07-2878.
Supported in part by a grant-in-aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (C.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the members of the Atopy (Allergy) Research Center and the Department of Immunology for helpful discussions. We also thank Dr Nobuhiro Nakano and Ms Mutsuko Hara for technical support, Drs Kyoko Takahashi and Jiang-Hu Piao for critical advice, and Ms Michiyo Matsumoto for secretarial assistance.

![Figure 3. Direct interaction between FOG-1 and GATA-1 is required for suppression of the β-chain promoter. (A) CV-1 cells were transfected with 500 ng each of reporter plasmids (pGL3-Basic; β-69/pGL3-Basic; or β M1234/pGL3-Basic [β-chain mutant promoter lacking 4 GATA motifs by nucleotide replacement]) and 100 ng each of expression plasmids (pCR-GATA-1; pCR-GATA-1 V205G [GATA-1 mutant lacking FOG-1 binding activity]; and/or pCR-FOG-1). Total amount of expression plasmids was adjusted to 200 ng by addition of the empty plasmid pCR3.1. The ratio of luciferase activity of each expression plasmid in the presence of reporter plasmid to that of the empty expression plasmid was represented as fold activation. (B) Transcriptional level of endogenously produced GATA-1 and FOG-1 in CV-1 cells. RT-PCR was performed with total RNA prepared from each transfectant. The expression level of GATA-1 V205G was similar to that of wild-type GATA-1 (data not shown). (C) Dose dependency of the FOG-1 effect on β-chain promoter activity. CV-1 cells were transfected with 100 ng of reporter plasmid β-69/pGL3-Basic with or without 100 ng of pCR-3.1 (empty vector), pCR-GATA-1, or various amounts of pCR-FOG-1 (10, 30, 100, or 300 ng). The ratio of luciferase activity of each construct in the absence of expression plasmid to that of pGL3-Basic was represented as relative luciferase activity. Protein levels of exogenously expressed GATA-1 and FOG-1 and control YY1 are analyzed by Western blotting (bottom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/1/10.1182_blood-2005-07-2878/2/m_zh80130697900003.jpeg?Expires=1765952424&Signature=mkmsz7Kf3pqLaA4-YGnGw2oF67ahBU6zj6FhI7uI25nRyST9Bgk3JF7MiVNcX3UubvWzqTtkI8BlZetOvqcEThOahWraVoqLwwlMu6WLYwQcXXvmyaguSg3fwDjIGyxhpBrv8ExPAJo9CfMFcmLMXO6vGcSGLLC4uqjdklNHNQ0jpwVFXv9LK0U-81~sMEMB9I-snqd4fWPGLz-aD~96b0rdLlogICStU0sI3pEDE7mIqVUzRyAnrNY8gp1kq1jrAkKJws6hT6-aMc-4AY0BBqtcbaANcxiJNTfFq4htrpxZGFbbs6DckcJU3jz0bLiaAmA8ncO39R43~GXj-ENmHg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal