Abstract
Although typically found in cancers, frameshift mutations in microsatellites have also been detected in chronically inflamed tissues. Allogeneic hematopoietic cell transplantation (HCT) may potentially produce chronic tissue stress through graft-versus-host reactions. We examined non-neoplastic epithelial tissues (colon, buccal) obtained 1 to 5061 days after human allogeneic HCT for the presence of genomic alterations at 3 tetranucleotide and 3 mononucleotide microsatellite loci. Novel bands indicative of microsatellite instability (MSI) at tetranucleotide repeats were detected in laser-microdissected colonic crypts and in buccal smears of 75% and 42% of patients who received an allograft, respectively. In contrast, no MSI was found in similar tissues from control subjects and from patients after intensive chemotherapy or in buccal cells from patients after autologous HCT. The MSI found in colon, which was often affected by graft-versus-host disease, was not due to loss of expression or nitrosylation of DNA repair proteins. MSI in clinically intact oral mucosa was more frequently found at later time points after HCT. MSI was also found in 3 posttransplant squamous cell cancers examined. Our data show that genomic alterations in epithelium regularly occur after allogeneic HCT and may be implicated in the evolution of posttransplantation diseases, including secondary cancer.
Introduction
Microsatellites are highly polymorphic repetitive sequences (1-6 nucleotides) of unknown function that are scattered throughout the genome.1 They are inherited stably and are unique to each individual which makes them ideal tools for forensic purposes as well as for documentation of engraftment following allogeneic hematopoietic cell transplantation (HCT).1,2 Ever since microsatellite length alterations were detected in hereditary nonpolyposis colorectal carcinoma and sporadic cancers, especially squamous carcinomas, microsatellite instability (MSI) has gained substantial attention and has been used as an indicator of genomic instability.1,3 Although typical in cancers, MSI has more recently also been found in non-neoplastic, chronically inflamed tissues from patients with ulcerative colitis and rheumatoid arthritis.4-6 The exact mechanism through which chronic inflammation produces MSI remains to be elucidated. To maintain genome integrity cells are equipped with several distinct but not completely independent DNA repair systems such as mismatch repair (MMR), base excision repeat (BER), and nucleotide excision repair (NER).7 Existing hypotheses postulate that either oxidative stress in inflamed tissues overwhelms the capacity for DNA repair, or that overproduction of free radicals inactivates the DNA repair pathways directly.6,8-10 The biologic meaning of the MSI found in chronic inflammatory settings is not yet clearly defined. Unlike ulcerative colitis, in which MSI has been associated with progression to cancer, MSI in rheumatoid arthritis does not indicate an increased cancer risk.4,5
After allogeneic HCT, epithelial tissues become injured through the preparative regimen and are then potentially attacked by alloreactive T cells. Graft-versus-host (GvH) reactions are complex processes involving release of cytokines, recruitment of inflammatory cells, cellular cytotoxicity, and production of free radicals, among others.11 The net effect of these alloantigenic reactions is tissue stress which we recognize clinically as GvH disease (GvHD), and this most frequently occurs in colon, skin, mouth, and liver. In addition, various delayed posttransplantation clinical syndromes such as pulmonary or musculoskeletal disease as well as secondary malignancies have been linked to chronic GvHD.12 On the basis of the concept that chronic tissue stress may cause genomic alterations, we examined genomic instability in epithelium after allogeneic HCT by MSI.
Patients, materials, and methods
Patients and sample collection
Patient information is summarized in the tables. Colon and tumor biopsies examined in this study were paraffin-embedded archival samples from the Pathology Department of the University of Freiburg. Colon biopsies were obtained endoscopically from patients after allogeneic HCT, after chemotherapy, or from patients with no history of chemotherapy, after informed consent. Colon endoscopy was performed for investigation of diarrhea, abdominal pain, abdominal bleeding, follow-up, or cancer screening (Table 1). Tumor biopsies were obtained from patients who developed a secondary epithelial malignancy after allogeneic HCT. Buccal smears were obtained, after informed consent, from patients after allogeneic HCT, from patients after autologous HCT, or from healthy volunteers. Buccal smears were performed after 3 mouth washes by gently but firmly twirling a sterile brush (Oribrush; Orifice Medical, Ystad, Sweden) on the inside surface of each cheek. At the time of buccal sampling no patient had clinical signs of mucositis or oral GvHD, except where otherwise indicated. Patients who received an autograft and healthy donors who gave buccal smears also donated blood or urine. Approval for these studies was obtained from the Freiburg University Medical Center institutional review board.
Microsatellite analysis of colon mucosa
. | . | . | . | . | . | . | . | . | . | . | Laser-microdissected crypts . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | . | Tetranucleotides . | . | . | . | |||
| HCT/chemo and patient ID . | Age, y/sex . | Disease . | History of chemo* . | Preparative regimen/last chemo . | Reason for colon biopsy . | D of sampling† . | Pathology of biopsy‡ . | Clinical GI-GvHD stage at sampling . | Overall max GvHD§ . | Whole colon section∥ . | THO-1 . | SEE33 . | D14S120 . | Mononucleotides¶ . | |||
| Allo | |||||||||||||||||
| C1 | 61/F | AML | Yes | FBM | Diar | 18 | N | 0 | 0 | S | S | S | S | S | |||
| C2 | 52/M | AML | Yes | BuCy | Diar | 20 | I° | I° | I° | S | S | MSI | S | S | |||
| C3# | 53/M | CML | No | BuCy | Diar | 30 | I° | I° | I° | S | MSI | NE | S | S | |||
| C4 | 19/M | ALL | Yes | TBI/VC | Diar | 42 | I° | II° | III° | S | MSI | MSI | S | S | |||
| C5 | 68/M | AML | No | FBM | Diar | 46 | III° | III° | III° | S | S | MSI | S | S | |||
| C6 | 59/M | MM | Yes | 2Gy/F | Diar | 46 | III° | III° | III° | NE | S | MSI | S | S | |||
| C7 | 65/M | AML | Yes | FBM | Diar | 49 | II° | II° | III° | S | S | S | S | S | |||
| C8 | 45/F | ALL | Yes | BuCy | Diar | 57 | III° | III° | IV° | S | MSI | MSI | S | S | |||
| C9 | 29/M | CML | Yes | BuCy | Diar | 61 | II° | II° | IV° | MSI | NE | MSI | S | S | |||
| C10 | 61/F | AML | Yes | FBM | Diar | 92 | I° | I° | III° | MSI | MSI | NE | NE | S | |||
| C11 | 20/M | ALL | Yes | TBI/VC | Diar | 148 | N | 0 | Ext | S | NE | S | S | S | |||
| C12 | 52/M | CML | No | FBM | Diar | 175 | I° | I° | I° | MSI | S | MSI | NE | S | |||
| C13 | 45/M | AML | Yes | BuCy | Diar | 182 | II° | II° | Ext | S | MSI | MSI | S | S | |||
| C14 | 36/M | NHL | Yes | BuCy | FU | 277 | N | 0 | 0 | MSI | MSI | S | MSI | S | |||
| C15 | 49/F | CML | No | FBM | Diar | 543 | I° | I° | Ext | S | MSI | MSI | NE | S | |||
| C16 | 45/F | MDS | No | BuCy | Diar | 1089 | Col | 0 | Ext | S | S | S | S | S | |||
| Chemo | |||||||||||||||||
| C17 | 52/F | EsCa | Yes | Vino | Diar | 7 | N | NA | NA | S | S | S | NE | S | |||
| C18 | 20/F | HD | No | HD15 | Diar | 10 | Col | NA | NA | S | NE | S | NE | S | |||
| C19 | 72/F | NHL | Yes | Chlor | FU | 240 | N | NA | NA | S | S | S | S | S | |||
| C20 | 66/F | Col Ca | No | 5-FU | FU | 1805 | N | NA | NA | NE | S | S | S | S | |||
| None | |||||||||||||||||
| C21 | 65/F | HepC | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C22 | 73/M | Healthy | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C23 | 48/M | Healthy | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C24 | 69/F | Divert | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C25 | 39/F | Col irr | NA | NA | A pain | NA | N | NA | NA | S | S | S | S | S | |||
| C26 | 57/F | Divert | NA | NA | A pain | NA | N | NA | NA | S | S | S | S | S | |||
| C27 | 83/F | BrCa | No | NA | Bleed | NA | N | NA | NA | S | S | S | NE | S | |||
| C3# | 53/M | CML | No | NA | A pain | NA | N | NA | NA | S | S | NE | S | S | |||
. | . | . | . | . | . | . | . | . | . | . | Laser-microdissected crypts . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | . | Tetranucleotides . | . | . | . | |||
| HCT/chemo and patient ID . | Age, y/sex . | Disease . | History of chemo* . | Preparative regimen/last chemo . | Reason for colon biopsy . | D of sampling† . | Pathology of biopsy‡ . | Clinical GI-GvHD stage at sampling . | Overall max GvHD§ . | Whole colon section∥ . | THO-1 . | SEE33 . | D14S120 . | Mononucleotides¶ . | |||
| Allo | |||||||||||||||||
| C1 | 61/F | AML | Yes | FBM | Diar | 18 | N | 0 | 0 | S | S | S | S | S | |||
| C2 | 52/M | AML | Yes | BuCy | Diar | 20 | I° | I° | I° | S | S | MSI | S | S | |||
| C3# | 53/M | CML | No | BuCy | Diar | 30 | I° | I° | I° | S | MSI | NE | S | S | |||
| C4 | 19/M | ALL | Yes | TBI/VC | Diar | 42 | I° | II° | III° | S | MSI | MSI | S | S | |||
| C5 | 68/M | AML | No | FBM | Diar | 46 | III° | III° | III° | S | S | MSI | S | S | |||
| C6 | 59/M | MM | Yes | 2Gy/F | Diar | 46 | III° | III° | III° | NE | S | MSI | S | S | |||
| C7 | 65/M | AML | Yes | FBM | Diar | 49 | II° | II° | III° | S | S | S | S | S | |||
| C8 | 45/F | ALL | Yes | BuCy | Diar | 57 | III° | III° | IV° | S | MSI | MSI | S | S | |||
| C9 | 29/M | CML | Yes | BuCy | Diar | 61 | II° | II° | IV° | MSI | NE | MSI | S | S | |||
| C10 | 61/F | AML | Yes | FBM | Diar | 92 | I° | I° | III° | MSI | MSI | NE | NE | S | |||
| C11 | 20/M | ALL | Yes | TBI/VC | Diar | 148 | N | 0 | Ext | S | NE | S | S | S | |||
| C12 | 52/M | CML | No | FBM | Diar | 175 | I° | I° | I° | MSI | S | MSI | NE | S | |||
| C13 | 45/M | AML | Yes | BuCy | Diar | 182 | II° | II° | Ext | S | MSI | MSI | S | S | |||
| C14 | 36/M | NHL | Yes | BuCy | FU | 277 | N | 0 | 0 | MSI | MSI | S | MSI | S | |||
| C15 | 49/F | CML | No | FBM | Diar | 543 | I° | I° | Ext | S | MSI | MSI | NE | S | |||
| C16 | 45/F | MDS | No | BuCy | Diar | 1089 | Col | 0 | Ext | S | S | S | S | S | |||
| Chemo | |||||||||||||||||
| C17 | 52/F | EsCa | Yes | Vino | Diar | 7 | N | NA | NA | S | S | S | NE | S | |||
| C18 | 20/F | HD | No | HD15 | Diar | 10 | Col | NA | NA | S | NE | S | NE | S | |||
| C19 | 72/F | NHL | Yes | Chlor | FU | 240 | N | NA | NA | S | S | S | S | S | |||
| C20 | 66/F | Col Ca | No | 5-FU | FU | 1805 | N | NA | NA | NE | S | S | S | S | |||
| None | |||||||||||||||||
| C21 | 65/F | HepC | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C22 | 73/M | Healthy | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C23 | 48/M | Healthy | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C24 | 69/F | Divert | NA | NA | TuScr | NA | N | NA | NA | S | S | S | S | S | |||
| C25 | 39/F | Col irr | NA | NA | A pain | NA | N | NA | NA | S | S | S | S | S | |||
| C26 | 57/F | Divert | NA | NA | A pain | NA | N | NA | NA | S | S | S | S | S | |||
| C27 | 83/F | BrCa | No | NA | Bleed | NA | N | NA | NA | S | S | S | NE | S | |||
| C3# | 53/M | CML | No | NA | A pain | NA | N | NA | NA | S | S | NE | S | S | |||
At the time of colon sampling all patients revealed complete donor chimerism in blood (blood chimerism analysis in patients C1 and C2 was done at day 30).
Chemo indicates chemotherapy; HCT, hematopoietic cell transplantation; GI, gastrointestinal; GvHD, graft-versus-host disease; AML, acute myeloid leukemia; Allo, allogeneic HCT; FBM, fludarabine, carmustine, melphalan (reduced intensity conditioning); Diar, diarrhea; S, stable microsatellites; BuCy, busulphan, cyclophosphamide; MSI, microsatellite instability; CML, chronic myeloid leukemia; NE, not evaluable; ALL, acute lymphatic leukemia; TBI/VC, total body irradiation (12 Gy), etoposide phosphate, cyclophosphamide; MM, multiple myeloma; 2Gy/F: low total body irradiation (2 Gy), fludarabine (reduced intensity conditioning); NHL, non-Hodgkin lymphoma; FU, follow-up; MDS, myelodysplastic syndrome; Col, colon; EsCa, esophagus cancer; Vino, vinorelbine; NA, not applicable; HD, Hodgkin disease; HD15, procarbazine, cyclophosphamide, etoposide phosphate, vincristine, bleomycin; Chlor, chlorambucil; Ca, cancer; 5-FU, 5-fluoruracil; HepC, hepatitis C; TuScr, tumor screening; Div, diverticulosis; irr, irritable; A pain, abdominal pain; BrCa, breast cancer.
History of multiple (≥ 3) myelosuppressive chemotherapies before HCT
Day after HCT or last chemotherapy
N indicates normal; Col, infectious colitis; I to IV, pathologic grading of GvHD
Occurrence of graft-versus-host disease (GvHD) any time after HCT, maximal grade; 0 indicates no GvHD; I-IV, grade acute GvHD; Ext, extensive chronic GvHD
Microsatellite PCR using as template DNA extracted from the whole colon section. In these cases, MSI denotes MSI in at least one marker and S no MSI in all markers
ZP3, BAT26, SRY
A gastric biopsy obtained from patient C3 before transplantation showed no MSI. This patient developed squamous cancer of his tongue 3.5 years after allogeneic HCT (see Table 3)
Tissue microdissection and DNA extraction
Colon and tumor biopsies were cut into 5-μm sections and examined after appropriate stainings in the Department of Pathology. Two kinds of molecular analyses were performed. First, whole sections were deparaffinized in 3 changes of xylene for 10 minutes each at 58°C and rinsed sequentially with 100% ethanol, and then DNA was isolated after overnight digestion at 55°C in TE-buffer containing proteinase K (10 mg/mL, 50 mM Tris-HCl solution pH 8.5, 1 mM EDTA pH 8.0), heating at 100°C for 20 minutes, and centrifugation. Second, serial sections were mounted on glass slides coated with polyethylene foil (PALM membrane slides; P.A.L.M. Microlaser Technologies, Benried, Germany) and used for laser-capture microdissection of selected areas. We found in our calibration experiments a microsatellite polymerase chain reaction (PCR) amplification efficacy of nearly 80% when 2 laser-microdissected colon crypts were used as template. Therefore, in colon biopsies 2 randomly chosen colon crypts were laser-capture microdissected by using the PALM LaserMicrobeam System (P.A.L.M.), collected into the lid of a 0.5-mL reaction tube, digested in 20 μL TE-buffer containing proteinase K (20 mg/mL) at 56°C for 12 hours, and after denaturation at 100°C for 10 minutes the lysate was used directly for individual PCR reactions. In tumor biopsies, areas of neoplastic cells and surrounding areas of normal-appearing tissue, as determined by an experienced pathologist, were microdissected and purchased for digestion and PCR analyses. The DNA from blood samples and buccal smears was extracted by using commercial kits (Qiagen QIAamp DNA Micro Kit; Qiagen GmbH, Hilden, Germany). DNA extraction and PCR amplification were separated into designated working areas.
Microsatellite PCR
Microsatellite analysis was performed using 3 tetranucleotide (THO-1, SEE33, D14S120) and 3 mononucleotide (ZP3, BAT26, SRY) markers. These microsatellites are located at different chromosomes and were selected because they are routinely used in our institution for chimerism analyses after allogeneic HCT. SRY is a marker located in the Y chromosome and therefore detectable only in men. The singleplex PCR reactions were performed with 6-FAM-labeled primers, as previously described in detail2 (primers and PCR conditions can be found in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). For each subject and microsatellite marker, each experiment included the following PCR reactions: DNA was isolated from blood obtained from the donor, DNA was isolated from blood obtained from the patient before transplantation (or paired blood or urine in the control groups), DNA was extracted from the whole colon section (or buccal smear), DNA lysate was obtained from 2 laser-microdissected crypts, and negative control (no DNA). After heat denaturation in the presence of Hi-Di formamide (Applied Biosystems, Weiterstadt, Germany) the PCR products together with an internal size standard (GeneScan ROX 500; Applied Biosystems) were loaded onto the ABI PRISM 310 Genetic Analyzer capillary electrophoresis system (Applied Biosystems), and allele analysis was done by the software package supplied with the instrument (GeneScan 310 Analysis), as previously described.2 With this analysis, we have never observed mistyping or formation of heteroduplexes in chimeric blood samples or artificial mixed samples of buccal cells obtained from different individuals. Signals with relative fluorescent units (RFUs) less than 150, blobs (peak area/peak height > 10) and spikes (peak area/peak height < 4.5) were not counted as microsatellite peaks. Stutter peaks and dye-associated peaks were easily identified as such and were excluded from the analysis.2,13 Quantification of MSI was done only in buccal samples and calculated by using the formula: MSI in % = 100 × (peak area of all novel bands) / (peak area of donor bands + recipient bands).
MSI definition
Only banding shifts or gains but not banding intensity changes, which are prone to false-positive results caused by unequal allelic amplification, were counted for MSI. At least 5 successful PCR reactions were performed for each marker to designate it as stable or unstable. Instability of a microsatellite marker in the allograft recipients was defined separately for each epithelial tissue examined (colon, buccal, or tumor) and was determined to have occurred when a band was found in the examined sample (colon, buccal, or tumor) that was not found in either the blood of the donor or the blood of the recipient before transplantation. MSI which was found in either the microdissected colon tissue or in whole colon tissue was enough to judge MSI in the colonic tissue of the allograft recipient. MSI in healthy donors was evaluated by comparing the buccal smear with paired blood or urine. MSI in the chemotherapy-treated patients was evaluated by comparing the buccal smear with paired blood and blood obtained before chemotherapy (not available when buccal sampling was done > 150 days after chemotherapy). MSI in the patients who received an autograft was evaluated by comparing the buccal smear with paired and pretransplantation blood. Colon biopsies from control subjects (chemotherapy treated or those with no relevant medical history) were examined retrospectively, and, because no paired blood samples were available, MSI analysis was based on replications of single-tissue biopsies. In these cases the occurrence of a band that was not detected in the other independent PCR reactions in 1 of 5 successful experiments, or the occurrence of 3 or more bands in a single experiment, was sufficient to determine MSI. Because these tests involve reproducibility issues and given that the stable microsatellite pattern in blood was unknown, we cannot formally exclude a bias against detecting MSI. However, it seems unlikely that MSI leads to an identical pattern of peaks in 5 independent experiments. Thus, a reproducible pattern of peaks in control biopsies would most likely indicate a stable microsatellite phenotype. In addition, colon biopsies from 4 “positive” patients with known hereditary nonpolyposis colorectal carcinoma always showed additional peaks that were indicative of MSI.
Cloning and sequencing
Novel bands found in the microsatellite analysis were excised from the agarose gel with the Qiaquick Gel extraction Kit (Qiagen), according to the manufacturer's procedures. The PCR product was cloned into the pCR2.1-TOPO vector with the use of the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA). The sequencing was performed with an ABI PRISM 3730XC capillary sequencer with the Big Dye terminator cycle sequencing ready reaction kit V 3.1 (Applied Biosystems).
Immunohistochemistry for DNA repair proteins and protein nitrosylation
Immunohistochemical staining of DNA repair proteins and 3-nitrotyrosine was performed on paraffin-embedded colon sections or cytospins of buccal cells by using the ChemMate Detection KIT (DAKO, Hamburg, Germany) and autostainer instruments (DAKO). Briefly, after deparaffinization, epitope retrieval and block of the endogenous alkaline phosphatase activity with levamisole, the tissues were incubated with the primary antibody, the biotinylated secondary antibody and alkaline phosphatase-conjugated streptavidin molecules and than the reaction was visualized by a Fast Red-type chromogen, as indicated in the instructions. The following antihuman antibodies were used: MLH1 (clone G168-15; BD Pharmingen, Heidelberg, Germany), MSH2 (clone FE11; Oncogene Research, San Diego, CA), MSH3 (clone H-300; Santa Cruz Biotechnology, Santa Cruz, CA), XPA (clone FL-273; Santa Cruz Biotechnology), APE1 (clone 2104; Novus Biological, Littleton, CO), 3-nitrotyrosine (clone 06-284; Upstate Inc, Lake Placid, NY). The negative control was incubated with normal serum, which resulted in no signal. Stainings of colon cancer and cholangiocarcinoma biopsies were used as positive controls. Tissue lymphocytes served as internal positive controls. The expression was classified as present or absent with no grading of staining intensity and widespread or limited according to the percentage of 100 evaluated colonic crypts showing immunoreactivity (> 80% versus < 80%).
Statistics
The occurrence of MSI was compared between groups using Fisher exact test. The following parameters were analyzed: age (< 55 years versus 55 years or older at transplantation), sex (male versus female), underlying disease (myeloid = acute myeloid leukemia [AML] or chronic myeloid leukemia [CML] or myelodysplastic syndrome [MDS] versus lymphoid = acute lymphatic leukemia [ALL] or non-Hodgkin lymphoma [NHL] or multiple myeloma [MM]), previous history of intensive chemotherapies before transplantation (< 3 versus ≥ 3), type of preparative regimen (myeloablative = busulfan/cyclophosphamide [BuCy] or TBI/VC versus FBM or 2 Gy/F), history of oral GvHD (yes versus no), stage of colon GvHD (0-I versus II-IV), maximum grade of overall GvHD at any time after HCT (acute GvHD 0-I or limited chronic GvHD versus acute GvHD II-IV or extensive chronic GvHD). The association between time after transplantation and the occurrence of MSI was additionally analyzed with Wilcoxon 2-sample test.
Results
Microsatellite instability in colon biopsies after allogeneic HCT
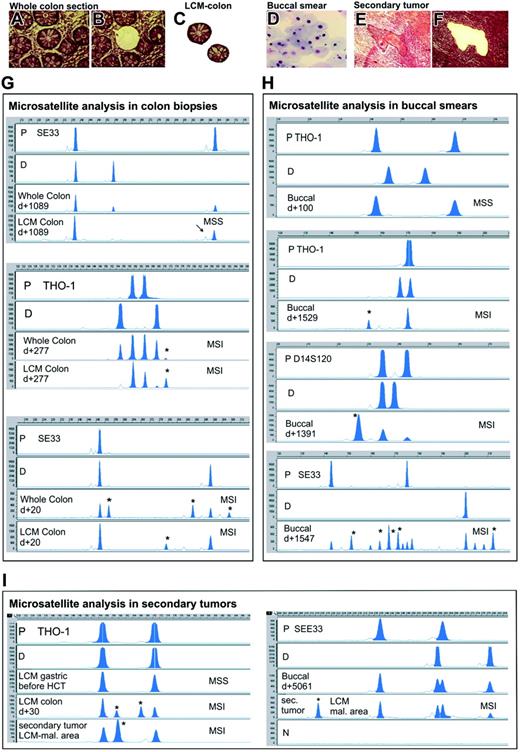
We examined 6 microsatellite loci by PCR in colon biopsies obtained 18 to 1089 days after allogeneic HCT. There were novel bands which were neither host nor donor specific in the laser-microdissected colonic crypts in 12 (75%) of 16 patients, which would indicate the regular occurrence of MSI (Table 1; Figure 1). In one patient in which a gastric biopsy before transplantation was available, stable microsatellites were found. In contrast, in the colon biopsy of this patient obtained 30 days after allografting because of intestinal GvHD, microsatellite alterations at THO-1 were present. Although MSI was detected in isolated colon crypts, it was frequently “masked” when the analysis was made on DNA extracted from the whole colon section (4 patients positive for MSI of 15, 27%). This is probably due to the dilution of the mutated DNA by unmutated DNA derived from recipient cells as well as from infiltrating hematopoietic donor cells (mixed chimerism). In contrast to the patients who received an allograft, we never found any indication of MSI in laser-microdissected crypts, or in whole colon sections from 4 patients after chemotherapy, or from 7 control subjects with no history of either colon malignancy or chemotherapy.
MSI in colon after allogeneic HCT was found only at tetranucleotide markers but not at mononucleotides. The most frequently unstable microsatellite markers were SEE33 (64%) and THO-1 (50%), whereas MSI at D14S120 was found in 8% of the patients. The altered alleles that were found in different crypts isolated from the same colon biopsies had sometimes the same length but most often they differed. The novel bands were smaller, between, or longer than the expected host- and donor-specific alleles. We sequenced the aberrant bands found in the SE33 microsatellite PCR of 2 patients and found them to contain the expected repeated motifs (Figure S1).
Twelve colon biopsies which were obtained from different allograft recipients had histologic signs of GvHD; MSI was detected in all of them except one (Table 1). We also analyzed MSI in 4 other colon biopsies with no GvHD signs (obtained from 4 other allograft recipients) and found MSI in only one of them. There was no statistical association between the underlying disease, the previous history of multiple (≥ 3) intensive chemotherapies before transplantation, the type of preparative regimen for HCT, the overall GvHD grade any time before sampling, or the GvHD stage in colon at sampling and the presence of MSI.
Microsatellite instability in buccal smears after allogeneic HCT
We additionally examined buccal cells for the presence of MSI (Table 2; Figure 1). At the time of buccal sampling (day 1-5061) no signs of mucositis or oral GvHD were present. MSI was found in the buccal smears of 10 (42%) of 24 patients who received an allograft. In contrast, no MSI was detected in the buccal smears obtained from 8 patients after autologous HCT (day 1-4100), from 7 patients after chemotherapy (day 1-930), or from 8 healthy control subjects. To evaluate more precisely, a possible direct effect of chemotherapy on MSI induction we isolated buccal cells (Table 2), blood mononuclear cells, and urine samples (data not shown) at short, frequent intervals after myelosuppressive or high-dose chemotherapy, but microsatellite alterations were never detected.
Microsatellite analysis of buccal mucosa
. | . | . | . | . | . | . | Tetranucleotides . | . | . | Mononucleotides . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCT/chemo and patient ID . | Age, y/sex . | Disease . | History of chemo* . | Preparative regimen/chemo . | D of sampling† . | Overall GvHD max‡ . | THO-1 . | SEE33 . | D14S120 . | BAT26 . | SRY . | ZP3 . | ||||
| Allo | ||||||||||||||||
| B1 | 57/M | ALL | Yes | TBI/VC | –1,1,5§,8,16 | 0 | S | S | S | S | S | S | ||||
| B2 | 34/M | AML | Yes | BuCy | 6,9§,13§,17 | 0 | S | S | S | S | S | S | ||||
| B3 | 66/M | AML | Yes | FBM | 10,13,17 | 0 | S | S | S | S | S | S | ||||
| B4 | 56/M | NHL | Yes | Flu/TT | 20 | 0 | S | S | S | S | S | S | ||||
| B5 | 60/F | AML | Yes | FBM | 34 | 0 | S | S | S | S | S | S | ||||
| B6 | 50/M | AML | No | BuCy | 48 | Illo | S | S | S | S | S | S | ||||
| B7 | 39/M | AML | Yes | BuCy | 84 | 0 | NE | NE | S | S | S | ND | ||||
| B8 | 44/M | AML | Yes | BuCy | 100 | 0 | S | S | S | S | S | S | ||||
| B9 | 44/F | AML | Yes | BuCy | 270 | Lim | S | S | S | ND | NA | ND | ||||
| B10 | 43/F | MDS | No | BuCy | 365 | Lim | S | S | S | S | NA | S | ||||
| B11 | 42/F | CML | No | FBM | 439 | Ext | S | MSI | S | S | NA | S | ||||
| B12 | 20/M | ALL | Yes | TBI/VC | 744 | Ext | S | S | S | S | S | ND | ||||
| B13 | 25/M | MDS | No | FBM | 1063 | 0 | MSI | MSI | ND | ND | ND | ND | ||||
| B14 | 53/F | AML | Yes | BuCy | 1068 | Ext | MSI | MSI | S | S | NA | S | ||||
| B15 | 63/M | NHL | Yes | BuCy | 1188 | Lim | S | S | S | S | S | ND | ||||
| B16 | 66/F | AML | No | FBM | 1234 | Ext | S | MSI | S | S | NA | S | ||||
| B17 | 26/F | AML | Yes | BuCy | 1373 | Ext∥ | S | MSI | S | S | NA | S | ||||
| B18 | 63/F | AML | Yes | FBM | 1391 | Ext | MSI | NE | MSI | S | NA | NE | ||||
| B19 | 39/F | AML | Yes | BuCy | 1510 | Ext∥ | MSI | NE | NE | NE | NA | NE | ||||
| B20 | 36/F | CML | Yes | FBM | 1529 | Ext | MSI | MSI | MSI | S | NA | S | ||||
| B21 | 19/M | ALL | Yes | TBI/VC | 1547 | 0 | MSI | MSI | ND | ND | ND | ND | ||||
| B22 | 43/F | AML | Yes | BuCy | 1821 | Ext∥ | S | MSI | S | S | NA | S | ||||
| B23 | 31/M | MPS | No | BuCy | 1845 | 0 | S | S | S | S | S | S | ||||
| B24 | 30/M | CML | No | TBI/C | 5061 | Ext∥ | S | S | S | S | S | S | ||||
| Auto | ||||||||||||||||
| B25 | 44/F | NHL | Yes | CarTh | –2,1,5§,8,16 | NA | S | S | S | S | S | S | ||||
| B26 | 68/F | MM | Yes | HDMEL | 41 | NA | S | S | S | S | S | S | ||||
| B27 | 58/M | MM | Yes | HDMEL | 150 | NA | S | S | S | S | S | S | ||||
| B28 | 54/M | MM | Yes | HDMEL | 240 | NA | S | S | S | S | S | S | ||||
| B29 | 32/M | NHL | Yes | BEAM | 485 | NA | S | S | S | S | S | S | ||||
| B30 | 57/M | MM | Yes | HDMEL | 540 | NA | S | S | S | S | S | S | ||||
| B31 | 47/M | HD | Yes | BEAM | 1650 | NA | S | S | S | S | S | S | ||||
| B32 | 30/M | Ca | Yes | VIC | 4100 | NA | S | S | S | S | S | S | ||||
| Chemo | ||||||||||||||||
| B33-B39 | 22-44F/M | AML/NHL/Ca | Yes | Various | 1-930 | NA | S | S | S | S | S | S | ||||
| None | ||||||||||||||||
| B39-B46 | 22-44F/M | Healthy | No | NA | NA | NA | S | S | S | S | S | S | ||||
. | . | . | . | . | . | . | Tetranucleotides . | . | . | Mononucleotides . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCT/chemo and patient ID . | Age, y/sex . | Disease . | History of chemo* . | Preparative regimen/chemo . | D of sampling† . | Overall GvHD max‡ . | THO-1 . | SEE33 . | D14S120 . | BAT26 . | SRY . | ZP3 . | ||||
| Allo | ||||||||||||||||
| B1 | 57/M | ALL | Yes | TBI/VC | –1,1,5§,8,16 | 0 | S | S | S | S | S | S | ||||
| B2 | 34/M | AML | Yes | BuCy | 6,9§,13§,17 | 0 | S | S | S | S | S | S | ||||
| B3 | 66/M | AML | Yes | FBM | 10,13,17 | 0 | S | S | S | S | S | S | ||||
| B4 | 56/M | NHL | Yes | Flu/TT | 20 | 0 | S | S | S | S | S | S | ||||
| B5 | 60/F | AML | Yes | FBM | 34 | 0 | S | S | S | S | S | S | ||||
| B6 | 50/M | AML | No | BuCy | 48 | Illo | S | S | S | S | S | S | ||||
| B7 | 39/M | AML | Yes | BuCy | 84 | 0 | NE | NE | S | S | S | ND | ||||
| B8 | 44/M | AML | Yes | BuCy | 100 | 0 | S | S | S | S | S | S | ||||
| B9 | 44/F | AML | Yes | BuCy | 270 | Lim | S | S | S | ND | NA | ND | ||||
| B10 | 43/F | MDS | No | BuCy | 365 | Lim | S | S | S | S | NA | S | ||||
| B11 | 42/F | CML | No | FBM | 439 | Ext | S | MSI | S | S | NA | S | ||||
| B12 | 20/M | ALL | Yes | TBI/VC | 744 | Ext | S | S | S | S | S | ND | ||||
| B13 | 25/M | MDS | No | FBM | 1063 | 0 | MSI | MSI | ND | ND | ND | ND | ||||
| B14 | 53/F | AML | Yes | BuCy | 1068 | Ext | MSI | MSI | S | S | NA | S | ||||
| B15 | 63/M | NHL | Yes | BuCy | 1188 | Lim | S | S | S | S | S | ND | ||||
| B16 | 66/F | AML | No | FBM | 1234 | Ext | S | MSI | S | S | NA | S | ||||
| B17 | 26/F | AML | Yes | BuCy | 1373 | Ext∥ | S | MSI | S | S | NA | S | ||||
| B18 | 63/F | AML | Yes | FBM | 1391 | Ext | MSI | NE | MSI | S | NA | NE | ||||
| B19 | 39/F | AML | Yes | BuCy | 1510 | Ext∥ | MSI | NE | NE | NE | NA | NE | ||||
| B20 | 36/F | CML | Yes | FBM | 1529 | Ext | MSI | MSI | MSI | S | NA | S | ||||
| B21 | 19/M | ALL | Yes | TBI/VC | 1547 | 0 | MSI | MSI | ND | ND | ND | ND | ||||
| B22 | 43/F | AML | Yes | BuCy | 1821 | Ext∥ | S | MSI | S | S | NA | S | ||||
| B23 | 31/M | MPS | No | BuCy | 1845 | 0 | S | S | S | S | S | S | ||||
| B24 | 30/M | CML | No | TBI/C | 5061 | Ext∥ | S | S | S | S | S | S | ||||
| Auto | ||||||||||||||||
| B25 | 44/F | NHL | Yes | CarTh | –2,1,5§,8,16 | NA | S | S | S | S | S | S | ||||
| B26 | 68/F | MM | Yes | HDMEL | 41 | NA | S | S | S | S | S | S | ||||
| B27 | 58/M | MM | Yes | HDMEL | 150 | NA | S | S | S | S | S | S | ||||
| B28 | 54/M | MM | Yes | HDMEL | 240 | NA | S | S | S | S | S | S | ||||
| B29 | 32/M | NHL | Yes | BEAM | 485 | NA | S | S | S | S | S | S | ||||
| B30 | 57/M | MM | Yes | HDMEL | 540 | NA | S | S | S | S | S | S | ||||
| B31 | 47/M | HD | Yes | BEAM | 1650 | NA | S | S | S | S | S | S | ||||
| B32 | 30/M | Ca | Yes | VIC | 4100 | NA | S | S | S | S | S | S | ||||
| Chemo | ||||||||||||||||
| B33-B39 | 22-44F/M | AML/NHL/Ca | Yes | Various | 1-930 | NA | S | S | S | S | S | S | ||||
| None | ||||||||||||||||
| B39-B46 | 22-44F/M | Healthy | No | NA | NA | NA | S | S | S | S | S | S | ||||
At the time of buccal sampling no signs of mucositis or oral GvHD were present (except where noted). At the time of buccal sampling all patients revealed complete donor chimerism in blood (blood chemerism analysis in patients B1, B2, and B3 was done at day 30).
Chemo indicates chemotherapy; HCT, hematopoietic cell transplantation; GvHD, graft-versus-host disease; ALL, acute lymphatic leukemia; Allo, allogeneic HCT; TBI/VC, total body irradiation (12 Gy), etoposide phosphate, cyclophosphamide; S, stable microsatellites; AML, acute myeloid leukemia; BuCy, busulphan, cyclophosphamide; FBM, fludarabine, carmustine, melphalan (reduced intensity conditioning); NHL, non-Hodgkin lymphoma; Flu/TT, fludarabine, thiotepa (reduced intensity conditioning); NE, not evaluable; ND, not done; NA, not applicable; MDS, myelodysplastic syndrome; CML, chronic myeloid leukemia; MSI, microsatellite instability; MPS, myeloproliferative syndrome; Auto, autologous HCT; CarTh, carmustine, thiotepa; MM, multiple myeloma; HDMEL, high-dose melphalan; BEAM, carmustine, cytosine arabinoside, etoposide phosphate, melphalan; HD, Hodgkin disease; Ca, cancer; VIC, etoposide phosphate, carboplatin, ifosfamide.
History of multiple (≥ 3) myelosuppressive chemotherapies before HCT
Day after HCT or last chemotherapy
Occurrence of graft-versus-host disease (GvHD) any time after HCT, maximal grade; 0 indicates no GvHD; I-IV, grade acute GvHD; Lim, limited chronic GvHD; Ext, extensive chronic GvHD
At the time of buccal sampling, signs of mucositis or oral GvHD were present
These subjects had a history of oral GvHD. Subjects B16 and B24 developed secondary squamous cancers (see Table 3)
There was no significant association between MSI in the buccal mucosa and patient age, sex, disease, type of conditioning, history of oral GvHD, or overall GvHD that occurred at any point before buccal sampling. Interestingly, we found a significant association between the time of buccal sampling after transplantation and the presence of MSI (P = .008, Wilcoxon 2-sample test). Buccal smears obtained within 1 year after allografting showed no evidence of MSI, whereas 71% of buccal samples obtained at later time points were MSI positive (P < .001, Fisher exact test).
Similar to the results seen with isolated colon crypts, MSI in buccal smears was evident only at tetranucleotide markers (SEE33, 35%; THO-1, 26%; D14S120, 10%). Both smaller and longer bands than the expected alleles were found. The calculated percentage of altered DNA in relation to the donor and recipient signals was 44% (range, 17%-210%). The median number of novel bands in the MSI+ buccal samples was 2 (range, 1-10). We made the assumption that in cases where only one novel band was found, the mutated DNA present in the sample should reflect the corresponding content of mutated cells. With this hypothesis in mind we found a median of 29% mutated cells from a total of 6 evaluable buccal samples, with some cases showing a predominance of altered DNA/cells compared with nonmutated DNA/cells (range, 17%-145%).
Microsatellite instability in secondary tumors after allogeneic HCT
We identified 3 patients who received an allograft who developed secondary invasive squamous cell cancer. In all cases, both laser-microdissected tumor areas and surrounding normal epithelium were MSI positive at tetranucleotide markers (Table 3; Figure 1). The novel THO-1 alleles in the tumor of patient C3 had the same length as the novel bands found in his colon biopsy obtained 3.5 years earlier. The SEE33 frameshifts in the cancer cells of patient B16 were found to be different from those in his buccal smear sampled approximately 1.5 years after tumor excision.
Microsatellite analysis of secondary epithelial cancers
. | . | . | Tetranucleotides . | . | . | . | . | . | Mononucleotides . | . | . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | THO-1 . | . | SEE33 . | . | D14S120 . | . | BAT26 . | . | SRY . | . | ZP3 . | . | ||||||||||
| Patient ID . | Time after HCT, y . | Pathologic diagnosis . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | ||||||||||
| C3 | 3.5 | Squamous Ca, tongue | MSI | MSI | NE | NE | MSI | NE | S | S | S | S | S | S | ||||||||||
| B16 | 2 | Squamous Ca, lip | MSI | S | MSI | MSI | MSI | NE | S | S | NA | NA | NE | NE | ||||||||||
| B24 | 10 | Squamous Ca, lip | MSI | MSI | MSI | NE | S | S | S | S | S | S | S | S | ||||||||||
. | . | . | Tetranucleotides . | . | . | . | . | . | Mononucleotides . | . | . | . | . | . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | THO-1 . | . | SEE33 . | . | D14S120 . | . | BAT26 . | . | SRY . | . | ZP3 . | . | ||||||||||
| Patient ID . | Time after HCT, y . | Pathologic diagnosis . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | Tumor . | Normal epith . | ||||||||||
| C3 | 3.5 | Squamous Ca, tongue | MSI | MSI | NE | NE | MSI | NE | S | S | S | S | S | S | ||||||||||
| B16 | 2 | Squamous Ca, lip | MSI | S | MSI | MSI | MSI | NE | S | S | NA | NA | NE | NE | ||||||||||
| B24 | 10 | Squamous Ca, lip | MSI | MSI | MSI | NE | S | S | S | S | S | S | S | S | ||||||||||
Clinical data of the patients are shown in Tables 1 and 2. Tumor indicates that laser-microdissected tumor area was analyzed; noral epith, that laser-microdissected nonmalignant epithelium adjacent to the tumor area was analyzed.
HCT indicates hematopoietic cell transplantation; Ca, cancer; MSI, microsatellite instability; NE, not evaluable; S, stable microsatellites; NA, not applicable.
Immunocytochemistry for DNA repair proteins and 3-nitrotyrosine
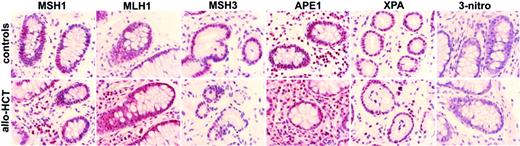
On the basis of the known relation between genomic instability and defects in the DNA repair system we evaluated the expression of DNA repair proteins. Colon biopsies from 4 allograft recipients who had intestinal GvHD and from 3 subjects who did not receive a transplant with no relevant medical history showed comparable expression for MLH1, MSH2, MSH3 (MMR proteins), APE-1 (BER protein), and XPA (NER protein) (Figure 2). Nitric oxide (NO) has been associated with GvH reactions after allogeneic HCT and has been shown to inhibit DNA repair proteins through nitrosylation. None of the colon specimens showed immunoreactivity for 3-nitrotyrosine, a reaction product of NO-derived ONOO2 with tyrosine residues (Figure 2). No expression of DNA repair proteins or 3-nitrotyrosine was detected in buccal cells consisting of nonproliferating peeled-off cells.
Discussion
We have performed microsatellite analysis in nonmalignant epithelial tissues and found frequent microsatellite alterations in the colon and oral mucosa of patients who underwent allogeneic HCT (Table 4). We have used cautious methods and controls to ensure the reliability of our findings. To ensure a precise estimation of the number and size of microsatellite alleles we applied denaturing capillary electrophoresis instead of gel electrophoresis. Only band shifts or gains, but not band intensity changes which are prone to false-positive results, were counted for MSI.14 Technical artifacts like stutter peaks, dye-associated peaks, blobs, and spikes, when present, were easily identified and excluded.13,14 Furthermore, sequencing in 2 cases confirmed that the detected novel bands represent actual microsatellite alleles with altered numbers of repeats and not unspecific amplification products generated by chance. Heteroduplexes between donor and recipient DNA could explain large aberrant bands but not the smaller ones found here. Contamination with exogenous DNA during the PCR reaction through the handling of the samples, and artifacts related to the processing of the samples (formalin-fixed material, laser microdissection, fluorescent compounds in the material) can be excluded as the cause of the aberrant bands found in the allograft recipients. First, contamination of the PCR reagents was always checked for by use of negative controls. Second and most importantly, examination of similar tissues from both patients who did not receive a transplant and patients who received an autograft performed under exactly the same conditions did not result in any aberrant bands.
Microsatellite instability in nonmalignant epithelial tissues (summary)
. | MSI+ in colon . | . | . | MSI+ in buccal mucosa . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group of patients . | No. . | D . | Patients with MSI+, % . | No. . | D . | Patients with MSI+, % . | ||||
| Allogeneic | 16 | 18-1089 | 75 | 24 | 1-5061 | 42 | ||||
| Autologous | ND | ND | ND | 8 | 1-4100 | 0 | ||||
| Chemotherapy | 4 | 7-1805 | 0 | 7 | 1-930 | 0 | ||||
| Healthy | 8 | NA | 0 | 8 | NA | 0 | ||||
. | MSI+ in colon . | . | . | MSI+ in buccal mucosa . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group of patients . | No. . | D . | Patients with MSI+, % . | No. . | D . | Patients with MSI+, % . | ||||
| Allogeneic | 16 | 18-1089 | 75 | 24 | 1-5061 | 42 | ||||
| Autologous | ND | ND | ND | 8 | 1-4100 | 0 | ||||
| Chemotherapy | 4 | 7-1805 | 0 | 7 | 1-930 | 0 | ||||
| Healthy | 8 | NA | 0 | 8 | NA | 0 | ||||
MSI+ indicates microsatellite instability positive; No., number of patients analyzed; D, day of tissue sampling after transplantation or chemotherapy; NA, not applicable; ND, not done. For explanations of groups, see Table 1.
Microsatellite instability in colon epithelium, buccal mucosa, and secondary tumors after allogeneic HCT. Top panels show representative photographs of tissues that were used for microsatellite analysis. An entire colon section before (A) and after (B) laser-capture microdissection of a crypt, 2 laser-microdissected colon crypts (C), a buccal smear (D), and a secondary squamous cell cancer before (E) and after (F) laser microdissection of a malignant area are shown (hematoxylin/eosin stain). Images were visualized using a Zeiss Axiovert 135 microscope and a C-Apochromat 40 ×/1.2 numeric aperture objective (Carl Zeiss, Jena, Germany). Images were acquired using a Zeiss AxioCam MR and Axioplan 2 imaging software. Images were processed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA). Panels G, H, and I show representative electropherograms of PCR microsatellite analysis in colon biopsies, buccal smears, and/or secondary tumors. MSS indicates microsatellite stable; MSI, microsatellite instability; P, patient-specific signals as determined from peripheral blood obtained before transplantation; D, donor-specific alleles; Whole Colon, bands found in the examination of a whole colon section; LCM Colon, bands found in examination of 2 laser-captured microdissected colon crypts; Buccal, bands found in the examination of buccal smears; sec. tumor and LCM mal. area, bands found in the examination of a laser-capture microdissected malignant area of a secondary tumor biopsy; N, negative control (no DNA); d+, day of sampling after transplantation; y-axis, relative fluorescent units; and x-axis, fragment length (base pairs). The analyzed microsatellite marker in each case is given. The asterisks indicate novel bands indicative for MSI. The arrow shows a representative stutter peak.
Microsatellite instability in colon epithelium, buccal mucosa, and secondary tumors after allogeneic HCT. Top panels show representative photographs of tissues that were used for microsatellite analysis. An entire colon section before (A) and after (B) laser-capture microdissection of a crypt, 2 laser-microdissected colon crypts (C), a buccal smear (D), and a secondary squamous cell cancer before (E) and after (F) laser microdissection of a malignant area are shown (hematoxylin/eosin stain). Images were visualized using a Zeiss Axiovert 135 microscope and a C-Apochromat 40 ×/1.2 numeric aperture objective (Carl Zeiss, Jena, Germany). Images were acquired using a Zeiss AxioCam MR and Axioplan 2 imaging software. Images were processed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA). Panels G, H, and I show representative electropherograms of PCR microsatellite analysis in colon biopsies, buccal smears, and/or secondary tumors. MSS indicates microsatellite stable; MSI, microsatellite instability; P, patient-specific signals as determined from peripheral blood obtained before transplantation; D, donor-specific alleles; Whole Colon, bands found in the examination of a whole colon section; LCM Colon, bands found in examination of 2 laser-captured microdissected colon crypts; Buccal, bands found in the examination of buccal smears; sec. tumor and LCM mal. area, bands found in the examination of a laser-capture microdissected malignant area of a secondary tumor biopsy; N, negative control (no DNA); d+, day of sampling after transplantation; y-axis, relative fluorescent units; and x-axis, fragment length (base pairs). The analyzed microsatellite marker in each case is given. The asterisks indicate novel bands indicative for MSI. The arrow shows a representative stutter peak.
We are aware of only one study in which microsatellite analysis was performed in epithelial tissues obtained after allogeneic HCT. Endler et al15 examined mouthwashes of allograft recipients and found a surprisingly frequent chimeric pattern, even after magnetic separation of epithelial cells. Furthermore, additional bands which were of neither host nor donor origin were found by gel electrophoresis and were explained by the researchers as being heteroduplexes generated during PCR between donor and recipient DNA.
What does the MSI found in non-neoplastic tissues after transplantation point out? Replication errors during DNA synthesis occur with a certain probability in all cells and may result in a change of the length of microsatellite loci, which are sequences particularly prone to DNA polymerase slippage because of their repetitive nature.1,16 The MMR system recognizes and corrects these genomic alterations.16 Whenever the DNA error cannot be repaired, apoptosis pathways are activated.17 Thus, cells with microsatellite alterations which escaped repair will not replicate and therefore should not become apparent among the large excess of the surrounding normal cells.18 However, if deficient DNA repair is coupled with a failure to elicit an apoptotic response, this association may result in a growth advantage sufficient to generate a detectable clonal population of cells that share genetic alterations. Therefore, the detection of MSI in a clinical sample, as are the ones in our study, demonstrates the presence of populations which were first induced to change the length of their microsatellites, then escaped DNA polymerase proofreading and finally selected for clonal outgrowth.1,3,17,18
Why does MSI occur? We found MSI in non-neoplastic epithelial tissues obtained from patients treated with myeloablative or reduced-intensity chemotherapy and allogeneic HCT but not from patients after myelosuppressive chemotherapy or myeloablative chemotherapy combined with autologous HCT. In addition, MSI in buccal cells was not evident at short intervals after myelosuppressive or myeloablative chemotherapy but was found many years after conditioning for HCT. Therefore, although chemotherapeutic agents alone may induce microsatellite alterations in individual cells in vitro,19 it is very likely that an “alloantigenic” effect is the driving force in producing detectable MSI in the patients who received an allograft. Additional work is needed to characterize in detail the nature of the alloantigenic stimulation that is operative in MSI generation. The observation that all colon biopsies with histologically detectable GvHD but one were MSI+, indicates a relation between organ GvHD and MSI. However, no GvHD signs were evident in one MSI+ colon biopsy or in the oral mucosa of any of the 10 subjects with MSI+ buccal smears. The time of sampling when GvH reactions have just appeared or already been censored or the presence of only subclinical GvH signs are only some of the reasons that could explain this lack of correlation. Analysis of further tissues which, unlike the colonic and the buccal mucosa, are not typical targets of GvHD might be informative in this regard. Interestingly, MSI in buccal smears was found significantly more frequently at later time points after transplantation, which would suggest that the duration of alloantigenic reactions might be of importance in generation of MSI.
How does MSI occur? Studies in chronic inflammatory diseases postulate that oxidative stress may lead to accumulation of genomic alterations either by overwhelming the capacity for DNA repair or by directly inactivating DNA repair pathways through epigenetic gene silencing or through damaging of DNA repair proteins.8,10,20 Although the MMR system is responsible for correcting DNA strand loops typical for MSI, studies suggest that the different DNA repair systems do not act independently and that MSI may be attributable to alterations in DNA repair pathways distinct from MMR.6 Immunostaining of colon sections after allogeneic HCT for key proteins of 3 different DNA repair systems (MMR, BER, NER) showed widespread expression comparable with the one found in colon sections from healthy individuals who did not receive a transplant. Therefore, silencing of DNA repair genes is very unlikely to be the underlying mechanism explaining the MSI found here, although components further downstream of the repair systems (eg, polymerases) should also be evaluated.7 NO produced by cytokine-stimulated (eg, TNF-α) epithelial cells has been found to participate in animal GvHD and enhanced plasma NO levels have been associated with GvHD severity in humans.21,22 NO has been found to inhibit DNA repair through protein nitrosylation and inactivation.9 Although we did not find increased levels of nitrosylated proteins in the colon biopsies after allogeneic transplantation, we cannot exclude that NO is a link between chronic alloantigenic stimulation, relaxation of DNA repair mechanisms, and MSI.
Alterations in noncoding microsatellites are unlikely to provide any growth advantage, ultimately leading to clonal outgrowth of the microsatellite-altered cells, which is required to detect MSI. How these clonal cells with novel repeat lengths are selected is unclear. Inactivation of p53 and the failure of cellular demise has been suggested as playing a mechanistic role in the occurrence of MSI in sporadic tumors and in chronically inflamed synovial tissue.3,5,17,23 Another hypothesis requiring further study is the role of the immune system in the selection of clones with microsatellite alterations. In hereditary nonpolyposis colorectal carcinoma, it has been suggested that cells with frameshift mutations avoid elimination by the immune system through abolishing expression of MHC proteins.24 If this is so, the donor-derived immune system in an allogeneic setting might attack the recipient-derived, normal epithelial cells, yet spare cells with an MSI phenotype, providing in this way a selective growth advantage for the cells with novel repeat lengths. This is clearly an area that requires additional investigation.
Immunohistochemistry for DNA repair proteins and 3-nitrotyrosine in colon biopsies. Expression of MLH1, MSH2, MSH3, XPA, and APE1 was similar in the colon biopsies obtained from subjects who did not receive a transplant with no relevant medical history (top) and from patients who received an allograft (bottom). There was no immunostaining for 3-nitrotyrosine in either group. Representative photographs are shown (× 600). Image acquisition performed as described for Figure 1.
Immunohistochemistry for DNA repair proteins and 3-nitrotyrosine in colon biopsies. Expression of MLH1, MSH2, MSH3, XPA, and APE1 was similar in the colon biopsies obtained from subjects who did not receive a transplant with no relevant medical history (top) and from patients who received an allograft (bottom). There was no immunostaining for 3-nitrotyrosine in either group. Representative photographs are shown (× 600). Image acquisition performed as described for Figure 1.
What does the MSI found in non-neoplastic epithelial tissues after allogeneic HCT mean? MSI has been mainly associated with cancer3 or cancer risk in chronic inflammatory diseases.4 Classical MSI in hereditary nonpolyposis colorectal carcinoma caused by inherited defects in MMR genes is characterized by mutations especially at mononucleotide and dinucleotide repeats, leading to the typical “ladder” effect in PCR microsatellite analysis. Interestingly, the form of MSI found in our study, namely tetranucleotide alterations detected as single aberrant sequences, is similar to the one frequently seen in sporadic squamous cancers with intact MMR systems and termed EMAST.3,23,25 Furthermore, squamous cell carcinomas are the most common secondary tumors after allogeneic HCT.12 However, our findings do not show an association between MSI phenotype and the risk of posttransplantation malignancy. First, the percentage of subjects with MSI phenotype we found is much higher than the expected incidence of posttransplantation malignancies. Second, MSI was frequently found in colon, but colon carcinoma is a very uncommon secondary malignancy. Third, the frameshift alterations found in posttransplantation tumors differed from those in the associated nonmalignant tissues. Despite this lack of correlation, it is quite possible that multiple MSI lesions could arise in a single subject, few, if any, of which ever accumulate the additional genetic abnormalities required to progress to a malignancy. Impaired DNA repair may indeed be the link between the MSI phenotype and pathogenesis of posttransplantation solid tumors.
MSI has been shown to be an indicator of genomic instability.1 It is very likely that mutations occur not only in nontranscribed microsatellites but also in coding regions of the genome.1 Mutations in coding genes might be responsible for the protean posttransplantation clinical syndromes and phenotypes such as scleroderma, Sjögren syndrome, musculoskeletal or pulmonary disease, and many others. In addition to serving as an indicator for genomic instability, mutations of microsatellites may directly contribute to evolution of posttransplantation diseases. A number of transcribed genes have been found to contain microsatellites within their coding regions, and changes in the lengths of these repetitive sequences have been shown to result in gene inactivation or modification of function.1,26 Examination of a larger number of patients, refinement of the selection of microsatellite markers, and correlation with clinical data may reveal important clinical associations.
In summary, we provide evidence of genomic alterations measured by MSI in non-neoplastic epithelial tissues of patients who underwent allogeneic HCT. When compared with data from control groups these results suggest that an “allogeneic” effect is substantially involved in the mutation process. Genome analyses in allograft recipients may indeed identify specific genomic alterations which might be responsible for or used as molecular biomarkers of posttransplantation diseases, including secondary cancer. Elucidating the ultimate mechanisms underlying the genomic instability following allogeneic HCT may prove to be of major therapeutic value.
Prepublished online as Blood First Edition Paper, December 20, 2005; DOI 10.1182/blood-2005-08-3431.
Supported by a grant from the Landesstiftung Baden-Württemberg (A.S., J.F.) and by the Beca Jorge Oster Fundacion Bunge y Born (M.W.).
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Prof R. Mertelsmann for supporting this study and to Dr M. Engelhardt for her support with patient material, helpful discussions, and contributions to the manuscript. We thank Prof N.C. Zoumbos (Patras University) and Dr R. Wäsch for critical discussions. We thank S. Krüger, Y. Metaxas, B. Weinhold, G. Theodorou, and A. Gaa for excellent laboratory assistance; Dr G. Ihorst for help in statistical analysis; and Dr Marie Follo for linguistic corrections.
Supplemental data
Sequence of a novel band found in the microsatellite analysis (SEE33 marker) of a colon biopsy obtained 57 days after allogeneic HCT (patient C8, Table 1). Sequencing of the novel band was performed as described in the Materials and Methods. The novel band is consisted of AAAG repeats. The sequence of the novel band was compared with the SEE33 GenBank Sequence (Accession V00481) given in the NCBI-sequence data base (http://www.ncbi.nlm.nih.gov).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal