Abstract
Integrated pathways are believed to determine hematopoietic cell fate and/or neoplastic transformation. Notch signaling has been shown to regulate T-cell differentiation and leukemogenesis. However, specific target genes and molecular partners are not fully elucidated. We show that Notch3 activation sustains aberrant SCL/Tal1 overexpression and phosphorylation in mature thymocytes. Furthermore, we define the role of SCL/Tal1 as a component of an activator complex, including phosphorylated Tal1 and Sp1, that specifically enhances cyclin D1 expression and demonstrate that Tal1/Sp1 specifically co-occupy the D1 promoter in vivo, only in the presence of pre-T-cell receptor (TCR). We therefore conclude not only that cyclin D1 is a target of the Tal1/Sp1 complex, but also that Notch3-dependent activation of pre-TCR/ERK signaling regulates SCL/Tal1 function.
Introduction
Signaling through the Notch family of transmembrane receptors is a crucial regulator of diverse developmental processes, including T-cell differentiation.1-3 Four Notch proteins (Notch1, Notch2, Notch3, and Notch4) have been described in mammalians cells. The 4 proteins are encoded by separate genes, but show substantial amino acid similarity and are expressed in an overlapping fashion in many cell types. Once activated by Delta and Jagged family ligand engagement, proteolytic cleavage events release the intracellular domain of Notch (Notch-IC), which translocates to the nucleus and binds their associated protein CBF1.1-3 This binding leads to derepression of the CBF1 transcription factor and allows induction of Notch-target genes. Consistent with its growth-promoting functions, abnormal Notch signaling is believed to be a driving force in several human cancers. Indeed, chromosomal abnormalities involving Notch genes and overexpression of Notch proteins have been observed in many malignancies. As a result of a chromosomal translocation, t(7;9) (q34;q34.3), NOTCH1 gene is rearranged, and the activated protein is expressed in a rare subset of T-cell acute lymphoblastic leukemia.4 This initial observation was corroborated by several reports showing that mice expressing constitutively active truncated Notch-IC proteins develop T-cell leukemias, confirming the causative nature of activation of these proteins in disease development.5-8 More recently, it has been reported that virtually 100% of human T-cell acute lymphoblastic leukemia (T-ALL), including tumors from all major molecular oncogenic and immunophenotypic subtypes, overexpress Notch3,9 whereas greater than 50% have activating mutations of Notch1.10
However, the molecular mechanisms by which aberrant Notch signaling causes T-cell leukemia are not fully understood. It is well known that T-cell development is a complex and multistep process that is critically dependent on combinatorial interaction among various effectors. It has been shown that Notch genes can collaborate with c-Myc, E2A-PBX1,11,12 and we have previously reported that Notch3 can down-regulate E2A to induce T-cell leukemia.13 Many studies over the past decade have implicated the overexpression of transcription factors, otherwise involved in normal T-cell differentiation, including SCL/Tal1, Hox11, and LMO2 via chromosomal rearrangements, in the pathogenesis of T-ALL.14 However, SCL/Tal1 as well Hox11 and LMO2 are frequently overexpressed in T-ALL samples in the absence of detectable cytogenetic abnormalities involving the chromosomal regions that contain these genes. These observations, coupled with the demonstration of biallelic expression of SCL/Tal1 and hypomethylation of Hox11 promoter sequences in T-ALL cases, support the hypothesis that mechanisms other than cis-acting chromosomal rearrangements can contribute to the aberrant expression of oncogenic transcription factors in T-ALL.15
Abnormal expression of SCL/Tal1 occurs in the majority of pediatric cases of T-ALL.15 More recently it has been shown that Tal1 contributes to leukemogenesis by repressing expression of the gene involved in T-cell differentiation and inducing a differentiation arrest in developing thymocytes.16 SCL/Tal1 heterodimerizes with class I bHLH proteins, including E12, E47, HEB, and E2-2, and may contribute to leukemia by interfering with E protein function. Consistent with the differentiation arrest, gene expression profile of premalignant SCL/Tal1 transgenic thymocytes reveals repression of several genes important for thymocytes differentiation, such as RAG2 and pre-Tα.16 Another potential E47 target gene is the cyclin-dependent kinase inhibitor p21CIP1/WAF1.17 However, no differences in p21CIP1/WAF1 expression levels were observed in tumors isolated from Tal1 transgenic mice.18 Thus, it is still uncertain whether Tal1 exerts its leukemogenic effects by interfering with regulatory programs involved in cellular proliferation and/or differentiation during normal thymocyte development. In the present study, we tested the possible role of Tal1 in an animal model of T-ALL,19 by studying its functional interaction with Notch3 and pre-T-cell receptor (TCR). We found that the SCL/Tal1 gene is a Notch3 target whose function is critically required for the proliferative boost deriving from Notch3-enforced expression. A novel Notch3/Tal1 target, namely cyclin D1, was also identified. We present a model that suggests how the combined signals generated from Notch3 and pre-TCR pathways may regulate SCL/Tal1 function and, in turn, T-cell proliferative response.
Materials and methods
Cell transfection
Dual-Luciferase/Renella Reporter Assay System (Promega, Madison, WI) reagent was used in accordance with the manufacturer's instructions. Expression vector for Notch3-IC was previously described.8 D1 and Tal1 promoter-luciferase reporter plasmids were generated using the following primers: forward, 5′-GCGGGTACCAGGAGCCTATCGTGTCTCAACC-3′, and reverse, 5′-GCGCTCGAGTCTGTAGCTCTCTGCTACTGC-3′ for cyclin D1; and F-5′-CTCTTCTGCTTCAAGGCAGG-3′ and R-GGCCGTTGTCTAGCCTCTTA-3′ for Tal1, and fragments of 1000 base pair (bp) and 1196 bp, respectively, were amplified from mouse genomic DNA. The amplified inserts were ligated in pGL3-Luciferase reporter plasmid. In the Luciferase experiments, pcDNA3 vector was used as an empty control vector and added for each sample to ensure an equal amount of total DNA. Tal1 cDNA was kindly provided by Prof G. Condorelli, University La Sapienza, Rome, Italy.
Flow cytometry and cell sorting
Thymocytes were prepared as single-cell suspensions, stained with labeled antibodies (Abs), and analyzed by flow cytometry. The analyses were performed using a FACScalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The CD8+ cells, including CD8 single-positive and CD8+CD4+ double-positive subsets, were positively selected by magnetic cell separation by using the MACS (magnetic cell sorter) system (Milteny Biotec, Auburn, CA) in accordance with the manufacturer's instructions. The purity of the isolated fractions was greater than 90%, as determined by flow cytometry analysis (Becton Dickinson). Anti-CD4-FITC, anti-CD8-PE, anti-CD122-FITC, and anti-CD45R/B220-PE Abs were purchased from Pharmingen (San Diego, CA).
N3-232T-cell cultures with IL-2 and anti-CD25 blocking antibodies
232N cells were cultured at 106 cells/wells in 12-well plates (Corning Costar, High Wycombe, United Kingdom). Anti-CD25 (IL-2 receptor α chain, p55) clone PC61 (Pharmingen) was added to the cultures 2 hours before the addition of IL-2 (100 U). Cells were collected 24 hours after IL-2 treatment.
Mice
Immunoblotting
A protocol for this procedure has been described.13 Blots were incubated with anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA), cyclin D1 (Santa Cruz Biotechnology), tubulin (Santa Cruz Biotechnology), ERK (Santa Cruz Biotechnology), and Sp1(Santa Cruz Biotechnology) for 2 hours at room temperature or with an anti-phospho-ERK (Cell Signaling Technology, Beverly, MA), anti-phospho-Tal1 (Santa Cruz Biotechnology), and anti-Tal1 (Santa Cruz Biotechnology) overnight at 4°C according to Cell Signaling Technology's instruction for detection of phosphor-ERK. The relative levels of pTal1 were quantified using Quantity-one software (Bio-Rad, Hercules, CA). Values were normalized to total Tal1 expression and represented as relative levels with respect to control cell population.
RNA analysis and RT-PCR
Total RNA was isolated from cell suspensions, in guanidine isothiocyanate and further processed for reverse transcriptase-polymerase chain reaction (RT-PCR), as described.13 Each sample was analyzed in 2 serial dilutions (1:10, 1:100) and at least in 2 independent experiments. PCR was performed at the opportune annealing temperature with the following primers: β-actin-f, 5′-GTGGGCCGCTCTAGGCACCAA-3′, and β-actin-r, CTCTTTGATGTCACGCACGATTTC; D1 F-5′-CGTGCAGAAGGAGATTGTGC-3′ and R-GTCTGCTTGTTCTCATCCGC-3′. The relative levels of RNA were quantified using Bio-Rad Quantity-one software. Values were normalized to β-actin expression and represented as relative levels with respect to control cell population. All RNAs were tested in triplicate samples and at least in 2 independent experiments.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) assays were performed with the Assay System (Upstate, Charlottesville, VA) reagent, and it was used in accordance with the manufacturer's instructions. An aliquot of the extracts was kept for isolation of input DNA, while samples were precleared and then incubated overnight at 4°C. PCR reactions were performed for 30 cycles using the primers, F1-AGGAGCCTATCGTGTCTCAACC-3′ and R1-CATCTATTCCTCCTCGCTGG-3′, 165 bp; F2-5′-CATCTTGAGCTGTTGCTGGA-3′ and R2-5′-AGCGTCCCTGTCTTCTTTCA-3′, 227 bp; F3-5′-AGAACAGGGTGTCCTTGCAC-3′ and R3-5′-CGGACTGCTTCTCTCCAAAC-3′, 200 bp; F4-GCATATCTACGAAGGCTGAGG-3′ and R4-5′-TCTGTAGCTCTCTGCTACTGCG-3′, 240 bp. All experiments described in this study were approved by the institutional review board of the Department of Experimental Medicine and Pathology of the University “La Sapienza,” Rome, and were conducted following the regulations established by the Italian animal care laws (D.L. 116, January 27, 1992).
Results
Tal1 is a target of Notch3 signaling
Both aberrant Tal1 and Notch3 expression have been demonstrated to play a role in T-cell leukemia development, confirming the causative nature of deregulation of these proteins in leukemogenesis.14,19 We previously demonstrated that Notch3 inhibits E2A function and promotes T-cell leukemia in a pre-TCR-dependent manner.9 To determine whether this function might involve the E2A inhibitor Tal1 protein, we monitored the Tal1 expression in lymphoma cells and thymocytes of Notch3-IC transgenic mice. As previously described,8,9,13 Notch3-IC Tg lymphomas display a significant proportion of CD4+CD8+ double-positive (DP) and CD8+ immature single-positive (SP) cells. The protein extracts and mRNA preparations were therefore obtained from positively selected CD8+ cells from wild-type or Notch3 tg thymocytes or splenic lymphoma cells, to monitor Tal1 expression by Western blot (WB) and RT-PCR, respectively. We found that lymphoma cells derived from Notch3-IC transgenic mice expressed high levels of Tal1 protein, and the increased protein expression was consistent with an increased mRNA expression, (Figure 1A-B). Previously published data suggested that the amino-terminal transactivation domain of Tal1 is positively regulated by S122 phosphorylation and that the functional properties of Tal1 can be influenced by signal transduction pathways that involve the MAP kinases,20 being Tal1 phosphorylated on serine residue 122 by the mitogen-activated protein (MAP) kinase ERK1.21 We previously reported that Notch3 induces activation of ERK1/2, a known downstream effector of the pre-TCR signaling.9,13 Thus, we assessed whether Notch3 activation could induce Tal1 phosphorylation. CD8+ lymphocytes from WT mice and from lymphomas of Notch3-IC transgenic mice were isolated, and Tal1 phosphorylation was assessed with a specific antibody against S122-phoshorylated Tal1. As shown in Figure 1A (middle), enforced Notch3-IC expression induced Tal1 phosphorylation. To further investigate the effect of activated Notch3 signaling on Tal1 expression and phosphorylation, we analyzed Tal1 expression in Notch3 transgenic thymocytes at 4 weeks of age when T-cell malignancy is not yet evident.8,9 Our analysis revealed that in CD8+/DP thymocytes, in which Tal1 expression is normally down-regulated, enforced Notch3-IC expression induced the increase of Tal1 protein, and in its phosphorylated form, when compared with the WT cells (Figure 1C). It is well established that Notch-driven leukemogenesis requires pre-TCR signaling, because mice lacking pre-TCR components are resistant to Notch-induced leukemia.8,9 Moreover, we previously demonstrated that the presence of pre-TCR is required to trigger the ERK/MAPK pathway, because ablation of pTα-/- in the Notch3-IC/pTα-/- double-transgenic mice strongly reduces Notch-mediated ERK1/2 activation.13 Thus, we verified whether an intact pre-TCR signaling contributed to the regulation of Tal1 expression and phosphorylation. Intriguingly, we found that Tal1-increased expression was also maintained in the Notch3-IC/pTα-/- double-transgenic mice, with respect to wild-type thymocytes (Figure 1C, top). In contrast, Notch3-mediated induction of phospho-Tal1 is strictly associated with the presence of a functional pre-TCR signaling (Figure 1C, bottom). Overall, these data show that, although Notch3 enhances Tal1 expression, the activated pre-TCR/ERK1/2 module has the ability to induce Tal1 phosphorylation.
Constitutive activation of Notch3 increases Tal1 expression. (A-B) Lymphoma cells derived from spleen of Notch3-IC Tg mice displayed high percentages of CD8 and CD8/CD4 cells; thus, Tal1 expression was analyzed by WB and RT-PCR in CD8+-selected thymocytes derived from either WT or Notch3-IC (N3Tg) mice (7-10 weeks). Each sample was analyzed in 2 serial dilutions as indicated (1:10, 1:100). β-Tubulin and β-actin, respectively, were used as loading control. (C) Immunoblot analysis of phosphorylated Tal1 (p-Tal1) or total Tal1 expression in whole-cell extracts of CD8+/DP WT, N3-IC Tg, and N3-IC Tg/pTα-/- thymocytes (4-week-old mice). Tubulin was used as loading control.
Constitutive activation of Notch3 increases Tal1 expression. (A-B) Lymphoma cells derived from spleen of Notch3-IC Tg mice displayed high percentages of CD8 and CD8/CD4 cells; thus, Tal1 expression was analyzed by WB and RT-PCR in CD8+-selected thymocytes derived from either WT or Notch3-IC (N3Tg) mice (7-10 weeks). Each sample was analyzed in 2 serial dilutions as indicated (1:10, 1:100). β-Tubulin and β-actin, respectively, were used as loading control. (C) Immunoblot analysis of phosphorylated Tal1 (p-Tal1) or total Tal1 expression in whole-cell extracts of CD8+/DP WT, N3-IC Tg, and N3-IC Tg/pTα-/- thymocytes (4-week-old mice). Tubulin was used as loading control.
Notch3-IC-increased Tal1 expression is mediated by an IL-2/IL-2R mechanism
It is known that the IL-2/IL-2R signal transduction system is a key element for T-cell proliferation. Moreover, constitutive expression of the IL-2 receptor α chain (CD25) on human adult T-cell leukemia cells suggests that it plays a major role in the development of this disease. We previously reported that premalignant thymocytes and lymphoma cells of Notch3-IC transgenic mice display an abnormal persistent surface expression of CD25.8
Here we show that both Notch3-IC transgenic and Notch3-IC/pTα-/- double-transgenic mice display increased expression of CD25 with respect to wild-type mice but also of CD122, the IL-2R β chain, though to a lower extent. Together these data suggest a role for Notch3 signaling in sustaining the expression of a functional IL-2 receptor, in a pre-TCR-independent manner (Figure 2A). The 1.1 kilobase (kb) region of the mouse Tal1 promoter contains several sequences that fully match the consensus-binding site of transcriptional activators that play a key role in IL-2/IL-2R signaling.22 Moreover, it has been previously reported that triggering of a chimeric IL-2 receptor can induce SCL gene expression in lymphoid cells.23
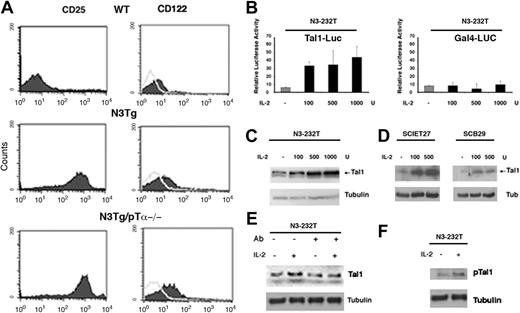
Tal1-sustained expression depends on IL-2/IL-2R pathway. (A) Fluorescence-activated cell sorting (FACS) analysis of CD25 and CD122 expression in thymocytes derived from WT Notch3-IC Tg and N3-IC Tg/pTα-/- mice. (B, left) Tal1-luciferase promoter activity in Notch3-overexpressing N3-232T cells. (B, right) GAL4-luciferase control promoter activity in N3-232T cells. Cells were transfected with either Tal1-promoter luciferase or GAL4-luciferase responsive promoter construct, 0.25 μg/well in 24-well dishes, and treated with the indicated amount of IL-2 for 12 hours; cells were harvested 24 hours after transfection for luciferase assay. All conditions were tested in triplicate samples, and SD is indicated. (C-D) Analysis of Tal1 protein expression by Western blot, in N3-232T, SCIET27, and SCB29 cells treated with the indicated amount of IL-2 for 24 hours; β-tubulin was used as loading control. (E) 232 cells were cultured with (+) and without (-) neutralizing antibodies anti-CD25 (5 μg/mL) alone or with added IL-2 (100 U). mAb was added 2 hours before addition of the cytokine, and the cells were collected after 24 hours. (F) Sample in lanes 1 and 2 of panel E were probed with p-Tal1 and β-tubulin antibodies.
Tal1-sustained expression depends on IL-2/IL-2R pathway. (A) Fluorescence-activated cell sorting (FACS) analysis of CD25 and CD122 expression in thymocytes derived from WT Notch3-IC Tg and N3-IC Tg/pTα-/- mice. (B, left) Tal1-luciferase promoter activity in Notch3-overexpressing N3-232T cells. (B, right) GAL4-luciferase control promoter activity in N3-232T cells. Cells were transfected with either Tal1-promoter luciferase or GAL4-luciferase responsive promoter construct, 0.25 μg/well in 24-well dishes, and treated with the indicated amount of IL-2 for 12 hours; cells were harvested 24 hours after transfection for luciferase assay. All conditions were tested in triplicate samples, and SD is indicated. (C-D) Analysis of Tal1 protein expression by Western blot, in N3-232T, SCIET27, and SCB29 cells treated with the indicated amount of IL-2 for 24 hours; β-tubulin was used as loading control. (E) 232 cells were cultured with (+) and without (-) neutralizing antibodies anti-CD25 (5 μg/mL) alone or with added IL-2 (100 U). mAb was added 2 hours before addition of the cytokine, and the cells were collected after 24 hours. (F) Sample in lanes 1 and 2 of panel E were probed with p-Tal1 and β-tubulin antibodies.
Given the persistent high expression of CD25 on the surface of transgenic and double-transgenic thymocytes, we hypothesized that Notch3-IC-overexpressing T cells were more sensitive to IL-2 and that the enhanced Tal1 expression may be mediated by increased activity of the IL-2/IL-2R signaling. To directly address this issue, we inserted 1169 bp of the Tal1 promoter upstream of the luciferase reporter gene and optimized a transactivation assay in Notch3-overexpressing N3-232T cells, an immortalized immature thymocyte cell line derived from Notch3-IC transgenic mice.8 High level of Tal1 promoter activity was achieved on IL-2 treatment in a dose-dependent manner (Figure 2B, left). This effect appears to be specific, because IL-2 treatment does not affect luciferase activity of a control promoter (Figure 2B, right). We also monitored the effect of IL-2 treatment on the expression of Tal1 protein in different cell lines. We found that Tal1 protein expression was strongly induced in a dose-dependent fashion in N3-232T cells, and in both TCR-β-deficient cell line SCIET27, lacking pre-TCR expression and derived from severe combined immunodeficient (SCID) thymocytes, and the TCR-β-transfected daughter cell line SCB2924 (Figure 2C-E). Importantly, after pretreatment of N3-232T cells with anti-CD25 blocking antibodies, the steady-state level and IL-2-induced increase of Tal1 expression was inhibited, further supporting a role for IL-2 signaling in the maintenance of Tal1 expression. Moreover, we found that IL-2-treated N3-232T cells display increased pTal1 protein that likely reflects the Tal1-increased expression (Figure 2F). Overall, these results suggest that the presence of pre-TCR is dispensable for both Notch3-dependent sustained surface expression of IL-2 receptor and for the increased expression of Tal1 in response to IL-2 treatment.
Colinearity of cyclin D1 expression and impaired expansion of mature T lymphocytes in Notch3-IC/pTα-/- double-transgenic mice
On the basis of the differential expression of CD25 and CD44, the CD4-CD8- double-negative (DN) thymocyte population can be subdivided into 4 developmental stages, termed DN1 to DN4. Up to DN3 stage (CD25+CD44-) the proliferation of thymocytes is driven by interleukins and c-Kit.25 A dramatic shift takes place at the DN3 stage. At this point thymocytes rearrange the β-chains of T-cell receptor and assemble the pre-TCR.26 The proliferation of thymocytes becomes cytokine independent, being instead sustained by the pre-TCR.26 Pre-TCR signaling drives the expansion of the DN4 (CD25-CD44-) and immature single-positive (ISP) cells, which finally differentiate into CD4+CD8+ DP thymocytes and arrest their proliferation. We previously reported that Notch3-enforced expression results in transcriptional activation of pTα gene and significant expansion of CD25-expressing DN thymocyte subsets, thus resulting in a significant increase of total thymocyte yield.8,13 In keeping with this, total thymocyte count was very reduced in Notch3-IC/pTα-/- double-transgenic when compared with both WT and Notch3-IC transgenic mice, despite the ability of enforced Notch3 expression to rescue the block of T-cell differentiation, due to the absence of pTα9 (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). The failure of Notch3-IC/pTα-/- double-transgenic mice to drive sufficient thymocyte expansion supports a model in which Notch3 and pre-TCR signals cooperate in a nonredundant manner to regulate proliferation and differentiation of thymocytes and further supports our previous observation that combined expression of Notch3 and pTα is required for leukemogenesis.9
Consistent with their growth-promoting function, abnormal expression of cyclins is believed to be a driving force in several cancers. Thus, we hypothesized that in the presence of enforced Notch3 expression and in the absence of pTα, cyclin(s) may represent the missing proliferative signal driving thymocyte expansion and mediating leukemia in Notch3 tg mice. To explore this hypothesis we compared cyclin protein levels in thymocytes from WT, Notch3-IC transgenic, and Notch3-IC/pTα-/- double-transgenic mice. We found that activation of Notch3 signaling led to strong up-regulation of cyclin D1 in Notch3-IC transgenic mice at both the mRNA and protein levels (Figure 3A-B). This finding was further strengthened by the observation that transfection of the constitutively active form of Notch3 resulted in induction of the D1 expression (Figure 3C). Then, we tested the connection between Notch3 and pre-TCR with regard to the cyclin D1 expression. As shown in Figure 3A-B, we found that cyclin D1 expression was down-modulated in thymocytes from Notch3-IC/pTα-/- double-transgenic mice with respect to Notch3-IC transgenic mice. Collectively, our results suggest that cyclin D1 represents a critical target of the Notch3/pre-TCR pathways operating in Notch3-induced leukemia.
Cyclin D1 promoter is activated through functional collaboration among Tal1 and Sp1 and depends on pre-TCR/ERK signaling
To directly address whether Tal1 regulates D1 expression, we inserted a 1000-bp D1 promoter upstream of the luciferase reporter gene and performed a luciferase assay in M31 T-cell line.27 Expression vector for Tal1 alone at several doses does not affect D1 promoter basal activity (Figure 3D). This observation was similar to what has been observed with c-Kit promoter activation by Tal1.28,29 It has been indeed reported that Tal1 regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. The 1.1-kb region of the mouse D1 promoter contains several sequences that fully match the consensus binding for Sp1 (Figure 4D). Thus, to assess the importance of Sp1 function for transcriptional regulation of D1 promoter by Tal1, we cotransfected Sp1 expression plasmid. Cotransfection of Tal1 and Sp1 induced a modest increase in luciferase activity (Figure 3D). In contrast, a significantly increased transactivation of D1 promoter was achieved on cotransfection of SCL/Tal1 with Sp1 and Notch3-IC. Therefore, D1 promoter activation by Tal1 relies on the formation of a complex with Sp1 and is increased by enhanced Notch3 signaling. Phosphorylation of Tal1 by ERK has been reported to increase its transcriptional activity.20 In keeping with this, when ERK activity in M31 T cells was prevented by the inhibitor of MEK1, PD98059, the effect of Sp1/Tal1 complex on D1 promoter activity was almost completely reversed (Figure 3D). Consistent with the fact that increased D1 expression relies on the phosphorylation of Tal1 protein, phospho-Tal1 (pTal1) is increased by enhanced Notch3 signaling (Figure 3C, right). To provide further evidence that in our system activation of the pre-TCR/ERK pathway regulates Tal1 phosphorylation, we performed additional experiments by using the SCB29 cell line. This cell line expresses endogenous pTα and is stably transfected with TCR-β, being thus suitable for analyzing pre-TCR signaling.24 Therefore, we analyzed the effect of the triggering of pre-TCR signaling by crosslinking of CD3. SCB.29 cells were incubated with anti-CD3 for 12 hours and examined for the presence of p-Tal1. Although p-Tal1 protein was detectable in control-untreated cells, a 3- to 4-fold increase of phosphorylated Tal1 was present in cells on pre-TCR activation (Figure 3F). When ERK activity in SCB.29 cells was prevented by the pharmacologic inhibitor of MEK1, PD98059, the effect of CD3 crosslinking on Tal1 phosphorylation was reversed (Figure 3F). To provide further evidence that the pre-TCR/ERK pathway regulates Tal1 phosphorylation and its transcriptional activity, we performed additional experiments in which SCB.29 cells were cotransfected with Tal1 and Sp1 expression plasmid. Cotransfection of Tal1 and Sp1 induced a modest increase in luciferase activity (Figure 3E). In contrast, a significantly increased transactivation of D1 promoter was achieved on crosslinking of CD3. Therefore, D1 promoter activation by Tal1 relies on the formation of a complex with Sp1 and is increased by enhanced ERK1/2 signaling. Together, these results indicate that Tal1-mediated D1 expression requires an ERK-dependent phosphorylation mechanism.
Notch3 increases D1 expression through modulation of Tal1 activity. (A-B) D1 expression analyzed by WB and RT-PCR from primary thymocytes derived from WT, Notch3-IC (N3-IC Tg), and N3-IC Tg/pTα-/- mice (4 weeks of age). β-Tubulin and β-actin, respectively, were used as loading control. (C) M31 cells were transfected with either the pCDNA3 or Notch3-IC plasmids as indicated. Transient transfection efficiency, as assessed by GFP expression of a cotransfected expression vector, was greater than 70%. At 48 hours after transfection, total cell extracts were analyzed by immunoblotting with indicated antibodies. (C, bottom) Densitometric analyses of Tal1 level in M31 cells transfected with Notch3-IC. (D) M31 cells were transfected with the D1-promoter luciferase reporter plasmid plus expression vectors for constitutively active Notch3-IC, Tal1, and SP-1. Twelve hours after transfection, cells were treated with 25 and 50 μM PD98059, MEKK1 inhibitor, as indicated. Luciferase activity was determined at 24 hours after transfection. (E) SCB.29 cells were cotransfected with D1-Luciferase promoter, Tal1 and Sp1 expression vectors. After 12 hours from transfection, cells were treated with anti-CD3 for the time indicated. Luciferase activity was determined at 36 hours after transfection. (F, left) SCB29 cells were treated with anti-CD-3 Ab for 12 hours, and p-Tal1 expression was analyzed by Western blot, Tal1 blot was used as a loading control. (F, right) Densitometric analyses of pTal1 level in SCB29 cells treated with anti-CD-3.
Notch3 increases D1 expression through modulation of Tal1 activity. (A-B) D1 expression analyzed by WB and RT-PCR from primary thymocytes derived from WT, Notch3-IC (N3-IC Tg), and N3-IC Tg/pTα-/- mice (4 weeks of age). β-Tubulin and β-actin, respectively, were used as loading control. (C) M31 cells were transfected with either the pCDNA3 or Notch3-IC plasmids as indicated. Transient transfection efficiency, as assessed by GFP expression of a cotransfected expression vector, was greater than 70%. At 48 hours after transfection, total cell extracts were analyzed by immunoblotting with indicated antibodies. (C, bottom) Densitometric analyses of Tal1 level in M31 cells transfected with Notch3-IC. (D) M31 cells were transfected with the D1-promoter luciferase reporter plasmid plus expression vectors for constitutively active Notch3-IC, Tal1, and SP-1. Twelve hours after transfection, cells were treated with 25 and 50 μM PD98059, MEKK1 inhibitor, as indicated. Luciferase activity was determined at 24 hours after transfection. (E) SCB.29 cells were cotransfected with D1-Luciferase promoter, Tal1 and Sp1 expression vectors. After 12 hours from transfection, cells were treated with anti-CD3 for the time indicated. Luciferase activity was determined at 36 hours after transfection. (F, left) SCB29 cells were treated with anti-CD-3 Ab for 12 hours, and p-Tal1 expression was analyzed by Western blot, Tal1 blot was used as a loading control. (F, right) Densitometric analyses of pTal1 level in SCB29 cells treated with anti-CD-3.
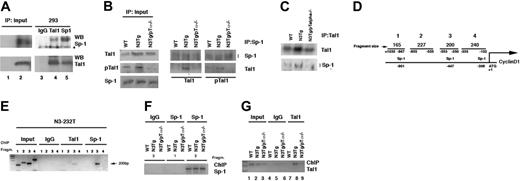
Binding of the Tal1 and SP-1 proteins to the endogenous D1 promoter as assessed by chromatin immunoprecipitation. (A) Physical association between the Sp1 and Tal1 transcription factors. HEK 293 cells were cotransfected with 8 μg mammalian expression vectors for Sp1 and Tal1. Cell lysates were immunoprecipitated with the indicated antibodies or affinity-purified mouse IgG. Immune complexes were analyzed by SDS 6% polyacrylamide gel for Sp1 and by SDS 10 % polyacrylamide gel for Tal1 analysis and sequential immunoblotting with Sp1 and Tal1 antibodies. Total extracts (60 μg) derived from 293 transfected cells were used as input control. (B) Total cell lysates (2 mg) derived from primary thymocytes obtained from the indicated mice (4 weeks of age) were immunoprecipitated with antibodies against either Sp1 (B) or Tal1 (C). Immune complexes were analyzed by immunoblotting with the goat polyclonal antibodies against Sp1 or Tal1. Immune complexes were analyzed by SDS 6% polyacrylamide gel for Sp1 and by SDS 10% polyacrylamide gel for Tal1 analysis. (B, left) Total extract (60 μg) derived from indicated mice was used as input control. (D) Schematic representation of D1 promoter; Sp1 binding sites are indicated. (E) N3-232 T cells were processed for chromatin immunoprecipitation with antibodies against both Sp1 and Tal1. The immunoprecipitates were analyzed by PCR with oligonucleotide primes specific for the indicated region of the mouse cyclin D1 promoter. (F) Primary T cells derived from the indicated mice were processed for chromatin preparation and then subjected to immunoprecipitation with antibodies against either Sp-1 or IgG control antibodies. The immunoprecipitates were analyzed by PCR with oligonucleotide primes specific for the indicated region of the mouse D1 promoter. (G) Primary T cells derived from the indicated mice (4 weeks of age) were processed for chromatin preparation and then subjected to immunoprecipitation with antibodies against either Tal1 or IgG control antibodies. The immunoprecipitates were analyzed by PCR with oligonucleotide primers specific for region 3 of the mouse cyclin D1 promoter.
Binding of the Tal1 and SP-1 proteins to the endogenous D1 promoter as assessed by chromatin immunoprecipitation. (A) Physical association between the Sp1 and Tal1 transcription factors. HEK 293 cells were cotransfected with 8 μg mammalian expression vectors for Sp1 and Tal1. Cell lysates were immunoprecipitated with the indicated antibodies or affinity-purified mouse IgG. Immune complexes were analyzed by SDS 6% polyacrylamide gel for Sp1 and by SDS 10 % polyacrylamide gel for Tal1 analysis and sequential immunoblotting with Sp1 and Tal1 antibodies. Total extracts (60 μg) derived from 293 transfected cells were used as input control. (B) Total cell lysates (2 mg) derived from primary thymocytes obtained from the indicated mice (4 weeks of age) were immunoprecipitated with antibodies against either Sp1 (B) or Tal1 (C). Immune complexes were analyzed by immunoblotting with the goat polyclonal antibodies against Sp1 or Tal1. Immune complexes were analyzed by SDS 6% polyacrylamide gel for Sp1 and by SDS 10% polyacrylamide gel for Tal1 analysis. (B, left) Total extract (60 μg) derived from indicated mice was used as input control. (D) Schematic representation of D1 promoter; Sp1 binding sites are indicated. (E) N3-232 T cells were processed for chromatin immunoprecipitation with antibodies against both Sp1 and Tal1. The immunoprecipitates were analyzed by PCR with oligonucleotide primes specific for the indicated region of the mouse cyclin D1 promoter. (F) Primary T cells derived from the indicated mice were processed for chromatin preparation and then subjected to immunoprecipitation with antibodies against either Sp-1 or IgG control antibodies. The immunoprecipitates were analyzed by PCR with oligonucleotide primes specific for the indicated region of the mouse D1 promoter. (G) Primary T cells derived from the indicated mice (4 weeks of age) were processed for chromatin preparation and then subjected to immunoprecipitation with antibodies against either Tal1 or IgG control antibodies. The immunoprecipitates were analyzed by PCR with oligonucleotide primers specific for region 3 of the mouse cyclin D1 promoter.
We observed that, despite a sustained Tal1 protein increased expression, the level of phospho-Tal1 and cyclin D1 is down-modulated in Notch3-IC/pTα-/- double-transgenic mice. Overall, our results allow us to hypothesize that pre-TCR-mediated Tal1 phosphorylation may represent the missing signals required for D1 expression in the Notch3-IC/pTα-/- double-transgenic mice.
It has been previously suggested that transcriptional synergy between Tal1 and Sp1 may result from direct interaction between them; thus, we determined whether Sp1 physically associates with Tal1 protein. As shown in Figure 4A, cotransfection of Tal1 and Sp1 in 293 HEK cells followed by immunoprecipitation reveals that a physical association between Sp1 and Tal1 takes place in transfected cells. To assess whether Tal1 and Sp-1 interaction occurs in vivo between the endogenous proteins, we performed a coimmunoprecipitation experiment using primary thymocytes derived from WT, Notch3-IC transgenic, and Notch3-IC/pTα-/- double-transgenic mice. Under these conditions Tal1 coprecipitated with anti-Sp-1 antibodies only in extracts derived from Notch3-IC transgenic mice, demonstrating a strong association between Tal1 and Sp1 (Figure 4B). In addition, anti-Sp1 antibodies efficiently brought down phospho-Tal1 protein only in Notch3 tg thymocytes, suggesting that only a phosphorylated Tal1 may form a complex with Sp1 (Figure 4B). Together, our results demonstrate that Tal1 and Sp1 directly interact in vitro and in vivo. Moreover, in vivo coprecipitation was consistently observed in extracts derived from Notch3-IC transgenic mice, in which both Tal1 expression and phosphorylation are increased, whereas Tal1/Sp1 interaction did not occur in extracts derived from Notch3-IC/pTα-/- double-transgenic mice in which Tal1 expression but not its phosphorylation is increased (Figures 1C and 4B). Finally, to ascertain whether this transcriptional complex occupies the D1 promoter in vivo, we performed a chromatin immunoprecipitation assay using formaldehyde crosslinked N3-232T cells.8,30 Chromatin extracts were subjected to immunoprecipitation with both anti-Tal1 and anti-Sp1 antibodies. Crosslink was reversed and DNA specifically retained was purified. These samples were subjected to PCR amplification with the use of specific oligos that target different regions of the D1 promoter. As shown in Figure 4E, both anti-Tal1 and anti-Sp1 antibodies recovered a 200-bp fragment of the D1 proximal promoter containing the Sp1 site. To assess that Tal1 and Sp1 regulate endogenous D1 promoter in vivo, and to address the relation among Notch3, pre-TCR, and Tal1/Sp1 complex, we performed chromatin immunoprecipitation using primary thymocytes derived from WT, Notch3-IC transgenic, and Notch3-IC/pTα-/- double-transgenic mice. As shown in Figure 4F, antibodies to Sp1 efficiently recovered the 200-bp fragment of the proximal region of the D1 promoter, in contrast no signal was present when the amplified region was the distal region (fragments 3 and 1, respectively, in Figure 4D) of the D1 promoter. Interestingly, Figure 4F shows that in WT, Notch3-IC transgenic, and Notch3-IC/pTα-/- double-transgenic thymocytes, Sp1 protein is associated with the D1 promoter, suggesting that this factor may form a constitutively associated factor to the D1 promoter. In contrast, Tal1 antibodies efficiently recovered the 200-bp proximal fragment only in the Notch3-IC transgenic thymocytes, in which Tal1 is phosphorylated by the pre-TCR-dependent ERK activation (Figure 4G, lane 8). These results demonstrate that Sp1 and Tal1 directly associate with the proximal D1 promoter region containing the functional Sp1 binding site and might explain the functional collaboration observed between Notch3 and the pre-TCR in sustaining D1 expression.
Discussion
Activation and overexpression of Notch signaling was reported to have a high relevance in T-cell leukemia.19,31,32 For this reason elucidation of the oncogenic pathways triggered by Notch activation is critically important to understand the role of this protein in human tumorigenesis. In the present work, we analyzed the function of Notch3 signaling through a combination of genetic crossing, in vivo studies, and molecular analysis to analyze the molecular pathway that impinges on Notch3 signaling in Notch-driven T-cell leukemia. We determined that Tal1 transcription factor is a downstream target of the Notch3 and pre-TCR pathway cooperation. We identified a role for Notch signaling in enhancing Tal1 phosphorylation and expression that is dependent on the IL-2/IL-2R signaling pathway. Although the mechanism by which Notch3 sustains the CD25 surface expression remains to be elucidated, the observation that Notch3, by sustaining the expression of IL-2 receptor, may cooperate with the endogenous IL-2 in increasing the expression of Tal1 represents an original finding. Overall, our data suggest that such cooperation, by allowing the formation of an activated transcriptional complex, can trigger the proliferation of lymphoma cells.
Notably, although expression of Tal1 is normally limited to the earliest T-cell precursors,33 molecular subtype of T-ALL defined by Tal1 overexpression is characterized by an immunophenotype that suggest that T-cell development is blocked at the late-stage cortical thymocyte population (CD4+CD8+),14 indicating that thymocytes have proceeded to a further step of differentiation. Thus, it is likely that Tal1-positive T-ALL requires the fine balance between negative but also positive regulatory mechanisms that control thymocyte development. Elevated Notch3 activity, coupled with increased Tal1 expression, could provide one such mechanism that needs to be activated for maintaining the leukemogenic phenotype. Our data indicate that enhanced Tal1 expression per se does not specify a particular cellular response. Indeed, we found that in Notch3-IC/pTα-/- double-transgenic mice, pre-TCR is dispensable for Notch-mediated Tal1-increased expression, but pre-TCR-dependent MAPK/ERK-triggered mitogenic signals are required for Notch3-induced leukemia, suggesting that combined signals generated from Notch3 and pre-TCR may regulate the cell proliferative response. The proliferative defect that we observed in Notch3-IC/pTα-/- double-transgenic mice prompted us to study whether the reduced proliferation could be ascribed to reduced cell cycle-related protein expression, namely, cyclin D1. Indeed, we found that both Tal1 phosphorylation and D1 expression are down-modulated in Notch3-IC/pTα-/- double-transgenic mice, indicating a close association within Tal1 phosphorylation and D1 expression, which suggests that the latter is a potential downstream target of phosphorylated Tal1. Moreover, we demonstrate that Tal1 requires the cooperation of Sp1, because enforced Tal1 expression induces D1 promoter activity only when the Sp1 partner is coexpressed. Finally, the study of the binding of Tal1 and Sp1 to the endogenous D1 promoter in vivo, reveals a constitutive binding of the Sp1 transcription factor to the proximal region of the D1 promoter. In a previous study, it was suggested that signals produced by pre-TCR up-regulate cyclin D3, which provide the essential proliferative drive needed for the expansion of the DN4 pool, the immediate precursors of DP thymocytes.34 In the same study, the researchers demonstrated that cyclin D3 is required for proliferation of both murine and human T-cell leukemias characterized by the expansion of the DP subset. In the current study, we found that T-cell leukemia developing in Notch3-IC transgenic mice, in which we previously observed that the pre-TCR-dependent expansion of DN2 and DN3 thymocytes subsets precedes the development of the disease,8 is characterized by the up-regulation of cyclin D1. This could represent a specific effect of constitutive activation of Notch3 signaling, because we previously demonstrated that the cooperation between Notch3 and pre-TCR triggers the ERK/MAPK pathway and we demonstrate here that this pathway is required to allow the formation of the Sp1/p-Tal1 complex, necessary to obtain the full transcriptional activation of cyclin D1.
Moreover, because we cannot exclude that different cyclins may have largely exchangeable functions when the early T-cell differentiation stages are perturbed by neoplastic transformation, a possibility is that the role of cyclin D3 in Notch3-induced leukemogenesis model is retained by the sustained D1 expression.
Note that, although we describe here a model in which increased phosphorylation of Tal1 depends on the increased expression of pTα/pre-TCR, it has been previously shown that Tal1 represses pTα expression by inhibiting the activity of a DNA-bound E2A transcription factor.33 However, we previously reported that in Notch3-IC transgenic mice activation of Notch signaling promotes increase of Id1 expression, which also is known to result in reduction of E2A DNA binding activity.13 Thus, it is likely that in Notch3 transgenic mice, because of the Id1 inhibitory function, Tal1 is not tethered to the pTα because of the reduced binding activity of E2A. Moreover, we previously showed that Notch3 is able to transcriptionally activate pTα, while inhibiting E2A activity,13 thus suggesting the involvement of alternative pathway(s).
Model depicting the pathways linking activated Notch3 with the induction of cyclin D1. Notch3-IC has been shown to activate the pTα promoter, thus promoting activation of pre-TCR signaling that in turn results in ERK1/2 activation. Signaling through the IL-2/IL-2R pathway acts to increases Tal1 expression. The induction of pTal1/Sp1 complex through a pre-TCR/ERK1/2-dependent pathway activates cyclin D1 expression.
Model depicting the pathways linking activated Notch3 with the induction of cyclin D1. Notch3-IC has been shown to activate the pTα promoter, thus promoting activation of pre-TCR signaling that in turn results in ERK1/2 activation. Signaling through the IL-2/IL-2R pathway acts to increases Tal1 expression. The induction of pTal1/Sp1 complex through a pre-TCR/ERK1/2-dependent pathway activates cyclin D1 expression.
On the basis of our previous and present data we can suggest a model (Figure 5) in which Notch3 directly activates pTα transcription and sustains pre-TCR signaling and, by increasing surface expression of CD25, triggers the IL-2/IL-2R pathway. Together these pathways cooperate to increase the expression and phosphorylation of Scl/Tal1. Also we identify a critical role for pre-TCR signaling in transmitting Notch3 mitogenic signals. Indeed, specifically we found that Notch3-induced pre-TCR/ERK signaling is required for Tal1 phosphorylation that in turn is required for cyclin D1 expression, highlighting downstream key components of the Notch3 leukemogenic pathway.
Prepublished online as Blood First Edition Paper, December 20, 2005; DOI 10.1182/blood-2005-07-2823.
Supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), the Ministero della Salute, and the Biologia e Medicina Molecolare (BEMM) Center of Excellence and by a Fondazione Italiana per la Ricerca sul Cancro (FIRC) fellowship (C.T.).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal