Abstract
AML1-ETO, a chimeric gene frequently detected in acute myelogenous leukemia (AML), inhibits the differentiation of myeloid progenitors by suppressing genes associated with myeloid differentiation and increases the replating ability of clonogenic myeloid progenitors. However, AML1-ETO alone cannot induce AML and thus additional genetic events are required for the onset of AML. The Wilms tumor gene (WT1), which has been identified as the gene responsible for Wilms tumor, is expressed at high levels in almost all human leukemias. In this study, we have generated transgenic mice (WT1-Tg) that overexpress WT1 in hematopoietic cells to investigate the effects of WT1 on AML1-ETO-associated leukemogenesis. AML1-ETO-transduced bone marrow (BM) cells from WT1-Tg mice exhibited inhibition of myeloid differentiation at more immature stages and higher in vitro colony-forming ability compared with AML1-ETO-transduced BM cells from wild-type mice. Most importantly, all of the mice that received a transplant of AML1-ETO-transduced BM cells from the WT1-Tg mice rapidly developed AML. These results demonstrate that AML1-ETO may exert its leukemogenic function in cooperation with the expression of WT1.
Introduction
Acute myeloid leukemia (AML) accounts for more than 80% of acute leukemia cases in adults. The (8;21)(q22;q22) translocation, which fuses AML1 (CBFA2/PEBP2α/RUNX1) on chromosome 21 with ETO (MTG8) on chromosome 8, is one of the most frequent chromosomal abnormalities detected in AML with maturation (FAB classification AML-M2), which accounts for approximately 40% of de novo AML.1,2 Studies of several murine models have demonstrated that AML1-ETO alone is not sufficient to develop leukemia. Mice carrying an AML1-ETO knock-in allele,3-5 AML1-ETO-transgenic (Tg) mice,6,7 and wild-type mice that received a transplant of BM cells transduced retrovirally with AML1-ETO were found not to develop leukemia.8-10 However, approximately 50% of AML1-ETO-Tg mice developed AML when additional random mutations were introduced by using N-ethyl-N-nitrosourea.7 These results indicate that AML1-ETO cannot induce AML by itself, and additional genetic events are required to induce AML.11
The Wilms tumor gene, WT1, has been identified as the gene responsible for Wilms tumor, a childhood renal cancer, and is defined as a tumor-suppressor gene.12,13 WT1 encodes a zinc finger transcription factor and plays an important role in tissue development, cell proliferation and differentiation, and apoptosis.14 We and others have previously reported high expression of wild-type WT1 in almost all leukemias, an inverse correlation between WT1 expression levels and prognosis, and an increased expression of WT1 at relapse in acute leukemia.15-22 WT1 is preferentially expressed in hematopoietic progenitors, and its expression is down-regulated as an early event when differentiation is induced.21-25 We have previously reported that constitutive WT1 overexpression inhibits the differentiation of myeloid progenitors and instead promotes their proliferation in response to granulocyte colony-stimulating factor (G-CSF)26,27 and that the growth of leukemic cells is inhibited by WT1 antisense oligomers.28 These results indicate that WT1 is involved in leukemogenesis.
In the present study, we generated transgenic (WT1-Tg) mice that overexpress WT1 in hematopoietic cells to investigate the effects of WT1 in AML1-ETO-associated leukemogenesis. We found that AML1-ETO-transduced BM cells from the WT1-Tg mice exhibited inhibition of myeloid differentiation at more immature stages and higher in vitro colony-forming ability compared with AML1-ETO-transduced BM cells from wild-type mice and that all of the mice that received a transplant of AML1-ETO-transduced BM cells from WT1-Tg mice rapidly developed AML.
Materials and methods
Generation of retrovirus
MSCV-IRES-EGFP was prepared as an empty control vector, as previously reported.29 A 2.5-kb XbaI-ClaI fragment containing the human AML1-ETO cDNA from pLNSX/AML1-ETO was inserted upstream of the internal ribosome entry site (IRES) element at the BamHI site of the MSCV-IRES-EGFP retrovirus vector. The retroviral constructs were lipo-transfected into GP+E86-based producer cells. Retrovirus titers, assessed by GFP expression in NIH3T3 cells that were transduced with the vectors, in the supernatant of the AML1-ETO- and empty control vector-producer cells were 0.7 × 106/mL and 1 × 106/mL, respectively.
Generation of WT1 transgenic mice
A 2.0-kb KpnI-BamHI fragment of the mouse tec promoter was ligated to the polylinker site of a PIND vector (Invitrogen, Carlsbad, CA). A 1.5-kb Sau3AI-fragment containing the mouse WT1(17AA+/KTS+) cDNA from pBluescript/mWT1(17AA+/KTS+) and the SV40 3′-untranslated region containing a poly(A) signal were subcloned into the BamHI and EcoRV sites of the PIND vector, respectively. We isolated a 4.5-kb WT1 transgene from the resulting PIND-tec-WT1(+/+)-SV40 poly(A) vector by digestion with XhoI and microinjected the transgene into the pronuclei of eggs from C57BL/6 × DBA/2 F1 mice, as previously reported.30 Three independent transgenic founders were identified by Southern blotting analysis and backcrossed onto C57BL/6-Ly5.2 mice for at least 5 generations to minimize strain differences. All mice were maintained in the Central Institute of Experimental Animals, Osaka University Graduate School of Medicine, according to institutional guidelines.
Hematopoietic cell isolation
For preparation of hematopoietic cells, the BM and spleen cells from WT1-Tg or wild-type mice aged 10 to 12 weeks were used. For isolation of c-kit+Sca-1+lineage-negative (Lin-) cells and c-kit+Sca-1-Lin- BM cells, BM cells were stained with biotinylated antibodies specific for lineage markers (Mac-1, Gr-1, Ter119, B220, CD3, CD4, and CD8). Lineage-positive cells were removed using streptavidin-coated Dynabeads (M-280; Dynal Biotech, Lake Success, NY) according to the manufacturer's instructions. Enriched lineage-negative cells were stained with FITC-conjugated anti-Sca-1 and PE-conjugated anti-c-kit antibodies. For isolation of Mac-1+Gr-1low and Mac-1+Gr-1high myeloid cells, BM cells were stained with PE-conjugated anti-Gr-1 and biotinylated anti-Mac-1 antibodies after being blocked with FcγIII/II antibodies. Biotinylated antibodies were visualized with streptavidin-PerCP. Stained cells were sorted using a FACS Vantage cell sorter (Becton Dickinson, San Jose, CA). For isolation of erythroid cells, BM cells were stained with biotinylated anti-Ter119 antibody after being blocked with FcγIII/II antibodies. For isolation of splenic B and T cells, low-density mononuclear cells were collected using Lymphocyte Separation Solution (Nacalai Tesque, Kyoto, Japan) and stained with biotinylated anti-B220 for B cells and anti-CD3, anti-CD4, and anti-CD8 antibodies for T cells. Stained cells were isolated using streptavidin-coated Dynabeads (M-280) according to the manufacturer's instructions. All antibodies were purchased from BD Pharmingen (San Diego, CA).
Quantitative real-time RT-PCR
The expression levels of WT1 in whole BM cells and subpopulations of hematopoietic cells were measured by real-time reverse transcriptase-polymerase chain reaction (RT-PCR) using a Prism 7700 thermal cycler and sequence detector (Applied Biosystems, Foster City, CA), as previously reported.31 Total RNA was extracted from isolated hematopoietic cells using an SV total RNA isolation kit (Promega, Madison, WI) or TRIZOL Reagent (Invitrogen) according to the manufacturer's instructions and converted to cDNA with oligo dT primer and Moloney murine leukemia virus (M-MLV) reverse transcriptase. The PCR reaction was performed for 40 cycles (denaturation at 95°C for 15 seconds and annealing and elongation at 61°C for 1 minute). To verify that there had been no contamination of the transgene DNA, RNA that had not been incubated with reverse transcriptase was also analyzed. Standard curves were drawn using serial dilutions of the WT1 and β-actin cDNAs, and the relative WT1 expression level was defined as (copy number of the WT1 transcripts divided by that of β-actin transcripts) × 108. All experiments were performed in duplicate and the average values were used.
Western blotting analysis
Western blotting analysis was done as previously reported.9,31 We detected AML1-ETO protein using goat polyclonal anti-AML-1 IgG (AML-1 N-20; Santa Cruz Biotechnology, Santa Cruz, CA) and WT1 protein using rabbit polyclonal anti-WT1 IgG (WT1 C-19; Santa Cruz Biotechnology). The primary staining was visualized with an appropriate horseradish peroxidase-conjugated secondary antibody and enhanced with a chemiluminescent substrate (Western Lighting; Perkin-Elmer Life Sciences, Boston, MA), according to the manufacturer's instructions. The band intensity was analyzed using NIH image software (version 1.62; Bethesda, MD).
Retroviral transduction into BM cells and BM transplantation
WT1-Tg and C57BL/6-Ly5.2 mice aged 10 to 16 weeks were used as a source of BM cells, and C57BL/6-Ly5.1 female mice aged 10 to 12 weeks were used as recipients for the BM transplantation. Four days after intravenous injection of 5-fluorouracil (FU, 150 mg/kg), the BM cells from 5-FU-treated mice were harvested, cultured for 2 days in DMEM with 15% fetal bovine serum (FBS), 6 ng/mL IL-3, 10 ng/mL IL-6, and 100 ng/mL stem cell factor (SCF), and then cocultured with irradiated (15 Gy) GP+E86 virus-producer cells for 2 days in the same medium with the addition of protamine sulfate (final concentration 5 μg/mL), as previously reported.29 Different numbers of BM cells (from 2 × 105 to 5 × 105) cocultured with virus producers were transplanted into the lethally irradiated (9.0 Gy) recipients. These mice were maintained for 4 weeks on water containing erythromycin (25 mg/100 mL), gentamicin (30 mg/100 mL), and fluconazole (2.5 mg/100 mL).
In vitro cell culture
GFP-positive BM cells were sorted using a FACSort flow cytometer (Becton Dickinson) and plated at 1 × 104 cells/plate in methylcellulose medium containing 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL SCF, and 3 U/mL erythropoietin (EPO) (Methocult M3434; Stem Cell Technologies, Vancouver, BC). In colony-replating assays, all the cells from the primary culture were collected and then replated at 1 × 104 cells/plate in new methylcellulose medium (M3434). In a single colony-replating assay, all the cells from a single colony were picked and replated in fresh methylcellulose medium (M3434). Lin- cells from colonies in at least their tenth generation were purified (see “Hematopoietic cell isolation”), and then cultured in DMEM with 10% FBS containing 5 ng/mL IL-3 and 100 ng/mL G-CSF. Splenocytes from leukemic animals were plated at 3000 cells/plate in methylcellulose medium (Methocult M3234; Stem Cell Technologies) containing 1 or more than 2 cytokines among 10 ng/mL IL-3, 100 ng/mL SCF, 100 ng/mL G-CSF, or 10 ng/mL GM-CSF. The number of colonies containing more than 50 cells was counted on day 8. All cytokines were purchased from PeproTech (Rocky Hill, NJ). All culturing was performed at 37°C in a humidified incubator under 5% CO2. Colony size was assessed by measuring the diameters of the colonies on 35-mm reversal films (Provia 100F; Fujifilm, Tokyo, Japan), using an Olympus IMT-2-21 microscope and a 4 ×/0.10 numeric aperture (NA) objective (both from Olympus, Tokyo, Japan). Images (Figures 3C, 6B left) were acquired using a Canon CanoScan 8400 color image scanner (Canon, Tokyo, Japan), then processed using Adobe Photoshop CS software (Adobe Systems, San Jose, CA).
Flow cytometric analysis
Single-cell suspensions were prepared from BM and peripheral blood (PB) and stained for fluorescence-activated cell sorter (FACS) analysis using PE-conjugated antibodies for c-kit or Gr-1 and biotinylated antibodies for Lin or Mac-1 after being blocked with FcγIII/II antibodies. Biotinylated antibodies were visualized with streptavidin-PerCP. We analyzed the stained cells using a FACSCalibur flow cytometer (Becton Dickinson) and Cell Quest software (Becton Dickinson).
Establishment of cell lines and WT1 antisense oligomer treatment
To establish cell lines, a single colony in leukemic cell culture was picked and seeded in DMEM with 10% FBS containing 5 ng/mL IL-3 and 50 ng/mL SCF. The cells were maintained for more than 6 months. Oligomer treatment was performed as previously reported.28,32 Cells at a density of 2 × 104 cells/mL were treated with WT1 antisense (WT1 AS) to suppress WT1 expression, or with random oligomers as a control, at concentrations of 200 μg/mL. Viable cells were counted after 72 hours using the trypan blue exclusion method. The sequence of WT AS was 5′-AGGGTCGAATGCGGTGGG-3′, and for the control, random 18-mer oligodeoxynucleotides were used.
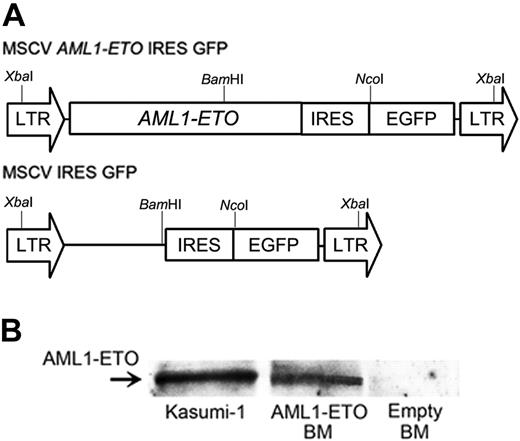
Retroviral transduction of AML1-ETO. (A) Schematic diagram of MSCV retroviral constructs (MSCV AML1-ETO IRES EGFP and MSCV IRES EGFP). XbaI, BamHI, and NcoI represent restriction enzyme sites. (B) Western blotting analysis for AML1-ETO expression. Lysates of FACS-sorted GFP-positive BM cells were examined for AML1-ETO fusion proteins by Western blotting analysis. AML1-ETO-expressing Kasumi-1 cells were used as a positive control.
Retroviral transduction of AML1-ETO. (A) Schematic diagram of MSCV retroviral constructs (MSCV AML1-ETO IRES EGFP and MSCV IRES EGFP). XbaI, BamHI, and NcoI represent restriction enzyme sites. (B) Western blotting analysis for AML1-ETO expression. Lysates of FACS-sorted GFP-positive BM cells were examined for AML1-ETO fusion proteins by Western blotting analysis. AML1-ETO-expressing Kasumi-1 cells were used as a positive control.
Southern blotting
High-molecular-weight DNA was isolated from BM or spleen cells using a PureGene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's instructions. For Southern blotting, 10 μg DNA was digested with XbaI or BamHI, separated on a 0.8% agarose gel, transferred to a Hybond-N+ membrane (Amersham, Arlington Heights, IL), and hybridized with a probe consisting of a 0.6-kb BamHI-NcoI fragment containing the IRES sequence from the MSCV-IRES-EGFP retrovirus vector. Autoradiographic images were obtained using x-ray film.
Kaplan-Meier analysis
Cumulative probability of survival after BM transplantation was estimated by the Kaplan-Meier method using Stat View software (SAS Institute, Cary, NC).
Results
Transduction of AML1-ETO into BM cells
We designed an AML1-ETO retroviral construct, in which human AML1-ETO was inserted into a murine stem cell retroviral vector (MSCV), which coexpressed GFP (Figure 1A). We used MSCV as the backbone because of its ability to transduce inserted genes into murine hematopoietic stem cells (HSCs) efficiently and to let HSCs and their progeny express them. We cocultured the BM cells from 5-FU-treated mice with retrovirus-producing cells, as previously reported.29 The gene transduction efficiencies, which were assessed by GFP expression in the BM cells, were 56.3% to 73.5%. To confirm the expression of AML1-ETO in the transduced BM cells, we performed Western blotting analysis with lysates from FACS-sorted GFP-positive cells and detected AML1-ETO fusion proteins (Figure 1B).
Generation of WT1-transgenic mice
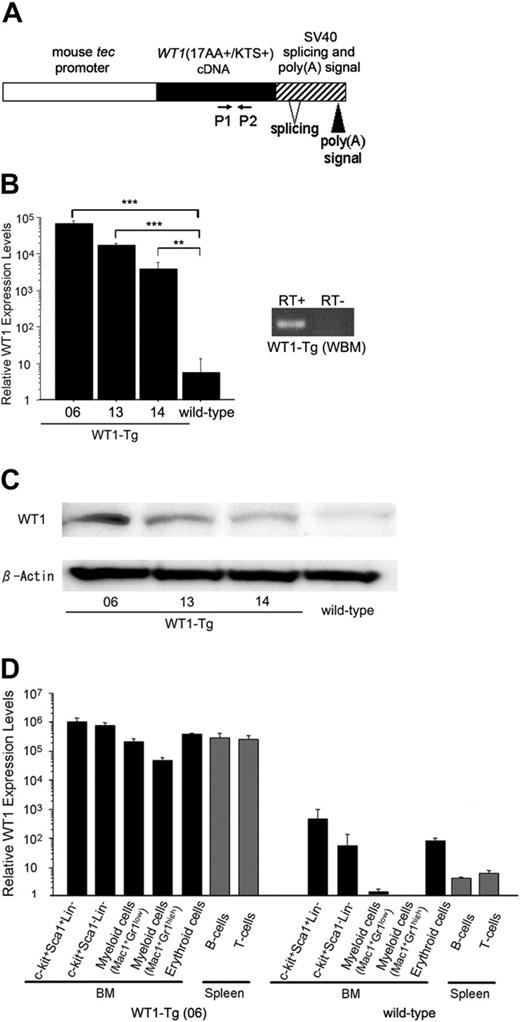
We generated transgenic (WT1-Tg) mice that expressed the mouse full-sized, nonspliced WT1 cDNA, WT1(17AA+/KTS+), under the control of the mouse tec promoter (Figure 2A). Tec is a cytoplasmic kinase that is preferentially expressed in hematopoietic progenitors.33 We assessed the expression of WT1 in the whole BM cells from each of the WT1-Tg lines by real-time RT-PCR and Western blotting analysis. The BM cells from the WT1-Tg mice expressed WT1 mRNA at a significantly higher level than those from the wild-type mice (Figure 2B). The relative WT1 protein expression levels in the WT1-Tg lines 06, 13, and 14, which were defined as the ratio of the intensity of the band for WT1 to that for β-actin by densitometry, were 45.2 ± 5.7-fold, 23.9 ± 3.6-fold, and 10.9 ± 1.4-fold, respectively, higher than those in the wild-type mice (Figure 2C). To characterize which population(s) of hematopoietic cells in the WT1-Tg mice expressed WT1, we isolated subpopulations of hematopoietic cells, including c-kit+Sca-1+Lin-, c-kit+Sca-1-Lin-, myeloid (Mac-1+Gr-1low and Mac-1+Gr-1high), and erythroid cells in the BM, and B and T cells in the spleen, and quantified their WT1 expression levels by real-time RT-PCR. The WT1 expression levels in all subpopulations of hematopoietic cells from the WT1-Tg mice were significantly higher than those in the comparable cells from the wild-type mice (Figure 2D).
Generation of WT1-transgenic mice. (A) Schematic model of the injected fragment used for generating WT1-transgenic mice. P1 and P2 represent the primers used for real-time RT-PCR. Three independent founders gave rise to lines 06, 13, and 14. (B) WT1 expression levels in the whole BM cells from the WT1-transgenic mice and wild-type littermates were measured by real-time RT-PCR. The mean values ± SD of duplicate measurements from 3 mice are shown. **P < .005; ***P < .001. (C) Western blotting analysis of WT1 expression in the BM cells from the WT1-Tg and wild-type mice to confirm WT1 transgene expression. (D) WT1 expression levels in subpopulations of hematopoietic cells from the WT1-Tg and wild-type mice were measured by real-time RT-PCR. Relative WT1 expression level was defined as (copy number of the WT1 transcripts divided by that of β-actin transcripts) × 108. The mean values ± SD of duplicate measurements from 3 mice are shown.
Generation of WT1-transgenic mice. (A) Schematic model of the injected fragment used for generating WT1-transgenic mice. P1 and P2 represent the primers used for real-time RT-PCR. Three independent founders gave rise to lines 06, 13, and 14. (B) WT1 expression levels in the whole BM cells from the WT1-transgenic mice and wild-type littermates were measured by real-time RT-PCR. The mean values ± SD of duplicate measurements from 3 mice are shown. **P < .005; ***P < .001. (C) Western blotting analysis of WT1 expression in the BM cells from the WT1-Tg and wild-type mice to confirm WT1 transgene expression. (D) WT1 expression levels in subpopulations of hematopoietic cells from the WT1-Tg and wild-type mice were measured by real-time RT-PCR. Relative WT1 expression level was defined as (copy number of the WT1 transcripts divided by that of β-actin transcripts) × 108. The mean values ± SD of duplicate measurements from 3 mice are shown.
Coexpression of WT1 enhances the colony-forming activity of AML1-ETO-expressing myeloid progenitors and inhibits their differentiation at more immature stages
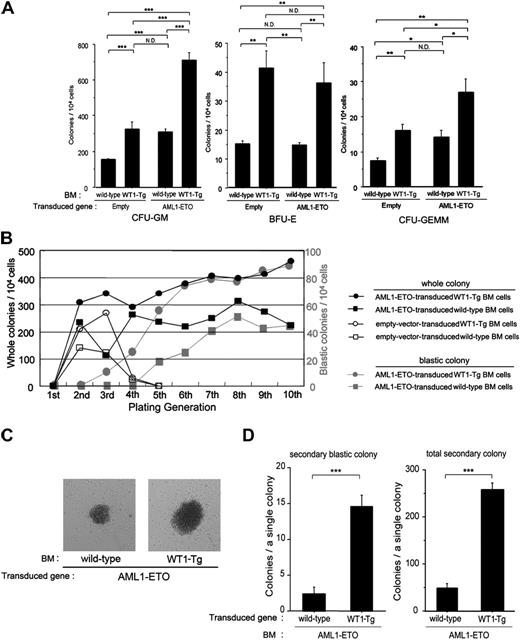
It has been reported that expression of AML1-ETO in hematopoietic progenitors inhibits the differentiation of myeloid progenitors, resulting in an expansion of myeloid clonogenic progenitors in vitro.3,4,11 To evaluate the effects of coexpression of WT1 in AML1-ETO-expressing progenitors, we transduced AML1-ETO into the BM cells from wild-type or WT1-Tg mice, FACS-sorted the GFP-positive cells in the BM cells, cultured the cells in methylcellulose containing SCF, IL-3, IL-6, and EPO, and counted the number of colonies after culturing for 8 days. We simultaneously performed similar experiments using an empty control vector. The number of first-generation colony-forming unit-granulocyte/macrophage (CFU-GM), burst-forming unit-erythroid (BFU-E), and colony-forming unit CFU-granulocyte, erythroid, macrophage, megakaryocyte (CFU-GEMM) colonies from the empty-vector-transduced WT1-Tg BM cells was significantly increased to approximately 2-fold more than those from the empty-vector-transduced wild-type BM cells (Figure 3A). On the other hand, while the number of first-generation CFU-GM and CFU-GEMM colonies from the AML1-ETO-transduced wild-type BM cells was also significantly increased to approximately 2-fold more than those from the empty-vector-transduced wild-type BM cells, the number of BFU-E colonies did not differ between them (Figure 3A). Strikingly, the number of first-generation CFU-GM and CFU-GEMM colonies from the AML1-ETO-transduced WT1-Tg BM cells was significantly increased to approximately 2-fold more and approximately 1.5-fold more, respectively, than those from the empty-vector-transduced WT1-Tg BM cells and the AML1-ETO-transduced wild-type BM cells (Figure 3A).
Next, we collected whole colonies, replated them in secondary methylcellulose cultures, and repeated the replating. Both AML1-ETO-transduced wild-type and WT1-Tg BM cells retained the replating activity and continued to produce myeloid colonies through repeated passages, but lost the replating activity of the erythroid colony by the third passage (data not shown). Furthermore, the number of colonies from AML1-ETO-transduced WT1-Tg BM cells gradually increased through repeated passages, whereas that from AML1-ETO-transduced wild-type BM cells remained virtually unchanged after the fourth passage (Figure 3B). On the other hand, empty-vector-transduced WT1-Tg BM cells retained a higher replating activity than empty-vector-transduced wild-type BM cells until the third passage, but the activity rapidly decreased on the fourth passage and was lost at the fifth passage (Figure 3B).
Coexpression of WT1 enhances colony-forming activity of AML1-ETO-expressing myeloid progenitors. (A) The number of first-generation colonies. The mean values ± SD of 5 independent assays for each group are shown. (B) Colony-replating assay. Empty-vector-transduced wild-type BM cells (□), empty-vector-transduced WT1-Tg BM cells (○), AML1-ETO-transduced wild-type BM cells (▪), AML1-ETO-transduced WT1-Tg BM cells (•). Gray squares and circles represent the numbers of blastic colonies in the AML1-ETO-transduced wild-type and WT1-Tg BM cell cultures, respectively. The mean values of 5 independent assays for each group are shown. (C) The eighth-generation blastic colonies from the AML1-ETO-transduced wild-type BM cells (left) and AML1-ETO-transduced WT1-Tg BM cells (right) are shown (original magnification, × 60). (D) The numbers of secondary colonies produced by a single blastic colony from the AML1-ETO-transduced wild-type BM cells and AML1-ETO-transduced WT1-Tg BM cells. Fifteen blastic colonies from each group were assayed and the mean values ± SD are shown. *P < .01; **P < .005; ***P < .001.
Coexpression of WT1 enhances colony-forming activity of AML1-ETO-expressing myeloid progenitors. (A) The number of first-generation colonies. The mean values ± SD of 5 independent assays for each group are shown. (B) Colony-replating assay. Empty-vector-transduced wild-type BM cells (□), empty-vector-transduced WT1-Tg BM cells (○), AML1-ETO-transduced wild-type BM cells (▪), AML1-ETO-transduced WT1-Tg BM cells (•). Gray squares and circles represent the numbers of blastic colonies in the AML1-ETO-transduced wild-type and WT1-Tg BM cell cultures, respectively. The mean values of 5 independent assays for each group are shown. (C) The eighth-generation blastic colonies from the AML1-ETO-transduced wild-type BM cells (left) and AML1-ETO-transduced WT1-Tg BM cells (right) are shown (original magnification, × 60). (D) The numbers of secondary colonies produced by a single blastic colony from the AML1-ETO-transduced wild-type BM cells and AML1-ETO-transduced WT1-Tg BM cells. Fifteen blastic colonies from each group were assayed and the mean values ± SD are shown. *P < .01; **P < .005; ***P < .001.
Conspicuous colonies consisting of tightly aggregated cells appeared in both the AML1-ETO-transduced wild-type and WT1-Tg BM cell cultures on the fifth and third passage, respectively (Figure 3B-C). These blastic colonies were formed mainly by myeloblasts, and the number of colonies gradually increased along with the passages (Figure 3B). Of interest, the size of these blastic colonies in the AML1-ETO-transduced WT1-Tg BM cell cultures (4.72 ± 1.03 mm diameter on 35-mm film; n = 10) was significantly larger than that in the AML1-ETO-transduced wild-type BM cell cultures (2.24 ± 0.80 mm diameter on 35-mm film; n = 10) (P < .001; Figure 3C).
Moreover, to examine how many secondary colonies were produced from a single blastic colony, we picked up and replated the blastic colonies in the eighth to tenth generation. One single blastic colony from both the AML1-ETO-transduced wild-type and WT1-Tg BM cells produced 2.4 ± 1.2 and 14.5 ± 3.5 secondary blastic colonies (P < .001), and 48.7 ± 8.5 and 257.2 ± 14.8 total secondary colonies (P < .001), respectively (Figure 3D). These results indicated that the coexpression of WT1 enhanced the colony-forming activity of AML1-ETO-expressing myeloid progenitors.
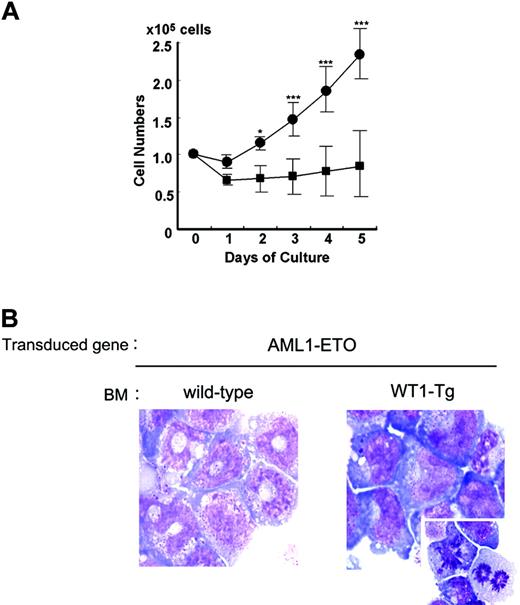
Next, we performed further experiments using myeloid progenitors to investigate the cooperative effects of WT1 on the expansion and differentiation of AML1-ETO-expressing myeloid progenitors. We collected colonies in their tenth or higher generation, removed differentiated cells from the whole colony cells, and cultured the purified lineage-negative (Lin-) cells in medium containing IL-3 and G-CSF. The Lin- cells from the AML1-ETO-transduced wild-type BM cell-derived colonies differentiated into myelocytes and metamyelocytes, but not into mature neutrophils, in response to G-CSF after culturing for 5 days (Figure 4A-B). In contrast, the Lin- cells from the AML1-ETO-transduced WT1-Tg BM cell-derived colonies did not differentiate into mature cells and instead proliferated in response to G-CSF (Figure 4A). Most of these cells were myeloblasts with finely reticulated chromatin and azurophilic cytoplasmic granules (Figure 4B).
These results indicate that AML1-ETO has the ability to inhibit the differentiation of myeloid progenitors and to change them into dysplastic ones with in vitro self-renewal ability and that coexpression of WT1 considerably enhanced the colony-forming activity of AML1-ETO-expressing myeloid progenitors and inhibited their differentiation at more immature stages.
Coexpression of WT1 inhibits the AML1-ETO-expressing myeloid progenitors at more immature stages and lets them expand. One hundred thousand lineage-negative (Lin-) cells of the AML1-ETO-transduced wild-type BM cell-derived colonies and AML1-ETO-transduced WT1-Tg BM cell-derived colonies were cultured in medium containing IL-3 and G-CSF for 5 days. (A) Proliferation of these cells in response to G-CSF + IL-3. Lin- cells of the AML1-ETO-transduced wild-type BM cell-derived colonies (▪) and Lin- cells of the AML1-ETO-transduced WT1-Tg BM cell-derived colonies (•). Numbers of viable cells were counted by the trypan blue exclusion method. The mean values ± SD of 5 independent assays for each group are shown. *P < .01; ***P < .001. (B) May-Giemsa staining of these cells after culturing for 5 days (original magnification, × 1000). Stainings were analyzed using a Zeiss AxioSkop2 Plus microscope and a 100 ×/1.30 NA oil objective (both from Carl Zeiss, Oberkochen, Germany), and images were processed using AxioVision software version 3.0 (Carl Zeiss) and Adobe Photoshop CS. (Left) Lin- cells of the AML1-ETO-transduced wild-type BM cell-derived colonies. Myelocytes and metamyelocytes can be seen. (Right) Lin- cells of the AML1-ETO-transduced WT1-Tg BM cell-derived colonies. Myeloblasts with finely reticulated chromatin, 2 or more nucleoli, gray-blue cytoplasm, a few azurophilic cytoplasmic granules, and myeloblasts undergoing cell division can be seen.
Coexpression of WT1 inhibits the AML1-ETO-expressing myeloid progenitors at more immature stages and lets them expand. One hundred thousand lineage-negative (Lin-) cells of the AML1-ETO-transduced wild-type BM cell-derived colonies and AML1-ETO-transduced WT1-Tg BM cell-derived colonies were cultured in medium containing IL-3 and G-CSF for 5 days. (A) Proliferation of these cells in response to G-CSF + IL-3. Lin- cells of the AML1-ETO-transduced wild-type BM cell-derived colonies (▪) and Lin- cells of the AML1-ETO-transduced WT1-Tg BM cell-derived colonies (•). Numbers of viable cells were counted by the trypan blue exclusion method. The mean values ± SD of 5 independent assays for each group are shown. *P < .01; ***P < .001. (B) May-Giemsa staining of these cells after culturing for 5 days (original magnification, × 1000). Stainings were analyzed using a Zeiss AxioSkop2 Plus microscope and a 100 ×/1.30 NA oil objective (both from Carl Zeiss, Oberkochen, Germany), and images were processed using AxioVision software version 3.0 (Carl Zeiss) and Adobe Photoshop CS. (Left) Lin- cells of the AML1-ETO-transduced wild-type BM cell-derived colonies. Myelocytes and metamyelocytes can be seen. (Right) Lin- cells of the AML1-ETO-transduced WT1-Tg BM cell-derived colonies. Myeloblasts with finely reticulated chromatin, 2 or more nucleoli, gray-blue cytoplasm, a few azurophilic cytoplasmic granules, and myeloblasts undergoing cell division can be seen.
AML1-ETO alone is not sufficient to induce leukemia
The mice that received a transplant of AML1-ETO-transduced wild-type BM cells showed abnormal myelopoiesis similar to myelodysplastic syndrome, in which immature myeloid cells significantly increased in the BM and spleen, but the mice did not develop leukemia after a long latency of approximately 10 months, as previously reported8-10,34 (Figure 5A; Tables 1, 2). Two of 8 mice died of anemia due to myelodysplasia.
Characteristics of the mice that received a transplant of AML1-ETO-transduced wild-type BM cells and AML1-ETO-transduced WT1-Tg BM cells
. | . | . | . | . | . | FACS analysis of peripheral blood, % (median) . | . | Weight, mg (median) . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transduced gene, BM . | n . | RBCs, × 1012/L (median) . | WBCs, × 109/L (median) . | Plts, × 109/L (median) . | GFP+ cells, % (median) . | Mac1+ Gr1low* . | Mac1+ Gr1high* . | Spleen . | Liver . | Phenotype . | ||
| Empty | ||||||||||||
| Wild type | 7 | 7.0-12.4 (8.8) | 3.3-17.2 (9.4) | 380-980 (560) | 30.5-91.8 (62.8) | 5.3-12.4 (10.2) | 12.5-80.5 (16.8) | 65-107 (82) | 750-1464 (1280) | Normal | ||
| WT1-Tg | 6 | 5.4-10.6 (8.9) | 4.0-12.1 (7.4) | 330-880 (550) | 31.4-73.7 (61.4) | 8.4-44.3 (18.1)† | 11.2-65.7 (41.2)† | 57-133 (120) | 1070-1556 (1395) | Granulocytosis | ||
| AML1-ETO | ||||||||||||
| Wild type | 8 | 4.5-10.9 (8.9) | 3.4-12.0 (6.5) | 200-900 (510) | 24.4-85.6 (50.8) | 15.4-58.2 (30.3)† | 8.1-51.0 (21.6) | 70-280 (168)† | 722-1900 (1520) | Dysplastic myelopoiesis | ||
| WT1-Tg | 10 | 2.8-9.1 (3.4)† | 3.1-35.1 (1.95)† | 120-370 (170)† | 67.4-96.1 (75.0) | 10.2-38.0 (21.4)† | 2.1-29.5 (3.5)† | 208-540 (345)† | 1335-2510 (2015)† | AML | ||
. | . | . | . | . | . | FACS analysis of peripheral blood, % (median) . | . | Weight, mg (median) . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transduced gene, BM . | n . | RBCs, × 1012/L (median) . | WBCs, × 109/L (median) . | Plts, × 109/L (median) . | GFP+ cells, % (median) . | Mac1+ Gr1low* . | Mac1+ Gr1high* . | Spleen . | Liver . | Phenotype . | ||
| Empty | ||||||||||||
| Wild type | 7 | 7.0-12.4 (8.8) | 3.3-17.2 (9.4) | 380-980 (560) | 30.5-91.8 (62.8) | 5.3-12.4 (10.2) | 12.5-80.5 (16.8) | 65-107 (82) | 750-1464 (1280) | Normal | ||
| WT1-Tg | 6 | 5.4-10.6 (8.9) | 4.0-12.1 (7.4) | 330-880 (550) | 31.4-73.7 (61.4) | 8.4-44.3 (18.1)† | 11.2-65.7 (41.2)† | 57-133 (120) | 1070-1556 (1395) | Granulocytosis | ||
| AML1-ETO | ||||||||||||
| Wild type | 8 | 4.5-10.9 (8.9) | 3.4-12.0 (6.5) | 200-900 (510) | 24.4-85.6 (50.8) | 15.4-58.2 (30.3)† | 8.1-51.0 (21.6) | 70-280 (168)† | 722-1900 (1520) | Dysplastic myelopoiesis | ||
| WT1-Tg | 10 | 2.8-9.1 (3.4)† | 3.1-35.1 (1.95)† | 120-370 (170)† | 67.4-96.1 (75.0) | 10.2-38.0 (21.4)† | 2.1-29.5 (3.5)† | 208-540 (345)† | 1335-2510 (2015)† | AML | ||
RBCs indicates red blood cells; WBCs, white blood cells; and Plts, platelets.
Mac-1+ Gr-1high populations represent mature segmented-form neutrophils, and Mac-1+ Gr-1low populations represent band-formed neutrophils.
Mac-1+ Gr-1low populations represent metamyelocytes and band-formed neutrophils.
GFP-positive gated-in population
P < .05 compared with the mice that received a transplant of empty vector-transduced wild-type BM cells
FACS analysis of the BM cells from the mice that received a transplant of AML1-ETO-transduced wild-type BM cells and AML1-ETO-transduced WT1-Tg BM cells
. | . | BM cells* . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Tranduced gene, BM . | n . | c-kit+ Lin-, % (median) . | c-kit+ Mac1–/low, % (median) . | Mac1+ Gr1low, % (median) . | Mac1+ Gr1high, % (median) . | |||
| Empty | ||||||||
| Wild type | 5 | 0.17-0.90 (0.77) | 3.43-5.43 (3.67) | 9.9-17.7 (15.6) | 23.1-38.2 (30.9) | |||
| WT1-Tg | 6 | 0.73-3.90 (2.00)† | 3.23-8.02 (5.20) | 11.5-35.2 (28.4)† | 37.2-49.0 (43.8)† | |||
| AML1-ETO | ||||||||
| Wild type | 6 | 5.85-13.5 (11.5)† | 22.7-35.4 (28.3)† | 22.0-39.1 (32.0)† | 20.4-32.1 (22.3) | |||
| WT1-Tg | 8 | 13.7-69.3 (38.7)† | 35.4-76.6 (60.5)† | 0.2-29.0 (13.9) | 0.2-12.3 (4.29)† | |||
. | . | BM cells* . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Tranduced gene, BM . | n . | c-kit+ Lin-, % (median) . | c-kit+ Mac1–/low, % (median) . | Mac1+ Gr1low, % (median) . | Mac1+ Gr1high, % (median) . | |||
| Empty | ||||||||
| Wild type | 5 | 0.17-0.90 (0.77) | 3.43-5.43 (3.67) | 9.9-17.7 (15.6) | 23.1-38.2 (30.9) | |||
| WT1-Tg | 6 | 0.73-3.90 (2.00)† | 3.23-8.02 (5.20) | 11.5-35.2 (28.4)† | 37.2-49.0 (43.8)† | |||
| AML1-ETO | ||||||||
| Wild type | 6 | 5.85-13.5 (11.5)† | 22.7-35.4 (28.3)† | 22.0-39.1 (32.0)† | 20.4-32.1 (22.3) | |||
| WT1-Tg | 8 | 13.7-69.3 (38.7)† | 35.4-76.6 (60.5)† | 0.2-29.0 (13.9) | 0.2-12.3 (4.29)† | |||
GFP-positive gated-in population
P < .05 compared with the mice that received a transplant of empty vector-transduced wild-type BM cells
AML1-ETO induces acute myeloblastic leukemia in cooperation with WT1
To investigate the cooperative effects of AML1-ETO and WT1 in in vivo hematopoiesis, we performed a transplantation assay.
The mice that received a transplant of empty-vector-transduced WT1-Tg BM cells appeared healthy but showed an increase in myelopoiesis in the BM and in mature granulocytosis in the PB and spleen, leading to mild splenomegaly (Table 1). Both Mac-1+Gr-1lo cells, representing myelocytes, metamyelocytes, and band-formed neutrophils, and Mac-1+Gr-1hi cells, representing mature segment-formed neutrophils, were more prominent in the GFP-positive BM cells from the mice that received a transplant of empty-vector-transduced WT1-Tg BM cells than those from the mice that received a transplant of empty-vector-transduced wild-type BM cells, and c-kit+Lin- cells were also significantly increased (2.38% ± 1.31% in the former, and 0.65% ± 0.39% in the latter; Figure 5A and Table 2).
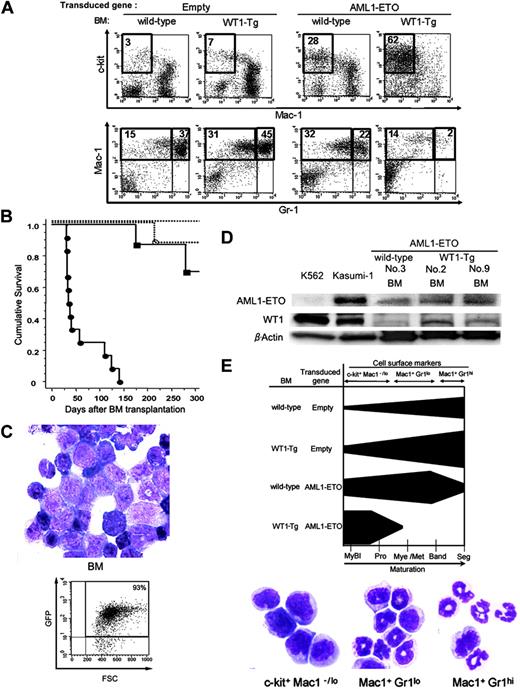
All of the mice that received a transplant of AML1-ETO-transduced WT1-Tg BM cells developed AML from 1 to 4 and a half months after bone marrow transplantation (BMT) (Figure 5B). These results were similar in the 3 founder lines, but it seemed that relatively longer periods were required to develop leukemia when we used the BM cells from WT1-Tg lines 13 and 14, in which WT1 was expressed at comparatively lower levels. These mice showed clinical characteristics such as severe anemia and elevated WBC counts (of which more than 40% were myeloblasts), thrombocytopenia, splenomegaly, and hepatomegaly (Table 1). The BM of the leukemic mice was hypercellular and occupied by immature myeloblasts with finely reticulated chromatin, 2 or more nucleoli, gray-blue cytoplasm, and delicate and azurophilic cytoplasmic granules, which were morphologically similar to human AML-M2 cells (Figure 5C). These leukemic cells also invaded the liver, spleen, kidney, and lung (data not shown). The leukemic cells were GFP positive and showed c-kit+Mac-1lo/-Gr-1- immature myeloblast phenotypes (Figure 5A). Approximately 45% to 60% of these leukemic cells also expressed CD34, while CD19 was expressed at very low frequencies (< 10%) (data not shown). The expression levels of AML1-ETO and WT1 proteins in the BM cells from the leukemic mice were lower than those in human t(8;21)-positive leukemic cells (Figure 5D).
Transplantation assay of the AML1-ETO-transduced BM cells. (A) Flow cytometric analysis of the GFP-positive BM cells from mice that received a transplant of empty-vector-transduced wild-type BM cells (left), empty-vector-transduced WT1-Tg BM cells (second from left), AML1-ETO-transduced wild-type BM cells (second from right), and AML1-ETO-transduced WT1-Tg BM cells (right). (B) Kaplan-Meier plot showing survival of mice after BM transplantation. Empty-vector-transduced wild-type BM cells (no symbol, n = 6), empty-vector-transduced WT1-Tg BM cells (○, n = 8), AML1-ETO-transduced wild-type BM cells (▪,n = 8), and AML1-ETO-transduced WT1-Tg BM cells (•, n = 12). (C) May-Giemsa staining (top) and FACS analysis of GFP expression (bottom) of BM cells from the leukemic mice (original magnification, × 1000). (D) Western blotting analysis of AML1-ETO and WT1 expression. Lysates of the BM cells from the mice that received a transplant of AML1-ETO-transduced wild-type or WT1-Tg BM cells were examined. K562 and Kasmi-1 cells were used as positive controls for WT1 and AML1-ETO, respectively. (E) (Top) Schematic representation of myelopoiesis in the BM from mice that received a transplant of empty-vector-transduced wild-type BM cells, empty-vector-transduced WT1-Tg BM cells, AML1-ETO-transduced wild-type BM cells, and AML1-ETO-transduced WT1-Tg BM cells. (Bottom) May-Giemsa staining of FACS-sorted BM cells from mice that received a transplant of empty-vector-transduced wild-type BM cells. MyBl indicates myeloblasts; Pro, promyelocytes; Mye/Met, myelocytes and metamyelocytes; Band, band-formed neutrophils; and Seg, segment-formed neutrophils. (C,E) Images were visualized and acquired as described in Figure 4B.
Transplantation assay of the AML1-ETO-transduced BM cells. (A) Flow cytometric analysis of the GFP-positive BM cells from mice that received a transplant of empty-vector-transduced wild-type BM cells (left), empty-vector-transduced WT1-Tg BM cells (second from left), AML1-ETO-transduced wild-type BM cells (second from right), and AML1-ETO-transduced WT1-Tg BM cells (right). (B) Kaplan-Meier plot showing survival of mice after BM transplantation. Empty-vector-transduced wild-type BM cells (no symbol, n = 6), empty-vector-transduced WT1-Tg BM cells (○, n = 8), AML1-ETO-transduced wild-type BM cells (▪,n = 8), and AML1-ETO-transduced WT1-Tg BM cells (•, n = 12). (C) May-Giemsa staining (top) and FACS analysis of GFP expression (bottom) of BM cells from the leukemic mice (original magnification, × 1000). (D) Western blotting analysis of AML1-ETO and WT1 expression. Lysates of the BM cells from the mice that received a transplant of AML1-ETO-transduced wild-type or WT1-Tg BM cells were examined. K562 and Kasmi-1 cells were used as positive controls for WT1 and AML1-ETO, respectively. (E) (Top) Schematic representation of myelopoiesis in the BM from mice that received a transplant of empty-vector-transduced wild-type BM cells, empty-vector-transduced WT1-Tg BM cells, AML1-ETO-transduced wild-type BM cells, and AML1-ETO-transduced WT1-Tg BM cells. (Bottom) May-Giemsa staining of FACS-sorted BM cells from mice that received a transplant of empty-vector-transduced wild-type BM cells. MyBl indicates myeloblasts; Pro, promyelocytes; Mye/Met, myelocytes and metamyelocytes; Band, band-formed neutrophils; and Seg, segment-formed neutrophils. (C,E) Images were visualized and acquired as described in Figure 4B.
We schematically summarized the cytologic and FACS results regarding myelopoiesis in the BM from the mice that received a transplant of empty-vector-transduced wild-type or WT1-Tg BM cells, and from the mice that received a transplant of AML1-ETO-transduced wild-type or WT1-Tg BM cells in Figure 5E.
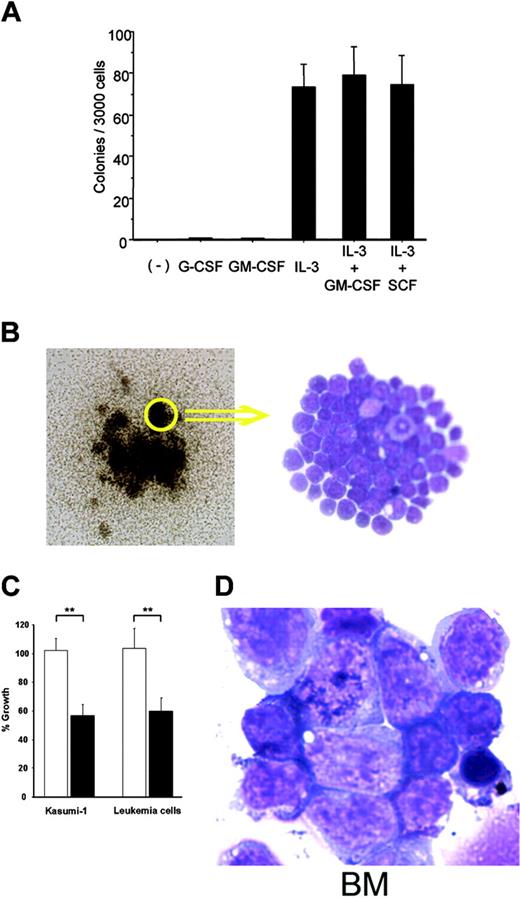
The inability of blast cells to differentiate into more mature cells is a hallmark of acute leukemia. To assess the differentiation ability of blast cells in leukemic mice, we cultured 3000 splenocytes from these mice in methylcellulose containing all possible combinations of IL-3, SCF, G-CSF, and GM-CSF. Sixty to 97 colonies per 3000 splenocytes were generated in the presence of IL-3, IL-3 + SCF, or IL-3 + GM-CSF, but not without growth factors, as is observed with most human myeloid leukemic cells35 (Figure 6A). These colonies formed tightly aggregated cell clusters that were entirely composed of immature myeloblasts (Figure 6B). Furthermore, we were able to establish 3 cell lines that grew in the presence of SCF and IL-3. To evaluate whether these leukemic cells required WT1 for their proliferation, we examined the effects of the suppression of WT1 expression by WT1 antisense oligomers (WT1 AS) on the growth of the 3 established leukemic cell lines and Kasmi-1. The growth of these cells was significantly inhibited by WT1 AS (Figure 6C).
Moreover, to test whether blast cells from the leukemic mice were transformed, we transplanted 1 × 107 splenocytes from these mice into lethally irradiated wild-type recipient mice. The mice that received the transplanted splenocytes developed fatal AML 2 to 6 weeks after the transplantation (Figure 6D). These results showed that blast cells in the leukemic mice were transplantable and thus transformed.
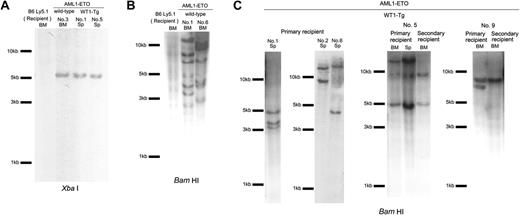
To evaluate the clonality of leukemic cells, we performed Southern hybridization analysis of integrated provirus using BM and spleen cells from primary and secondary leukemic recipients. The DNA was digested with XbaI, which has one cutting site within the LTRs or with BamHI, which has one cutting site within the AML1-ETO gene of MSCV AML1-ETO IRES GFP provirus, and probed the digested DNA with IRES sequences. XbaI digestion of the DNA from the BM or spleen cells from the mice that received a transplant of AML1-ETO-transduced wild-type or WT1-Tg BM cells yielded the single expected 5-kb band, indicating that the integrated retrovirus vectors were intact (Figure 7A). When the DNA was digested with BamHI, BM cells from the mice that received a transplant of AML1-ETO-transduced wild-type BM cells yielded many strong bands (Figure 7B). On the other hand, leukemic cells from primary and secondary recipients that received a transplant of AML1-ETO-transduced WT1-Tg BM cells yielded 2 to 3 and 1 to 2 predominant bands, respectively (Figure 7C). These results indicated that BM cells from the mice that received a transplant of AML1-ETO-transduced wild-type BM cells were polyclonal, whereas the leukemic cells from the mice that received a transplant of AML1-ETO-transduced WT1-Tg BM cells were monoclonal or oligoclonal.
Discussion
Leukemia-associated fusion proteins generally function as aberrantly activated signaling or transcriptional regulators that directly interfere with the differentiation, self-renewal, and/or proliferation of normal hematopoietic cells.36 AML1-ETO is a chimeric gene frequently detected in AML.1,2 In the present study, and in several previously reported investigations using murine models, it was demonstrated that AML1-ETO was not able to induce leukemia independently.3-10,34 However, it was shown that AML1-ETO inhibited the differentiation of myeloid progenitors, resulting in an increase in their replating activity and an expansion of clonogenic myeloid progenitors in vitro,3,4 and induced dysplastic myelopoiesis in vivo, in which immature myeloid cells significantly increased in the BM and spleen.5,8,10 These results indicated that AML1-ETO-expressing hematopoietic progenitors remained responsive to normal in vivo homeostatic controls, were at preleukemic stages, and required additional genetic events to complete full leukemic transformation. The effects of AML1-ETO expression on the differentiation of myeloid progenitors were examined in myeloid cell lines that retained the ability to terminally differentiate.37 The expression and transcriptional activity of AML1, C/EBPα, and PU.1, which are important transcription factors in normal myeloid differentiation, were down-regulated and/or inactivated by AML1-ETO, resulting in the inhibition of myeloid differentiation.38-42 However, the mechanisms by which AML1-ETO promoted the proliferation of myeloid progenitors is not well understood. Linggi et al recently demonstrated that AML1-ETO directly repressed the p14ARF promoter and reduced the expression level of the p14ARF tumor-suppressor gene,43 which is expressed in response to various types of activated oncogenes, including Ras, c-myc, Abl, and E2F-1, and which antagonizes the MDM2-mediated destabilization of p53, resulting in induction of G1 and G2 arrest.44-49 The repression of p14ARF mediated by AML1-ETO may impair p53-mediated growth arrest and/or apoptosis in response to the activated oncogenes and contribute to prolonging the survival of myeloid progenitors, allowing them to become susceptible to other genetic alterations, and eventually leading to leukemia.
Transformation assays of the myeloblasts from the leukemic mice. (A) Colony assays of splenocytes from leukemic mice in methylcellulose medium-containing cytokines. The mean values ± SD of 3 independent assays are shown. (B) Leukemic blast cell colonies formed in methylcellulose containing IL-3 and GM-CSF (left; original magnification, × 60) and May-Giemsa staining of cells from these colonies (right; original magnification, × 400) are shown. (C) Growth inhibition of leukemic blast cells by WT1 antisense oligomers. Leukemic blast cells and AML1-ETO-expressing Kasmi-1 cells were treated with WT1 AS or random oligomers, and the numbers of viable cells were counted at 72 hours after the start of treatment. Open and closed columns represent random oligomers and WT1 AS, respectively. The percent growth was defined as the ratio of the number of viable cells treated with WT1 AS or random oligomers to that of cells treated with PBS. **P < .005. (D) May-Giemsa-stained BM cells from mice that received a transplant of the splenocytes from the leukemic mice (original magnification, × 1000). (B right, D) Images were visualized and acquired as described in Figure 4B.
Transformation assays of the myeloblasts from the leukemic mice. (A) Colony assays of splenocytes from leukemic mice in methylcellulose medium-containing cytokines. The mean values ± SD of 3 independent assays are shown. (B) Leukemic blast cell colonies formed in methylcellulose containing IL-3 and GM-CSF (left; original magnification, × 60) and May-Giemsa staining of cells from these colonies (right; original magnification, × 400) are shown. (C) Growth inhibition of leukemic blast cells by WT1 antisense oligomers. Leukemic blast cells and AML1-ETO-expressing Kasmi-1 cells were treated with WT1 AS or random oligomers, and the numbers of viable cells were counted at 72 hours after the start of treatment. Open and closed columns represent random oligomers and WT1 AS, respectively. The percent growth was defined as the ratio of the number of viable cells treated with WT1 AS or random oligomers to that of cells treated with PBS. **P < .005. (D) May-Giemsa-stained BM cells from mice that received a transplant of the splenocytes from the leukemic mice (original magnification, × 1000). (B right, D) Images were visualized and acquired as described in Figure 4B.
We have previously reported the following: (1) almost all leukemias, including t(8;21)-positive AML, expressed wild-type WT1 at high levels,20-22 (2) the growth of WT1-expressing leukemia cells was suppressed by WT1 antisense oligomers,28 (3) WT1 overexpression in 32Dcl3 murine myeloid progenitors and human hematopoietic progenitors inhibited their differentiation and promoted their proliferation in response to G-CSF.26,27 These results indicated that WT1 is involved in leukemogenesis. A variety of genes involved in proliferation, differentiation, and apoptosis have been identified as targets of WT1.14 Han et al50 recently showed that WT1 caused an increase in the expression level of c-myc, which is one of the early growth response genes and plays an essential role in cell proliferation and oncogenesis, by up-regulation of its promoter.51-53 The increased expression of c-myc in the constitutive WT1-overexpressing hematopoietic progenitors may let them enter the cell cycle and promote G1- to S-phase progression, resulting in an increase in their proliferation.
In the present study, we showed that WT1-Tg BM cells produced more myeloid colonies than wild-type BM cells and that the mice that received a transplant of WT1-Tg BM cells showed an increase in myelopoiesis. These results indicated that WT1 overexpression in hematopoietic cells expanded the pool of hematopoietic progenitors, especially of myeloid progenitors, resulting in an increase in myelopoiesis. WT1 is expressed in hematopoietic stem/progenitor cells and is down-regulated with the differentiation of these cells in normal hematopoiesis, but when down-regulation of WT1 expression is impaired by an as yet undetermined mechanism in the hematopoietic progenitors, constitutive WT1 overexpression lets hematopoietic progenitors expand and thus makes them susceptible to genetic alterations that subsequently lead to leukemia.
These findings led us to investigate whether AML1-ETO induced leukemia in cooperation with WT1 overexpression as one of the putative additional genetic events. We showed that AML1-ETO-expressing dysplastic myeloid progenitors enhanced the colony-forming activity and inhibited the differentiation at more immature stages in cooperation with WT1 overexpression and that the mice that received a transplant of AML1-ETO-transduced WT1-Tg BM cells rapidly developed AML similar to AML-M2. The results presented here are consistent with the general postulate that the development of leukemia is a stepwise process in which the synergistic effects of several genetic alterations lead to leukemic transformation (which features increased cell survival, promoted self-renewal and proliferation, inhibition of differentiation, and genomic instability) and ultimately cause leukemia.54 According to this model, in the first step, the expression of AML1-ETO expanded the dysplastic myeloid progenitors and inhibited their differentiation into mature myeloid cells. In the second step, the constitutive WT1 overexpression significantly expanded these dysplastic progenitors, inhibited their differentiation at more immature stages, and enhanced AML1-ETO-involved leukemogenesis. Finally, these synergistic effects allowed hematopoietic progenitors to be transformed.
Proviral integration in leukemic mice. Southern hybridization analysis of genomic DNA from leukemic mice. All blots were hybridized with the IRES probe. (A) DNA was digested with XbaI, which has one recognition site within the LTRs of the MSCV AML1-ETO IRES GFP provirus. (B) The genomic DNA of BM cells from the mice that received a transplant of the AML1-ETO-transduced wild-type BM cells and wild-type C57BL/6 Ly5.1 recipient mice was digested with BamHI, which has one recognition site within the AML1-ETO gene of the MSCV AML1-ETO IRES GFP provirus. (C) The genomic DNA of BM and spleen cells from the primary leukemic mice and secondary recipients was digested with BamHI. BM indicates bone marrow cells; Sp, spleen cells.
Proviral integration in leukemic mice. Southern hybridization analysis of genomic DNA from leukemic mice. All blots were hybridized with the IRES probe. (A) DNA was digested with XbaI, which has one recognition site within the LTRs of the MSCV AML1-ETO IRES GFP provirus. (B) The genomic DNA of BM cells from the mice that received a transplant of the AML1-ETO-transduced wild-type BM cells and wild-type C57BL/6 Ly5.1 recipient mice was digested with BamHI, which has one recognition site within the AML1-ETO gene of the MSCV AML1-ETO IRES GFP provirus. (C) The genomic DNA of BM and spleen cells from the primary leukemic mice and secondary recipients was digested with BamHI. BM indicates bone marrow cells; Sp, spleen cells.
The integrated provirus numbers and integrated sites did not appear to influence the disease, because all of the leukemic mice demonstrated similar clinical and phenotypic characteristics and short latency of disease development. However, the possibility that spontaneous mutations occurred within a short time was not excluded because there were dominant clones that proliferated in the secondary recipients.
The interferon consensus binding protein (ICSBP) gene, which encodes growth inhibitory signaling molecules, is down-regulated in 66% and 79% of patients with AML and CML, respectively, and ICSBP-deficient mice show CML-like disease.55,56 Schwieger et al showed that AML1-ETO induced myeloblastic transformation under conditions of ICSBP deficiency.9 However, this transformation was granulocytic sarcoma-like, not lethal AML. TEL-PDGFβR fusion protein, which is generated by the (5;12) translocation and constitutively activates tyrosine kinase, is mainly associated with chronic myelomonocytic leukemia (CMML), and patients with t(5;12)-positive CMML sometimes develop AML with t(8;21).57,58 Grisolano et al demonstrated that AML1-ETO cooperated with TEL-PDGFβR to cause AML.10 FLT3 is a receptor tyrosine kinase expressed in immature hematopoietic cells, and length mutation of the FLT3 gene is detected in approximately 30% of patients with AML, especially in 40% of t(15;17)-positive AML.59 It has been shown that FLT3 length mutation mediates proliferation and survival of hematopoietic progenitors, but does not affect their differentiation.59 In a recent study, Schessl et al demonstrated that AML1-ETO induced AML and acute lymphoblastic leukemia (ALL) in cooperation with FLT3 length mutation.34 We demonstrated that constitutive WT1 overexpression in the hematopoietic cells increased hematopoietic progenitors and induced myeloproliferation. Therefore, WT1 may alter signal transduction for proliferation and differentiation of hematopoietic progenitors and thereby play oncogenic roles associated with leukemogenesis.
In conclusion, we showed that AML1-ETO was able to induce AML in cooperation with WT1 overexpression.
Prepublished online as Blood First Edition Paper, December 27, 2005; DOI 10.1182/blood-2005-04-1656.
Supported in part by a Grant-in Aid from the Ministry of Education, Science, Sports, and Culture, Japan.
S.N. designed research, performed research, analyzed data, and wrote the paper; N.H. designed research and performed research; T.S., K.K., M.Y., S.-i.N., Y. Hoshida, T.N., Y. Harada, N.T., A.T., M.K., K.A., and I.K. performed research; Y. Oka wrote the paper; Y. Oji designed research and performed research; and H.S. designed research, wrote the paper, and directed this study in chief.
S.N. and N.H. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The human AML1-ETO cDNA was a generous gift from M. Ohki (National Cancer Center of Japan, Tokyo, Japan). The mouse WT1(17AA+/KTS+) cDNA was a generous gift from H. Nakagama (National Cancer Center of Japan). The mouse tec promoter was a generous gift from H. Mano (Jichi Medical School, Tochigi, Japan). We thank H. Enomoto and T. Naka (Osaka University Graduate School of Medicine, Osaka, Japan) for technical advice, C. Tanaka (Osaka University Graduate School of Medicine) for technical assistance, and also thank T. Kurosawa and M. Tajima, and other staff of the Central Institute of Experimental Animals, Osaka University Graduate School of Medicine, for general advice regarding animal experimentation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal