Primary CNS lymphoma is an aggressive form of non-Hodgkin lymphoma whose growth is restricted to the central nervous system. We used cDNA microarray analysis to compare the gene expression signature of primary CNS lymphomas with nodal large B-cell lymphomas. Here, we show that while individual cases of primary CNS lymphomas may be classified as germinal center B-cell, activated B-cell, or type 3 large B-cell lymphoma, brain lymphomas are distinguished from nodal large B-cell lymphomas by high expression of regulators of the unfolded protein response (UPR) signaling pathway, by the oncogenes c-Myc and Pim-1, and by distinct regulators of apoptosis. We demonstrate that interleukin-4 (IL-4) is expressed by tumor vasculature as well as by tumor cells in CNS lymphomas. We also identify high expression in CNS lymphomas of several IL-4-induced genes, including X-box binding protein 1 (XBP-1), a regulator of the UPR. In addition, we demonstrate expression of the activated form of STAT6, a mediator of IL-4 signaling, by tumor cells and tumor endothelia in CNS lymphomas. High expression of activated STAT6 in tumors was associated with short survival in an independent set of patients with primary CNS lymphoma who were treated with high-dose intravenous methotrexate therapy.
Introduction
Primary CNS lymphoma refers to non-Hodgkin lymphoma confined to the CNS, a disease whose prognosis is markedly worse than most other localized extranodal lymphomas of indistinguishable histology.1,2 Several clinical features of primary CNS lymphoma suggest that these tumors exhibit a distinct biologic phenotype: (1) a short duration of response to combined modality therapy; (2) a unique proclivity to grow only in the CNS; and (3) a heightened responsiveness to methotrexate, the most important agent used in the treatment of this disease.3
Most primary CNS lymphomas are large B-cell lymphomas. Systemic large B-cell lymphomas exhibit 3 major subclasses of gene expression: one that resembles the pattern of expression of germinal center B cells (GCBs) and is associated with a more favorable outcome, a second that resembles the gene expression pattern of activated B cells (ABCs), and a third that is termed type 3.4,5 A variety of evidence suggests that primary CNS lymphomas have been exposed to a germinal center microenvironment based upon shared genetic properties including a high frequency of mutations in the immunoglobulin variable gene region and expression of bcl-6.6-8 Molecular distinctions between primary CNS lymphoma and nodal lymphomas of large B-cell histology have not previously been defined.
Dissemination of lymphoma cells within the CNS, occurring as a secondary event, is also associated with poor prognosis.9 CNS metastases of lymphoma are a devastating complication of non-Hodgkin lymphoma. Unlike malignant gliomas, which are highly vascular tumors, both primary and metastatic CNS lymphomas exhibit angiotropism10 ; the brain is the only tissue environment in which large B-lymphoma cells accumulate densely around tumor vessels, suggesting that the CNS vasculature may contribute significantly to the pathogenesis of this type of lymphoma (Figure 1A). Insights into the molecular pathogenesis of CNS lymphomas are likely relevant to the devastating complication of systemic lymphoma relapse within the CNS.
Patients, materials, and methods
Study subjects
Samples from patients with nodal or CNS lymphomas were obtained under a protocol approved by the UCSF institutional review board. Informed consent was provided according to the Declaration of Helsinki. Specimens were obtained from the UCSF Brain Tumor Research Center Tumor Bank, M. D. Anderson Neuropathology, Massachusetts General Hospital Neuropathology, and the NCI AIDS and Cancer Specimen Tumor Bank. All specimens were reviewed by a single reference hematopathologist at UCSF.
Purification of total RNA from frozen lymphoma specimens or from nonneoplastic brain
Lymphoma cell nuclei accounted for at least 70% of all cells in each tumor used in this analysis as determined by histopathologic analysis of hematoxylin and eosin-stained sections used in parallel. Depending on tumor size, between 3 and 16 14-μm-thick tissue sections were prepared using a cryostat. Samples were placed in RLT buffer plus 1% β-ME, homogenized, and RNA prepared according to the animal tissue protocol of the RNeasy handbook (Qiagen, Valencia, CA). RNA was quantified based on absorbance at 260 nm. Quality of RNA was assured by measuring OD 260:280 ratios.
Angiotropism and gene expression in primary CNS lymphoma. (A) Angiotropic growth pattern in primary CNS lymphoma. Hematoxylin and eosin stain were used. Specimens used for gene expression analysis in this study were more highly cellular, consisting of at least 70% tumor cells. (B) Distinctions in gene expression between CNS lymphoma and nodal large B-cell lymphoma. Transcriptional profiles of 23 specimens of primary CNS lymphomas were compared with 3 specimens of nonneoplastic brain and with 9 consecutive specimens of large B-cell lymphoma obtained from lymph nodes. Only genes differentially expressed with a false discovery rate less than 0.01 are shown. The samples are clustered within molecular subtypes and ordered by the subtype. The image reveals distinct patterns of gene expression between CNS lymphomas and nodal lymphomas. Each column represents an individual tumor, and each row represents a single gene. Red indicates up-regulation; green, down-regulation; and black, similar to the median of the reference pool. (C) Assignment of PCNSL and nodal DLBCL tumors to germinal center (GCB), activated B cell (ABC), and type 3 subclasses. PCNSL tumors were distributed equally among the 3 subclasses. Hierarchic clustering was based upon the expression of 38 established marker genes of ABC and GCB subtypes. (D) Mean expression of 4 marker genes distinguishes CNS lymphoma as determined by quantitative real-time PCR in cases of primary CNS lymphomas (blue), large B-cell lymphomas from lymph nodes (red), and secondary CNS lymphomas (metastatic CNS lymphomas; yellow). Y-axis corresponds to percent expression of control gene. Error bars depict standard deviation of mean value. P values refer to the differences in expression between primary CNS lymphomas and nodal large B-cell lymphoma: XBP-1 (P < .001), c-Myc (P < .001), PIM-1 (P < .001), and E2F-5 (P < .001). CD10 expression was similar in the 3 types of lymphomas. Gene expression values obtained by microarray analysis were in agreement with those obtained by quantitative RT-PCR. (The minimum Pearson correlation is 0.65 corresponding to the P value of < .001 for the test of the hypothesis of no association between microarray and Taqman data for these marker genes.)
Angiotropism and gene expression in primary CNS lymphoma. (A) Angiotropic growth pattern in primary CNS lymphoma. Hematoxylin and eosin stain were used. Specimens used for gene expression analysis in this study were more highly cellular, consisting of at least 70% tumor cells. (B) Distinctions in gene expression between CNS lymphoma and nodal large B-cell lymphoma. Transcriptional profiles of 23 specimens of primary CNS lymphomas were compared with 3 specimens of nonneoplastic brain and with 9 consecutive specimens of large B-cell lymphoma obtained from lymph nodes. Only genes differentially expressed with a false discovery rate less than 0.01 are shown. The samples are clustered within molecular subtypes and ordered by the subtype. The image reveals distinct patterns of gene expression between CNS lymphomas and nodal lymphomas. Each column represents an individual tumor, and each row represents a single gene. Red indicates up-regulation; green, down-regulation; and black, similar to the median of the reference pool. (C) Assignment of PCNSL and nodal DLBCL tumors to germinal center (GCB), activated B cell (ABC), and type 3 subclasses. PCNSL tumors were distributed equally among the 3 subclasses. Hierarchic clustering was based upon the expression of 38 established marker genes of ABC and GCB subtypes. (D) Mean expression of 4 marker genes distinguishes CNS lymphoma as determined by quantitative real-time PCR in cases of primary CNS lymphomas (blue), large B-cell lymphomas from lymph nodes (red), and secondary CNS lymphomas (metastatic CNS lymphomas; yellow). Y-axis corresponds to percent expression of control gene. Error bars depict standard deviation of mean value. P values refer to the differences in expression between primary CNS lymphomas and nodal large B-cell lymphoma: XBP-1 (P < .001), c-Myc (P < .001), PIM-1 (P < .001), and E2F-5 (P < .001). CD10 expression was similar in the 3 types of lymphomas. Gene expression values obtained by microarray analysis were in agreement with those obtained by quantitative RT-PCR. (The minimum Pearson correlation is 0.65 corresponding to the P value of < .001 for the test of the hypothesis of no association between microarray and Taqman data for these marker genes.)
RNA amplification and labeling
Total sample RNA (1 μg) was linearly amplified through one round of modified in vitro transcription exactly in parallel with Universal Human Reference RNA (Stratagene, La Jolla, CA).11,12 Pearson coefficient for the amplification steps was at least 0.75 and for self-hybridization was at least 0.95.13 Amplified RNA was converted to amino-allyl modified cDNA and coupled to N-hydroxysuccinimidyl esters of Cy3 or Cy5 (Amersham-Pharmacia, Piscataway, NJ) and then hybridized to a microarray slide at 65°C for 12 to 16 hours. The slide was then washed and immediately scanned with Axon Imager 4000B using GenePix Pro3 software (both from Axon Instruments, Union City, CA).
Expression microarray preparation
The 20 862 cDNAs used in these studies were from Research Genetics (Huntsville, AL). On the basis of unigene build 166, these clones represent 19 740 independent loci. Briefly, clones were polymerase chain reaction (PCR)-amplified using universal primers and precipitated, dissolved in 3 × SSC, and printed on poly-l-lysine-coated slides. Slides were postprocessed using standard protocols. Microarrays from 5 different print runs were used in this study. Identity of clones described in this paper was verified by resequencing. Hybridization, washing, scanning, and primary data analysis were performed as described.
Microarray data preprocessing
The images were quantified using GenePix Pro 3 software, and the background-corrected ratios of the median signal of Cy5 and Cy3 channels were formed for each spot and log-transformed (base 2) with the replicate spots averaged. Each array was normalized by applying subarray median normalization.14 A spot was retained if it was present in at least 80% of the experiments and has shown at least 2-fold change in at least one experiment.
Missing values were imputed by using the median value for a given gene computed over 5 samples most highly correlated with the sample containing a missing value for that gene. The process was repeated until no more missing values remained. Finally, spots for which the average value among CNS lymphomas was less than the average value among normal brain samples were removed from the analysis. Thus, all spots remaining had to have at least twice the overexpression relative to the normal brain, considering that there were not more than 30% of normal brain cells present in the CNS samples.
Data analysis
Data were analyzed using freely available R statistical software version (V. 1.715 ). The genes were tested for significant association with outcome using 2-tailed t test with equal variance. The unadjusted P values were assessed by repeatedly shuffling the labels and recomputing the absolute value of the t statistic. Thus, the P value was assigned by computing the proportion of times the permutation-based statistic exceeded the observed one. The adjustment of the P value was done using the false discovery rate approach under an assumption of independence.16
Clustering of genes and lymphoma samples in our dataset was done using agglomerative hierarchic clustering using Euclidean metric and Ward method.
Differentiation analysis of primary CNS lymphoma
We applied a nearest centroid method17,18 with Pearson correlation as a similarity measure to assign the 32 primary CNS and lymph node large B-cell lymphomas in our dataset to corresponding subtypes (ABC, GCB, type 3). We identified 38 matching genes among the 100 genes used to assign lymphoma subclass as defined by Rosenwald et al.5 In the case of multiple probes, values were averaged. We further centered and scaled each gene in our dataset as well as the Rosenwald et al5 dataset by subtracting the median and dividing by the standard deviation estimated for that gene in each dataset. To verify that this normalization removed dataset-specific effects, we applied the hierarchic clustering to the samples combined from the 2 datasets; we did not observe significant coclustering by dataset. In contrast, clustering prenormalized samples clearly separated samples by dataset. In order to make sure that normalization did not remove signal pertinent to the subtypes of interest and that 38 of the 100 original genes are sufficient for subtype determination, we applied the nearest centroid procedure described next to the 80 validation samples in the Rosenwald et al5 series. We did so using the 160 training samples to define centroids and determined that the resulting type assignments closely reproduced the original listed, with only 8 of 80 samples misclassified. Thus, we proceeded with the analysis of the normalized data in our samples.
To define the centroid for each of the subtypes in the Rosenwald et al5 samples, we calculated the mean value for each gene in the 3 subtypes. We further computed a Pearson correlation from each of our lymphoma samples to the 3 centroids and assigned each sample to the subtype corresponding to the highest Pearson correlation.
Immunohistochemistry, immunofluorescence, and in situ RNA hybridization
Frozen sections (each 8 μM) were cut and immunostained for XBP-1, IL-4, or VWF by the biotin-streptavidin peroxidase method. Anti-XBP-1 was from Santa-Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-IL-4 was from Pharmingen (San Diego, CA); polyclonal anti-IL-4 was from Santa-Cruz Biotechnology (each was used at 1:50-1:100 dilution and gave similar results). IL-4 immunoreactivity was blocked by competition with recombinant human IL-4 (R and D Systems, Minneapolis, MN) (Tables S1-S2, available on the Blood website; see the Supplemental Tables link at the top of the online article). Monoclonal anti-VWF was from DAKO (Carpinteria, CA). Anti-phospho-STAT6 (P-STAT6) was from Upstate Biotechnology (Lake Placid, NY). Immunohistochemistry for P-STAT6 was performed as described.19 Confocal immunofluorescence microscopy was performed using a Zeiss LSM 510 Meta microscope (Carl Zeiss, Heidelberg, Germany) using multiphoton 488-nm and 543-nm laser excitation. Images were acquired using a 63 ×/1.4 NA Plan-Apochromat oil immersion objective (Nikon, Melville, NY). Anti-IgG goat anti-mouse FITC secondary antibody was used to recognize anti-VWF and anti-IgG goat anti-rabbit Alexa 594 was used to recognize anti-IL-4. DAPI counterstain was used to visualize cell nuclei. Brightness and contrast of images were modified for publication. For in situ hybridization, 12-μm frozen sections were fixed in 4% paraformaldehyde and adjacent sections subjected to in situ hybridization using sense and antisense IL-4 riboprobes as described.20 The IL-4 gene was isolated using human IL-4 gene-specific oligonucleotides and reverse-transcriptase (RT)-PCR, cloned into Bluescript vector and verified by sequencing. IL-4 riboprobes were generated in an in vitro transcription reaction using the pBluescript IL-4 plasmid.
Quantitative RT-PCR
A set of representative genes identified by array analysis as well as IL-4 were selected for validation using commercially available Assays-on-Demand probe-primer sets (Applied Biosystems, Foster City, CA) by real-time quantitative PCR analysis. Briefly, 500 ng RNA was reverse transcribed with 1.25 units of murine leukemia virus reverse transcriptase (Invitrogen, Frederick, MD) using 5-μM random primers (Invitrogen) in the presence of 7.5 mM MgCl2 and 1 unit of ANTI-Rnase (Ambion, Austin, TX) at 25°C for 10 minutes, 48°C for 40 minutes, and 95°C for 5 minutes. Quantitative-PCR analysis was performed on an AB Prism 7900 sequence detection system (Applied Biosystems). Quantitative detection of specific nucleotide sequences was based on the fluorogenic 5′ nuclease assay, as summarized.21 Relative expression was calculated as described.22 The cDNA equivalent of 3 to 5 ng RNA was measured in triplicate by real-time PCR using QPCR master mix with final concentrations of 5.5 mM MgCl2, 200 μM dNTPs, and 0.5 units Hotstart Amplitaq Gold (Applied Biosystems) in 20-μL volume for 384-well plates or 50-μL volume for 96-well plates. For normalization, cDNA equivalent to 3 to 5 ng input RNA was measured for a control gene GUS (β-glucuronidase); data are presented as a percentage of the control gene.
Results
Molecular features of primary CNS lymphoma
We used cDNA microarrays to define the features of gene expression in tumors from immunocompetent patients with primary CNS lymphoma that are most distinct from systemic large B-cell lymphoma. We analyzed the gene expression pattern of 23 primary CNS lymphoma diagnostic specimens—all of large B-cell histology—and compared these with 9 consecutive cases of large B-cell lymphoma isolated from peripheral lymph nodes.
More than 460 genes exhibited highly significant differential expression between the nodal and CNS lymphomas (false discovery rate < 0.01) (Figure 1B; Table 1). CNS lymphomas were distinguished by activation of genes involved in apoptosis, in cell proliferation, in the unfolded protein response pathway, and by distinct molecular interactions with the stromal environment. For example, CNS lymphomas exhibited significantly higher expression of cathepsin-D, while nodal lymphomas expressed higher levels of matrix metalloproteinase-2, ADAM-12, and E-cadherin. Expression of CD11c, a marker of monocytes and dendritic cells, was also higher in systemic large B-cell lymphomas obtained from lymph nodes.
The most significantly differentially expressed genes in CNS and nodal large B-cell lymphomas
Gene function and accession no. . | Name . | FDR . | RAWP . | CNS/LN ratio . |
|---|---|---|---|---|
| Genes with higher level of expression in PCNSL | ||||
| Cell cycle, proliferation, and growth | ||||
| W87741 | v-Myc myelocytomatosis viral oncogene homolog | 0.0056 | < .001 | 2.74 |
| AA464689 | Myc-induced nuclear antigen | 0.0062 | < .001 | 1.54 |
| AA669443 | Eukaryotic translation initiation factor 5 | 0.0043 | < .001 | 2.02 |
| H56918 | Eukaryotic translation initiation factor 4A | 0.0043 | < .001 | 5.58 |
| AA086475 | Cullin 5 | 0.0077 | < .001 | 2 |
| AA455521 | E2F5 transcription factor 5 | 0.0077 | < .001 | 4.2 |
| H38804 | Mitotic checkpoint protein BUB 3 | 0.0089 | .005 | 2.55 |
| AA453663 | Pim-1 | 0.035 | .005 | 2.46 |
| AA284634 | Janus kinase 1 | 0.056 | .009 | 1.86 |
| Unfolded protein response | ||||
| W90128 | X-box binding protein 1 | 0.0043 | < .001 | 4.59 |
| AA707661 | Activating transcription factor 6 | 0.001 | < .001 | 1.87 |
| Programmed cell death | ||||
| W01536 | Programmed cell death 4 | 0.0043 | < .001 | 5.41 |
| W70189 | Fas (TNFRSF6)-associated factor 1 | 0.0043 | < .001 | 2.22 |
| N94588 | Caspase-8 and FADD-like apoptosis regulator | 0.064 | .01 | 2.2 |
| Invasion, adhesion, stromal environment | ||||
| AA425451 | Integrin alpha E (CD103) | 0.036 | .005 | 1.79 |
| N20475 | Cathepsin-D | 0.0056 | < .001 | 2.35 |
| Genes with lower level of expression in PCNSL | ||||
| Transcription factor | ||||
| AA001604 | Zinc finger 36 | 0.0056 | .002 | 0.56 |
| AA701046 | Myeloid/lymphoid or mixed lineage leukemia | 0.0043 | .001 | 0.49 |
| H84982 | Checkpoint suppressor 1 | 0.015 | < .001 | 0.36 |
| Signal transduction | ||||
| AA455591 | EphB3 | 0.0043 | .001 | 0.56 |
| H67666 | Ribosomal S6 kinase, 90 kDa, polypeptide 3 | 0.0099 | < .001 | 0.43 |
| AA148524 | Discoidin domain receptor family member 2 | 0.0043 | < .001 | 0.47 |
| Invasion, adhesion, stromal environment | ||||
| AA099554 | Disintegrin and metalloprotease domain 12 | 0.009 | < .001 | 0.42 |
| AA936799 | Matrix metalloprotease 2 | 0.0056 | < .001 | 0.15 |
| AA430540 | Collagen type IV alpha 2 | 0.01 | < .001 | 0.5 |
| R75636 | Collagen type V alpha 1 | 0.015 | .001 | 0.54 |
| H16637 | Vascular cell adhesion molecule 1 | 0.034 | .005 | 0.49 |
| H97778 | E-cadherin | 0.039 | .006 | 0.48 |
| N64384 | Integrin alpha X (CD11C) | 0.025 | .003 | 0.32 |
Gene function and accession no. . | Name . | FDR . | RAWP . | CNS/LN ratio . |
|---|---|---|---|---|
| Genes with higher level of expression in PCNSL | ||||
| Cell cycle, proliferation, and growth | ||||
| W87741 | v-Myc myelocytomatosis viral oncogene homolog | 0.0056 | < .001 | 2.74 |
| AA464689 | Myc-induced nuclear antigen | 0.0062 | < .001 | 1.54 |
| AA669443 | Eukaryotic translation initiation factor 5 | 0.0043 | < .001 | 2.02 |
| H56918 | Eukaryotic translation initiation factor 4A | 0.0043 | < .001 | 5.58 |
| AA086475 | Cullin 5 | 0.0077 | < .001 | 2 |
| AA455521 | E2F5 transcription factor 5 | 0.0077 | < .001 | 4.2 |
| H38804 | Mitotic checkpoint protein BUB 3 | 0.0089 | .005 | 2.55 |
| AA453663 | Pim-1 | 0.035 | .005 | 2.46 |
| AA284634 | Janus kinase 1 | 0.056 | .009 | 1.86 |
| Unfolded protein response | ||||
| W90128 | X-box binding protein 1 | 0.0043 | < .001 | 4.59 |
| AA707661 | Activating transcription factor 6 | 0.001 | < .001 | 1.87 |
| Programmed cell death | ||||
| W01536 | Programmed cell death 4 | 0.0043 | < .001 | 5.41 |
| W70189 | Fas (TNFRSF6)-associated factor 1 | 0.0043 | < .001 | 2.22 |
| N94588 | Caspase-8 and FADD-like apoptosis regulator | 0.064 | .01 | 2.2 |
| Invasion, adhesion, stromal environment | ||||
| AA425451 | Integrin alpha E (CD103) | 0.036 | .005 | 1.79 |
| N20475 | Cathepsin-D | 0.0056 | < .001 | 2.35 |
| Genes with lower level of expression in PCNSL | ||||
| Transcription factor | ||||
| AA001604 | Zinc finger 36 | 0.0056 | .002 | 0.56 |
| AA701046 | Myeloid/lymphoid or mixed lineage leukemia | 0.0043 | .001 | 0.49 |
| H84982 | Checkpoint suppressor 1 | 0.015 | < .001 | 0.36 |
| Signal transduction | ||||
| AA455591 | EphB3 | 0.0043 | .001 | 0.56 |
| H67666 | Ribosomal S6 kinase, 90 kDa, polypeptide 3 | 0.0099 | < .001 | 0.43 |
| AA148524 | Discoidin domain receptor family member 2 | 0.0043 | < .001 | 0.47 |
| Invasion, adhesion, stromal environment | ||||
| AA099554 | Disintegrin and metalloprotease domain 12 | 0.009 | < .001 | 0.42 |
| AA936799 | Matrix metalloprotease 2 | 0.0056 | < .001 | 0.15 |
| AA430540 | Collagen type IV alpha 2 | 0.01 | < .001 | 0.5 |
| R75636 | Collagen type V alpha 1 | 0.015 | .001 | 0.54 |
| H16637 | Vascular cell adhesion molecule 1 | 0.034 | .005 | 0.49 |
| H97778 | E-cadherin | 0.039 | .006 | 0.48 |
| N64384 | Integrin alpha X (CD11C) | 0.025 | .003 | 0.32 |
False discovery rate (FDR) corresponds to the expected proportion of falsely called genes among the genes declared to be differentially expressed. RAWP corresponds to the permutation-based unadjusted P value.
In addition, primary CNS lymphomas were distinguished from nodal lymphomas by high expression of the oncogenes Pim-1 and c-Myc. Mina53, a recently described Myc-induced nuclear antigen involved in cell proliferation, was also expressed at higher levels in primary CNS lymphoma, supporting a role for c-Myc pathway in the pathogenesis of brain lymphomas.23 Mina53 has not previously been reported to be expressed in lymphoma.
CNS lymphomas are also characterized by high expression of XBP-1 and activating transcription factor 6 (ATF-6), each is a basic leucine-zipper transcription factor that regulates the UPR.24 ATF-6 has been shown to induce expression of XBP-1.25 Both XBP-1 and ATF-6 regulate expression of an overlapping subset of endoplasmic reticulum chaperone genes as well as other target genes, including c-Myc in the UPR in mouse embryonic fibroblasts (MEFs).26
We used data described by Alizadeh et al4 and by Rosenwald et al5 and their subtype assignments to subclassify the primary CNS lymphomas in our dataset. We used 38 shared unique gene identifiers, which are present on our array platform and are among the 100 genes used by Rosenwald et al5 to classify large B lymphomas as ABC, GCB, and type 3. After recentering and scaling genes in our and Rosenwald et al's5 datasets, we first verified that 38 genes are sufficient to recover original subtypes in the Rosenwald et al5 dataset. We thus proceeded to classify the lymphomas in our study using the Rosenwald et al5 classification into ABC, GCB, and type 3 subtypes. We found that all 3 molecular subtypes of systemic diffuse large B-cell lymphoma (DLBCL) were represented in the nodal lymphomas used in our study: 7 of 9 lymph node lymphomas were assigned to the GCB subtype; 1 of the nodal lymphomas, to the ABC subtype; and 1, to the type 3 subtype. PCNSL cases (all large B-cell lymphomas) were assigned among the 3 subtypes equally with 8 tumors each assigned to the ABC and GCB subgroups and 7 tumors assigned to the type 3 subgroup.
In addition, we noted overlapping expression of germinal center genes such as bcl-6 and activation genes such as cyclin D2 among specimens classified as ABC type (Figure 1C). This observation is consistent with the findings of recent immunohistochemical studies that have detected coexpression of a germinal center marker bcl-6 and an activation marker MUM-1 in the majority of primary CNS lymphomas, suggesting an overlapping state of differentiation (Figure 1C)7,27
We used quantitative real-time PCR to validate the cDNA microarray data, focusing on several genes likely to be significant in the pathogenesis of CNS lymphoma (Figure 1D). There was strong agreement between the quantitative real-time PCR data and the cDNA microarray results. In addition, we analyzed expression of CNS lymphoma marker genes in 3 consecutive cases of secondary CNS lymphoma—systemic large B-cell lymphomas that had metastasized to brain. We found that secondary CNS lymphomas also exhibited high levels of c-Myc, Pim-1, and XBP-1 compared with nodal large B-cell lymphomas. This finding is consistent with the hypothesis that a factor(s) in the brain tumor microenvironment induces some but not all of the transcriptional characteristics of primary CNS lymphoma.
XBP-1 was among the most significantly differentially expressed genes in CNS lymphomas (FDR < 0.004). We used immunohistochemistry to characterize its pattern of expression and found evidence for the strongest XBP-1 protein expression in dense populations of tumor cells closely associated with the vasculature in CNS lymphomas (Figure 2). Large B-cell lymphoma cells in lymph nodes exhibited lower XBP-1 immunoreactivity overall, and there was no relationship between XBP-1 immunoreactivity and the tumor vasculature (10/10 specimens analyzed with at least 4 fields/specimen at × 100 power). These findings suggest that XBP-1 expression in primary CNS lymphoma is at least partially regulated by paracrine interactions with the CNS vasculature.
Interleukin-4 expression and lymphoma angiotropism
We then investigated how interactions with the tumor vasculature might regulate XBP-1 gene expression. The multifunctional cytokine IL-4, an important regulator of B-cell activation, proliferation, aggregation, and survival, is the only cytokine known to regulate expression of XBP-1 in B cells and was thus a logical candidate.24,28-31 This cytokine has also been demonstrated to regulate angiogenesis.32-35 IL-4 was not among the 20 862 cDNAs on the expression microarray chip used in this study. Immunohistochemical analysis revealed expression of IL-4 by endothelial cells (Figure 3) as well as high levels by angiotropic tumor cells in CNS lymphoma tumors, including one case of secondary CNS lymphoma. In a parallel analysis, IL-4 immunoreactivity was lower or undetectable in the tumor vessels of nodal large B-cell lymphomas. The vasculature of adjacent nonneoplastic brain did not exhibit IL-4 immunoreactivity. In addition, we did not detect significant IL-4 immunoreactivity by tumor vessels or tumor cells in 4 different glioblastoma specimens or in 2 cases each of breast and lung carcinomas metastatic to brain. IL-4 protein expression by tumor endothelia was confirmed by confocal microscopy (Figure 3).
XBP-1 expression. Immunohistochemical localization of XBP-1 expression (brown color) in tumor cells surrounding the CNS tumor vasculature in a hematoxylin-stained tumor. (A) Transverse view; arrow points to a blood vessel. (B) Longitudinal view of vessel; XBP-1 immunoreactivity of tumor cells distinguishes them from endothelial cells. (C) Intense XBP-1 immunoreactivity exhibited by a dense population of tumor cells growing around a tumor vessel at the infiltrating edge of a different CNS lymphoma tumor specimen. (D) There was minimal XBP-1 immunoreactivity in a representative specimen of large B-cell lymphoma isolated from a lymph node. When assessed by immunohistochemistry, the pattern of BCL-6 expression was diffuse within CNS lymphomas, not differentially related to the tumor vasculature (not shown). Original magnification × 200 for all panels.
XBP-1 expression. Immunohistochemical localization of XBP-1 expression (brown color) in tumor cells surrounding the CNS tumor vasculature in a hematoxylin-stained tumor. (A) Transverse view; arrow points to a blood vessel. (B) Longitudinal view of vessel; XBP-1 immunoreactivity of tumor cells distinguishes them from endothelial cells. (C) Intense XBP-1 immunoreactivity exhibited by a dense population of tumor cells growing around a tumor vessel at the infiltrating edge of a different CNS lymphoma tumor specimen. (D) There was minimal XBP-1 immunoreactivity in a representative specimen of large B-cell lymphoma isolated from a lymph node. When assessed by immunohistochemistry, the pattern of BCL-6 expression was diffuse within CNS lymphomas, not differentially related to the tumor vasculature (not shown). Original magnification × 200 for all panels.
IL-4 gene expression by tumor vessels was demonstrated by in situ hybridization in 5 of 5 primary CNS lymphomas as well as in 1 case of lymphoma metastatic to brain (Figure 4). This is the first demonstration of IL-4 production by endothelial cells. There was no evidence for IL-4 expression by tumor vasculature in all 7 cases of nodal large B-cell lymphomas analyzed by in situ hybridization, suggesting that vascular expression of IL-4 occurs selectively in CNS lymphoma. We also detected expression of IL-4 by lymphoma cells as evidenced by in situ hybridization, suggesting an autocrine role for this cytokine in stimulation of growth and survival, both in lymphoma as well as endothelial cell types in this tumor (Figure 4). While in situ hybridization revealed a diffuse pattern of IL-4 gene expression by tumor vessels as well as endothelia, immunohistochemical analysis demonstrated that IL-4 protein accumulation was most intense in the vessel microenvironment.
We confirmed IL-4 gene expression in individual primary CNS lymphoma specimens by quantitative real-time PCR and noted a trend between higher relative IL-4 expression in CNS lymphoma tumors associated with shorter survival (not shown); however, patients whose CNS lymphoma tumors were included in this study received different treatments, a variable that makes survival analysis in this dataset somewhat tenuous.
A number of IL-4 target genes were expressed at significantly higher levels in CNS lymphomas compared with nodal lymphomas including cathepsin-D, Src-like adapter protein, caspase-8, and FADD-like apoptosis regulator (FLICE), as well as XBP-1 (Table 1; Figure 1C).30 These results suggest that autocrine and paracrine IL-4 production in CNS lymphomas has a significant role in the biology of this tumor, possibly through regulation of the activity of Janus kinase 1, which we demonstrated to be expressed at significantly higher levels in CNS lymphomas compared with nodal lymphomas (Table 1).
IL-4 has been shown to potentiate antiapoptotic mechanisms in B lymphocytes as well as resistance to chemotherapy in cancer cells.36,37 IL-4-dependent gene expression is mediated by the activation of the signaling and transcription factor STAT6. To provide evidence for STAT6 activation in CNS lymphoma tumors, we performed an immunohistochemical analysis of phosphorylated STAT6 (P-STAT6) in diagnostic CNS lymphoma specimens. There was reproducible expression of P-STAT6 both by the tumor endothelia and by lymphoma cells, with evidence for particularly strong activation in the vessel microenvironment, supporting a role for STAT6 as a mediator of IL-4 signaling in CNS lymphoma (Figure 5). We did not detect activated STAT6 expression by tumor endothelia in 8 of 8 nodal DLBCLs; focal expression of activated STAT6 expression by tumor cells was detected in one of these specimens.
In addition, we characterized STAT6 activation in an independent set of 20 cases of primary CNS lymphoma derived from a population of immunocompetent patients with B-cell PCNSL, all of whom were treated with a high-dose methotrexate-containing regimen (3-8 g/m2) and evaluated at the University of California, San Francisco (radiation was reserved for patients with relapsed disease refractory to intravenous methotrexate). Tumors that contained at least one focus in which dense populations of lymphoma cells scored positive for nuclear P-STAT6 expression were associated with early disease progression and short overall survival in comparison with tumors of low cellular density or tumors that exhibited sparse or absent P-STAT6 expression (Figure 5D-E). Taken together, these results provide independent lines of evidence that support the conclusion that expression of the interleukin-4 signaling pathway is associated with tumorigenesis and adverse prognosis in primary CNS lymphoma.
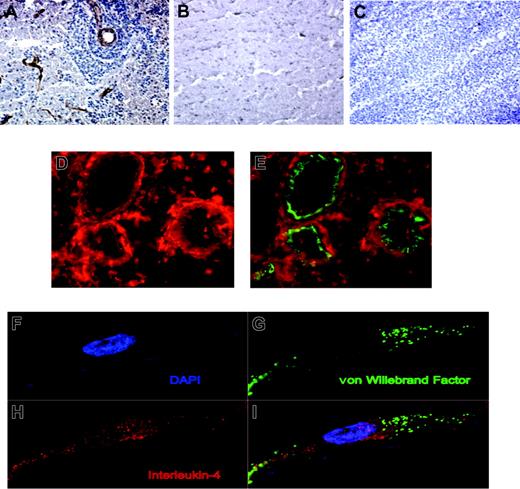
IL-4 expression in primary CNS lymphoma. (A) Immunohistochemical localization of IL-4 (brown color) to vessels associated with angiotropic CNS lymphoma cells at the infiltrating edge of a primary CNS lymphoma. (B) IL-4 immunoreactivity was not detected in the vessels of normal brain. (C) IL-4 immunoreactivity was absent or less prominent in association with the vessels of large B-cell lymphoma isolated from lymph nodes. Original magnification × 200 for panels A-C. (D-E) Dual-color immunofluorescence demonstrates colocalization of IL-4 (D-E; Alexa 594 red immunofluorescence) and von Willebrand factor (E; FITC immunofluorescence) on tumor vessels in primary CNS lymphomas. Original magnification × 400 for panels D-E. (F-I) Endothelial cells in CNS lymphoma are immunoreactive for IL-4 as demonstrated by confocal microscopy. Diffuse cytoplasmic and perinuclear IL-4 expression by an endothelial cell did not exhibit significant spectral overlap with von Willebrand factor expression, which appeared to be localized to specialized Weibel-Palade bodies. Dimensions of displayed images are 68.3 μm × 31.8 μm each.
IL-4 expression in primary CNS lymphoma. (A) Immunohistochemical localization of IL-4 (brown color) to vessels associated with angiotropic CNS lymphoma cells at the infiltrating edge of a primary CNS lymphoma. (B) IL-4 immunoreactivity was not detected in the vessels of normal brain. (C) IL-4 immunoreactivity was absent or less prominent in association with the vessels of large B-cell lymphoma isolated from lymph nodes. Original magnification × 200 for panels A-C. (D-E) Dual-color immunofluorescence demonstrates colocalization of IL-4 (D-E; Alexa 594 red immunofluorescence) and von Willebrand factor (E; FITC immunofluorescence) on tumor vessels in primary CNS lymphomas. Original magnification × 400 for panels D-E. (F-I) Endothelial cells in CNS lymphoma are immunoreactive for IL-4 as demonstrated by confocal microscopy. Diffuse cytoplasmic and perinuclear IL-4 expression by an endothelial cell did not exhibit significant spectral overlap with von Willebrand factor expression, which appeared to be localized to specialized Weibel-Palade bodies. Dimensions of displayed images are 68.3 μm × 31.8 μm each.
IL-4 in situ hybridization. (A) In situ hybridization with an antisense riboprobe against IL-4 demonstrates gene expression by CNS lymphoma cells. (B) There was no significant hybridization with a sense riboprobe against IL-4 in a parallel section from the same tumor. (C) Tumor cells and vessels in a breast carcinoma metastatic to brain did not exhibit significant hybridization with an antisense riboprobe against IL-4. Original magnification × 200 for panels A-C. (D) In situ hybridization with an antisense riboprobe against IL-4 reveals increased gene expression in a representative CNS lymphoma tumor vessel. (E) There was no significant hybridization with a sense riboprobe against IL-4 in a parallel section from the same tumor. (F) Vessels in normal brain did not exhibit significant hybridization with an antisense riboprobe against IL-4. Original magnification × 400 for panels D-F.
IL-4 in situ hybridization. (A) In situ hybridization with an antisense riboprobe against IL-4 demonstrates gene expression by CNS lymphoma cells. (B) There was no significant hybridization with a sense riboprobe against IL-4 in a parallel section from the same tumor. (C) Tumor cells and vessels in a breast carcinoma metastatic to brain did not exhibit significant hybridization with an antisense riboprobe against IL-4. Original magnification × 200 for panels A-C. (D) In situ hybridization with an antisense riboprobe against IL-4 reveals increased gene expression in a representative CNS lymphoma tumor vessel. (E) There was no significant hybridization with a sense riboprobe against IL-4 in a parallel section from the same tumor. (F) Vessels in normal brain did not exhibit significant hybridization with an antisense riboprobe against IL-4. Original magnification × 400 for panels D-F.
Three common patterns of P-STAT6 expression in PCNSL. (A) Low cellular density; absent P-STAT6 expression. (B) Low cellular density; positive P-STAT6 expression. (C) High cellular density; strong P-STAT6 nuclear expression both by tumor cells and on vascular endothelia. Patients whose tumors exhibited foci of high cellular density with positive P-STAT6 expression (C) had significantly worse outcomes than patients with tumors that were negative for P-STAT6 or tumors that were of low cellular density that scored positive for P-STAT6. Original magnification × 200 for panels A-C. Patients with intense P-STAT6 expression (C; red) experienced early progression (D) and short overall survival (E) when treated initially with high-dose methotrexate-based regimens. Green refers to tumors with sparse or absent P-STAT6 expression. (Whole-brain radiation was reserved for patients with methotrexate-refractory disease.) (Ten patients in each group; P values were calculated by log-rank test.)
Three common patterns of P-STAT6 expression in PCNSL. (A) Low cellular density; absent P-STAT6 expression. (B) Low cellular density; positive P-STAT6 expression. (C) High cellular density; strong P-STAT6 nuclear expression both by tumor cells and on vascular endothelia. Patients whose tumors exhibited foci of high cellular density with positive P-STAT6 expression (C) had significantly worse outcomes than patients with tumors that were negative for P-STAT6 or tumors that were of low cellular density that scored positive for P-STAT6. Original magnification × 200 for panels A-C. Patients with intense P-STAT6 expression (C; red) experienced early progression (D) and short overall survival (E) when treated initially with high-dose methotrexate-based regimens. Green refers to tumors with sparse or absent P-STAT6 expression. (Whole-brain radiation was reserved for patients with methotrexate-refractory disease.) (Ten patients in each group; P values were calculated by log-rank test.)
Discussion
Here, we identify several molecular features of primary CNS lymphoma, some of which are distinct from large B-cell lymphoma isolated from lymph nodes. While individual primary CNS lymphomas may be assigned to ABC, GCB, or a type 3 subclass of differentiation, our data suggest some overlap in expression of GCB and ABC markers in this disease. This observation is consistent with recent immunohistochemical data that demonstrate that a majority of primary CNS lymphomas express both bcl-6 (a GCB marker) as well as MUM-1 (an ABC marker), suggesting an overlapping histogenetic time slot of B-cell differentiation.27
We demonstrate for the first time that tumor cells as well as endothelia in CNS lymphomas express IL-4, a B-cell growth and survival factor, and provide evidence that IL-4 signal transduction is associated with tumor progression in CNS lymphoma. Several IL-4 target genes including XBP-1 are among the most significantly differentially expressed genes in CNS lymphomas. In addition, we demonstrate that increased expression of activated STAT6 by tumor cells and endothelia in CNS lymphoma predicts early progression and short survival in an independent set of patients all treated with high-dose methotrexate-based chemotherapy. We propose that detection of nuclear expression of P-STAT6 in dense populations of tumor cells may be a clinically useful biomarker to delineate prognosis in the evaluation of formalin-fixed diagnostic specimens in primary CNS lymphoma.
CNS lymphomas express high levels of ATF-6 and XBP-1, both transcriptional regulators of the UPR; XBP-1 is an established IL-4 target gene in B cells. XBP-1 and the UPR have been shown to be essential in the support of tumor growth under conditions of hypoxic stress as well as glucose deprivation.38,39 Our data suggest that production of IL-4 in CNS lymphomas may support tumor progression through several potential mechanisms. For example, production and secretion of IL-4 by endothelial cells may facilitate expression of XBP-1 and activation of the UPR stress response pathway in dense populations of tumor cells that accumulate around blood vessels. CNS lymphomas exhibited significantly higher expression of caspase-8 and FADD-like apoptosis regulator (FLICE), an IL-4 target gene and inhibitor of death receptor-mediated apoptosis.40 In addition, we detected a significant correlation between high IL-4 RNA levels and decreased expression in CNS lymphomas of F1Aα, an apoptotic mediator and death-receptor binding protein (data not shown).41 We did not detect a correlation between intratumoral IL-4 expression and subclass of CNS lymphoma as GCB, ABC, and type 3.
While the IL-4 target gene bcl-6, a marker of GC origin, was detectable in CNS lymphomas both by gene expression and by immunohistochemistry, its level of expression was similar overall compared with the nodal lymphomas. There are also conflicting data on the significance of bcl-6 as an immunohistochemical marker of prognosis in CNS lymphoma with 2 of 3 published studies finding no significant correlation of bcl-6 with survival.7,27,42
The pathophysiology of tumor angiotropism in CNS lymphoma may recapitulate a normal adaptation of the brain microvasculature to facilitate the rapid induction of B-cell proliferation and aggregation in the setting of antigenic stimulation and vascular inflammation. Such an adaptation may provide a physiologic advantage given that the brain is devoid of a lymphoid environment. Production of IL-4 by activated CNS endothelia may also contribute significantly to B-cell lymphoma retention in the CNS, one of the cardinal features of CNS lymphoma.
We demonstrate that both primary and metastatic CNS large B-lymphoma tumor cells express high levels of c-Myc, which has been associated with adverse prognosis in systemic DLBCL.43,44 It is intriguing that c-Myc is also a UPR-target gene regulated both by XBP-1 as well as by ATF-6 in MEFs.26 This observation suggests a possible link between the expression of the UPR and other genes in the STAT3 pathway and could provide an explanation for high coexpression of XBP-1 as well as c-Myc and Pim-1 genes, which we observed both in primary as well as metastatic CNS large B-cell lymphomas.
Our results suggest several potential novel therapeutic interventions to treat CNS lymphomas. Disruption of the IL-4 signaling pathway involving Janus kinase 1 and STAT6 could potentially facilitate apoptosis in primary CNS lymphomas. Also, Pim-1 kinase may be a potential drug target in CNS lymphomas. In addition, we have shown that primary CNS lymphomas express high levels of the transcription factors XBP-1 and ATF-6, both regulators of the UPR. There is increasing evidence that pharmacologic blockade of the UPR may constitute a novel and effective approach in the targeting of tumors that are dependent on this survival pathway.39,45
Prepublished online as Blood First Edition Paper, January 17, 2006; DOI 10.1182/blood-2005-03-0897.
Supported by an NCI Research Career Award and by grants from the American Society of Clinical Oncology and UCSF Mt Zion Health Fund (J.L.R.) and by RO1 NCI CA101042-01 (C.H.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to J. Tsai for technical support and to D. Louis for his gift of 3 lymphoma cases; we extend thanks to L. Lanier for helpful discussions and J. Cyster and D. Bredt for critical reading of the paper.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal