Abstract
Dendritic cells (DCs) are recognized as the most potent antigen-presenting cells of the immune system with the unique ability to initiate and maintain primary immune responses. In order to better characterize the functional and phenotypic features of DCs, a subtractive cDNA library to identify differentially expressed genes in monocyte-derived DCs (MDCs) was constructed. Using this approach, we found that the epithelial transcription factor ESE-3, which was previously shown to be exclusively expressed in cells of epithelial origin, is differentially expressed in MDCs. This was further confirmed by reverse transcriptase–polymerase chain reaction (RT-PCR) and Western blot analyses. The expression of ESE-3 is up-regulated upon maturation of MDCs and inhibited by treating the cells with IL-10 or IFN-γ. Knockdown experiments using siRNA suggest that ESE-3 plays an important role during MDC development. Our results might help to improve the phenotypic characterization of DCs and lead to a better understanding of the cellular mechanisms involved in antigen presentation and T-cell stimulation.
Introduction
Dendritic cells (DCs) are the most potent antigen-presenting cells and play an important role in initiating and maintaining primary immune responses. DCs reside in peripheral tissues in an immature state in which they are able to take up and process antigens. Upon stimulation with inflammatory cytokines, bacterial compounds, or T-cell interaction they undergo a maturation process involving up-regulation of costimulatory and major histocompatibility complex (MHC) class II molecules on the cell surface, leading to the increased ability to migrate to the draining lymph nodes and to stimulate naive T cells.
DCs can be divided into various subsets distinguished by their phenotypic and functional properties that can be found in different organs and peripheral blood.1 In vitro, CD34+ precursor cells or CD14+ peripheral-blood monocytes can be induced to differentiate into DCs using different cytokine combinations.2,3 Although DCs can be generated from monocytes, the phenotypic and functional properties between these 2 cell types vary considerably.
The identification of differentially expressed genes during differentiation of DCs from monocytes is an important step toward a better characterization of this cell population and understanding of pathways involved in DC development. This aims at a more specific phenotypic classification of subpopulations and a better understanding of the cellular mechanisms involved in antigen presentation and T-cell stimulation in order to use this knowledge for in vitro manipulation of DCs for the specific induction of T-cell responses in patients.
The Ets transcription factors constitute an important family of transcriptional regulators during development.4-10 They are characterized by a conserved winged helix-turn-helix DNA binding domain (Ets domain) that enables them to bind the core DNA sequence -GGA(A/T). Several members of this family are proto-oncogenes involved in cancer development and tumor invasiveness. While most Ets transcription factors are ubiquitously expressed, there are exceptions such as the epithelial-specific subfamily of these transcription factors, ESE. So far, 3 members of this family are known, ESE-1/ELF3/ESX, ESE-2/ELF5, and ESE-3/EHF.11-13 Their expression is limited to epithelial cells, but not all epithelial cells express these transcription factors. Functionally, ESE-3 has been shown to be a transcriptional repressor of several genes positively regulated by MAP kinase signaling with a dependency on a high-affinity binding site.14 Moreover, linkage analyses have suggested involvement of ESE-3 in the susceptibility of asthma.15
In our study, we demonstrate that ESE-3 is differentially expressed in human DCs and might be critically involved in their development.
Materials and methods
Dendritic-cell generation
DCs were generated from peripheral blood–adhering monocytes by either magnetic cell sorting or plastic adherence as described previously.16 In some experiments, maturation of the cells was induced by addition of TNF-α (10 ng/mL; R&D Systems, Wiesbaden, Germany), LPS (100 ng/mL; Sigma-Aldrich, Deisenhofen, Germany), CD40L (500 ng/mL; Bender MedSystems, Vienna, Austria), and IFN-γ (100 U/mL; R&D Systems), Pam3Cys (5 μg/mL; EMC microcollections, Tübingen, Germany), Poly(I:C) (50 μg/mL; Sigma-Aldrich), or R848 (2 μg/mL; InvivoGen, San Diego, CA) for 24 hours. For the inhibition of DC development, IL-10 (10 ng/mL; R&D Systems) was used. IL-6 (100 ng/mL) was purchased from R&D Systems. The generation of CD34+-derived DCs was performed as described previously.17
Direct isolation of cells from peripheral blood and bone marrow
Magnetic cell sorting (MACS) technology was used to isolate various cell populations from peripheral blood and bone marrow according to the manufacturer's instructions (Blood Dendritic Cell Isolation Kit, CD1c Dendritic Cell Isolation Kit, BDCA-4 Cell Isolation Kit, CD19 Microbeads, CD3 Microbeads, CD235a Microbeads, CD33 Microbeads, CD61 Microbeads; all from Miltenyi Biotec, Bergisch Gladbach, Germany). Granulocytes were isolated by combined Histopaque-1119 (Sigma-Aldrich) and Ficoll/Paque (Biochrom, Berlin, Germany) density gradient centrifugation. Erythrocytes were lysed by incubation with a buffer containing 155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.3. Macrophages were generated by culturing CD14+ monocytes in RP10 medium (RPMI 1640 with glutamax-I, supplemented with 10% inactivated FCS, and antibiotics [Invitrogen, Karlsruhe, Germany]) supplemented with GM-CSF (100 ng/mL, Leukine Liquid Sargramostim; Berlex Laboratories, Richmond, CA) for 24 hours (macrophages day 1) or 48 hours (macrophages day 2).
For activation of the cells, B cells were incubated with CD40L (100 ng/mL) and IL-4 (20 ng/mL; R&D Systems); T cells, with phorbol 12-myristate 13-acetate (PMA; 25 ng/mL; Sigma-Aldrich) and ionomycin (1 μg/mL; Sigma-Aldrich); and granulocytes, with LPS (10 ng/mL), for 24 hours.
Immunostaining
Immunostaining was performed as described previously16 and analyzed on a FACSCalibur cytometer (BD Biosciences, Heidelberg, Germany). A proportion of 1% false-positive events was accepted in the negative control samples.
siRNA construction and transfection of DCs
siRNA directed against ESE-3 was synthesized using the siRNA Construction Kit (Ambion, Austin, TX) as described by the manufacturer. The DNA oligonucleotides used were as follows: antisense: 5′-aagtggaacggccagtgcagtcctgtctc; sense: 5′-aaactgcactggccgttccaccctgtctc.As a control, siRNAagainst firefly luciferase was used as described by Elbashir et al18 (GL2antisense: aacgtacgcggaatacttcgacctgtctc; GL2sense: 5′-aatcgaagtattccgcgtacgcctgtctc).
The ready-to-use siRNA was stored at -20°C. For transfection, freshly isolated CD14+ monocytes were resuspended in X-VIVO 20 medium in a final concentration of 1 to 2 × 107/mL. Subsequently, 200-μL cell suspension was transferred to a 4-mm cuvette, mixed with 5 to 100 ng siRNA, and electroporated using an Easyject Plus unit (Peqlab, Erlangen, Germany) with voltage of 300 V, capacitance of 150 μF, resistance of 1540 Ω, and pulse time of 231 ms. After electroporation, the cells were immediately transferred into RP10 medium supplemented with GM-CSF (100 ng/mL) and IL-4 (20 ng/mL).
Reverse transcriptase–polymerase chain reaction (RT-PCR)
Total RNA was isolated from cell lysates using QIAGEN RNeasy Mini anion-exchange spin columns (QIAGEN, Hilden, Germany) according to the instructions of the manufacturer. Up to 200 ng total RNA was subjected to a 20-μL cDNA synthesis reaction using SuperScript RTII (Invitrogen). Oligo(dT) was used as primer. cDNA (1 μL) was used in a 15-μL PCR amplification reaction. Gene-specific primers used were ESE3F: 5′-tgacctgttccagtccacac; ESE3R: 5′-tgtctgggttcaagaggatg; GAPDHF: 5′-gaaggtgaaggtcggagtc; GAPDHR: 5′-gaagatggtgatgggatttc; β2microglobulin (β2m)F: gggtttcatccatccgacat; and β2mR: gatgctgcttacatgtctcga.
Real-time quantitative RT-PCR
Total RNA was isolated from cell lysates using QIAGEN RNeasy Mini anion-exchange spin columns according to the instructions of the manufacturer. Up to 1 μg total RNA was subjected to a 20-μL cDNA synthesis reaction using SuperScript RTII (Invitrogen). Oligo(dT) was used as primer.Approximately 100 ng cDNA was used in the real-time quantitative RT-PCR using Taqman technology on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Primers and probes used were ESE3-FP: 5′-agcatgagtttgcaggagttcac; ESE3-RP: 5′-ttgttcagtcttgacaatgacattgt; ESE3-probe: 5′-FAM-actactgcactggccgttccacttcagat-TAMRA; GAPDH-FP: 5′-ccacatcgctcagacaccat; GAPDH-RP: 5′-ggcaacaatatccactttaccagagt; GAPDH-probe: 5′-FAM-accaaatccgttgactccgaccttca-TAMRA.
Cytokine determination
Cytokine concentrations in supernatants from DC cultures were measured with commercially available 2-site sandwich enzyme-linked immunosorbent assays (ELISAs) from Beckmann Coulter (Hamburg, Germany) according to the manufacturer's instructions.
Polyacrylamide gel electrophoresis (PAGE) and Western blotting
Nuclear extracts were prepared from DCs as described previously.17,19 For the detection of ESE-3, approximately 20 μg nuclear extracts was separated on a 12% sodium dodecyl sulfate (SDS)–polyacrylamide gel and transferred onto nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). Ponceau S staining of the membrane was performed to confirm that equal amounts of protein were present in every lane. The blot was probed with a rat monoclonal antibody against ESE-3 (5A5.5, a kind gift of A. Tugores). For analysis of nuclear amount of RelB, a rabbit polyclonal antibody (C-19; Santa Cruz Biotechnology, Santa Cruz, CA) was used.
Immunohistochemistry
To study the localization of ESE-3 in Langerhans cells, 5-μm cryosections of normal skin were fixed for 2 minutes with paraformaldehyde-lysine-periodate solution, blocked with normal donkey serum, and incubated overnight with a mix of goat anti–ESE-3 (N-20; Santa Cruz Biotechnology) at a 1:30 dilution and with undiluted monoclonal anti-CD1a (Immunotech, Marseille, France). Fluorochrome labeling was performed with a mix of Cy3-donkey antigoat serum and Cy5-donkey antimouse serum (Dianova, Hamburg, Germany), both diluted 1:500. Nuclei were stained with YOPRO (Molecular Probes, Leiden, The Netherlands) for 5 minutes. The sections were analyzed with a confocal laser scanning microscope (Leica TCS SP; Leica Microsystems, Bensheim, Germany), equipped with a PL Fluotar 40 ×/1.00-0.50 numeric aperture oil objective. Images were taken using Leica PowerScan Software TCS SP1.
Results
ESE-3 is differentially expressed in monocyte-derived DCs
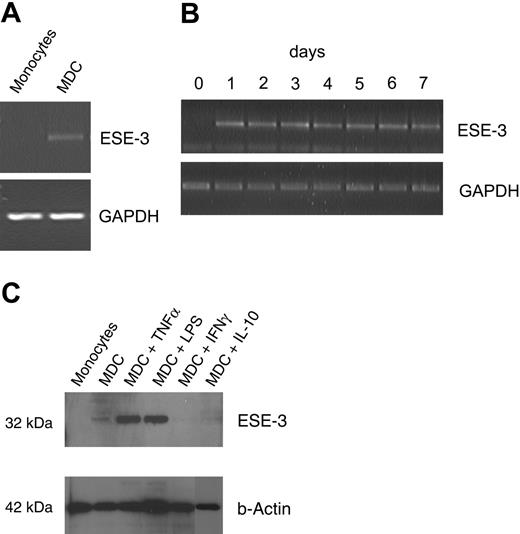
In this study, we used a PCR-based subtraction technique to identify genes differentially expressed in DCs during their development from peripheral-blood monocytes as described previously.20 One of the identified clones encoded for the epithelial-specific transcription factor ESE-3. The differential expression in MDCs but not monocytes (isolated with the Monocyte Isolation Kit; Miltenyi Biotec) was primarily verified by RT-PCR (Figure 1A). ESE-3 transcription could be detected already 24 hours after culturing peripheral-blood monocytes with GM-CSF and IL-4 (Figure 1B). The differential expression of ESE-3 in MDCs but not monocytes was further confirmed by Western blotting with nuclear extracts using the rat monoclonal antibody 5A5.514 (Figure 1C lanes 1-2). Interestingly, treatment of cells with IL-10, which inhibits DC development and antigen-presenting function,21 resulted in the inhibition of ESE-3 expression. Surprisingly, like IL-10, addition of IFN-γ to the culture medium inhibited the induction of this transcription factor.
Epithelium-specific transcription factor ESE-3 is expressed in DCs. Untouched CD14+ peripheral-blood monocytes isolated by magnetic depletion and monocyte-derived DCs (MDCs) were analyzed for the expression of ESE-3 by RT-PCR using a gene-specific primer pair (A-B) and Western blotting of nuclear extracts using the ESE-3–specific monoclonal rat antibody 5A5.5 (C). For maturation of DCs, TNF-α, LPS, and IFN-γ were added to the cell cultures 24 hours before harvesting the cells. IL-10 was added from the first day of cell culture.
Epithelium-specific transcription factor ESE-3 is expressed in DCs. Untouched CD14+ peripheral-blood monocytes isolated by magnetic depletion and monocyte-derived DCs (MDCs) were analyzed for the expression of ESE-3 by RT-PCR using a gene-specific primer pair (A-B) and Western blotting of nuclear extracts using the ESE-3–specific monoclonal rat antibody 5A5.5 (C). For maturation of DCs, TNF-α, LPS, and IFN-γ were added to the cell cultures 24 hours before harvesting the cells. IL-10 was added from the first day of cell culture.
In contrast, treatment of MDCs with factors that mediate DC activation, such as TNF-α or LPS, led to a distinct up-regulation of ESE-3 mRNA (data not shown) as well as protein (Figure 1C).
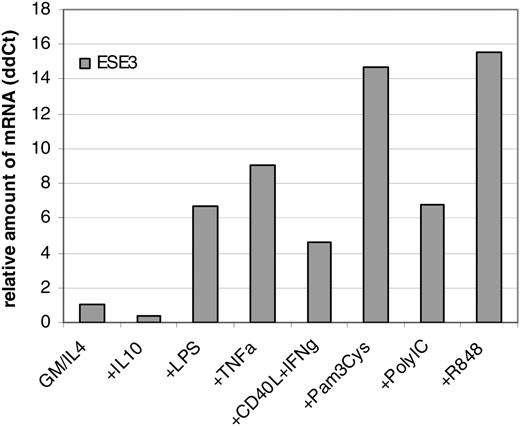
In order to further quantify the changes of ESE-3 expression, real-time quantitative RT-PCR was performed. For this purpose, CD14+ peripheral-blood monocytes were incubated for 6 days in RP10 medium supplemented with GM-CSF and IL-4 as well as substances known to inhibit DC generation from monocytes such as IL-10.21 Moreover, several compounds promoting maturation of DCs, such as LPS, TNF-α, CD40L/IFN-γ, IFN-α, and toll-like receptor (TLR) ligands Pam3Cys (TLR2L), Poly(I:C)(TLR3L), and R848 (TLR7L), were included in the analysis. The quantification was performed in relation to GAPDH expression. In line with previous results, the inhibiting cytokine IL-10 reduced the relative amount of ESE-3 mRNA (Figure 2). Maturation of DCs led to an up-regulation of ESE-3 transcription (Figure 2).
Real-time quantitative RT-PCR of differentially stimulated MDCs. CD14+ peripheral-blood monocytes were incubated for 6 days in RP10 medium supplemented with GM-CSF and IL-4. IL-10 was added from the first day of culture; stimulating substances were added 24 hours before harvesting of the cells. The amount of mRNA was analyzed in relation to GAPDH.
Real-time quantitative RT-PCR of differentially stimulated MDCs. CD14+ peripheral-blood monocytes were incubated for 6 days in RP10 medium supplemented with GM-CSF and IL-4. IL-10 was added from the first day of culture; stimulating substances were added 24 hours before harvesting of the cells. The amount of mRNA was analyzed in relation to GAPDH.
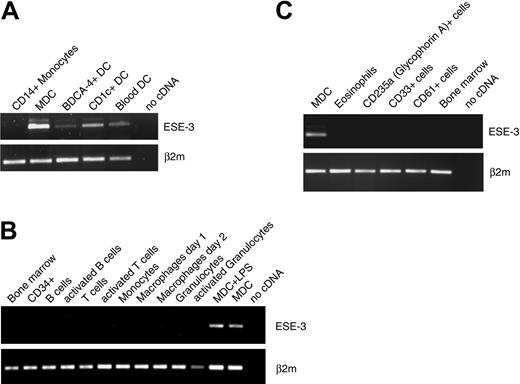
In the next set of experiments, we analyzed the expression of ESE-3 in DCs generated from CD34+ progenitor cells or DC subpopulations (BDCA4+, CD1c+) directly isolated from buffy coat preparations with MACS technology. ESE-3 transcripts were found in all analyzed cell populations by RT-PCR (Figure 3A). Further RT-PCR analyses revealed that DCs are the only hematopoietic cells that express ESE-3. ESE-3 transcripts were not detected in isolated CD34+ peripheral-blood progenitor cells; bone marrow; purified resting or activated B cells, T cells, and granulocytes; or macrophages (Figure 3B). In addition, analyses of eosinophils and also erythroid cells (glycophorin A/CD235a+), myeloid precursor cells (CD33+), and megakaryocytes (CD61+) isolated directly from the bone marrow revealed no ESE-3 transcript in these cells (Figure 3C).
DCs are the only hematopoietic cells that express ESE-3. The expression of ESE-3 was analyzed by RT-PCR in (A) DC populations directly isolated from buffy coat preparations; in (B) bone marrow, isolated CD34+ peripheral-blood progenitor cells, and purified resting or activated B cells, T cells, and granulocytes, as well as macrophages; and in (C) eosinophils, erythroid cells (CD235a+), myeloid precursor cells (CD33+), and megakaryocytes (CD61+).
DCs are the only hematopoietic cells that express ESE-3. The expression of ESE-3 was analyzed by RT-PCR in (A) DC populations directly isolated from buffy coat preparations; in (B) bone marrow, isolated CD34+ peripheral-blood progenitor cells, and purified resting or activated B cells, T cells, and granulocytes, as well as macrophages; and in (C) eosinophils, erythroid cells (CD235a+), myeloid precursor cells (CD33+), and megakaryocytes (CD61+).
ESE-3 is expressed in Langerhans cells
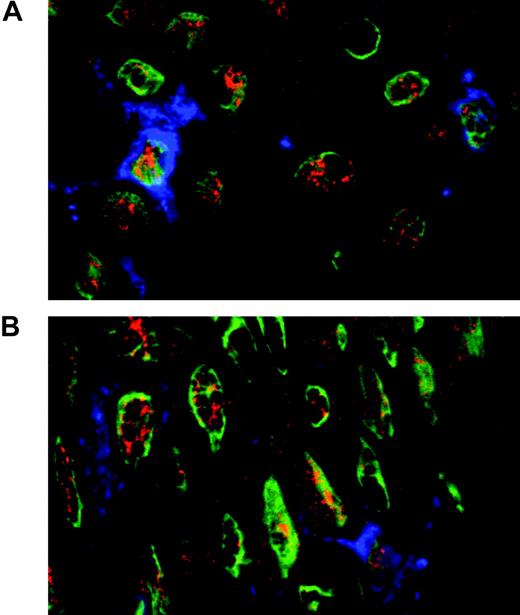
In order to further extend our analysis of ESE-3 expression in DC populations, immunohistologic staining of normal skin was performed. To identify Langerhans cells, CD1a staining was included. As shown in Figure 4, nuclear ESE-3 expression can be detected in CD1a+ Langerhans cells. ESE-3 protein is also present in CD1a- keratinocytes.
Down-regulation of ESE-3 during DC development leads to an impaired DC phenotype
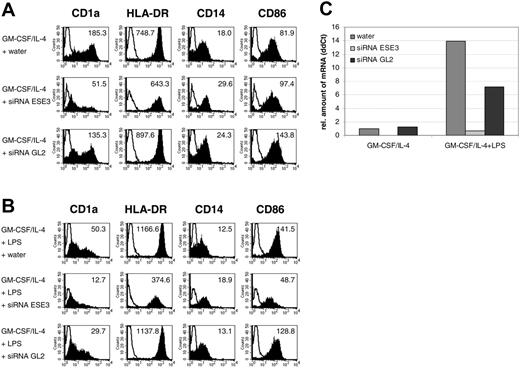
In order to determine the possible role of ESE-3 during development of MDCs, CD14+ peripheral-blood monocytes were isolated directly from buffy coat preparations and transfected with 5 nM ESE-3–specific siRNA. As a control, cells were electroporated with siRNA specific for firefly luciferase (GL2) or mock transfected with water. After 48 hours of culturing the cells in RP10 medium supplemented with GM-CSF and IL-4, the phenotype of the cells was analyzed by fluorescence-activated cell sorter (FACS) staining. Cells transfected with siRNA against ESE-3 showed a reduced expression of DC marker CD1a compared with mock-transfected cells as well as with cells transfected with irrelevant GL2 siRNA (Figure 5A). Furthermore, down-regulation of ESE-3 resulted in decreased expression of HLA-DR and CD86 molecules and in reduced down-regulation of CD14 on the surface of cell populations additionally incubated with maturation stimuli such as LPS (Figure 5B) for additional 24 hours. Analyses of the mRNA level of ESE-3 by quantitative real-time RT-PCR revealed that the average down-regulation was approximately 50% to 100% (Figure 5C). The viability of the cells decreased extremely after 48 hours; therefore, it was not possible to cultivate the cells for a longer period of time.
ESE-3 is expressed in Langerhans cells. Immunohistologic staining of normal skin using antibodies against CD1a (blue) for the detection of Langerhans cells and ESE-3 (red). The nuclei are stained with YOPRO (green).
ESE-3 is expressed in Langerhans cells. Immunohistologic staining of normal skin using antibodies against CD1a (blue) for the detection of Langerhans cells and ESE-3 (red). The nuclei are stained with YOPRO (green).
Of interest, the inhibitory effects on DC phenotype were not mediated by reduced amount of nuclear RelB protein (data not shown), a transcription factor of the NF-κB family important for DC development.22
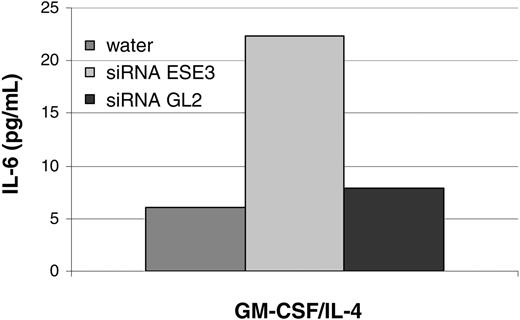
In order to further analyze the effects of ESE-3 down-regulation, secretion of cytokines known to be important for DC function was analyzed by ELISA. While no effect on IL-12 (p70), TNF-α, and IL-10 production was detected (data not shown), IL-6 production was increased in cells transfected with siRNA against ESE-3 (Figure 6).
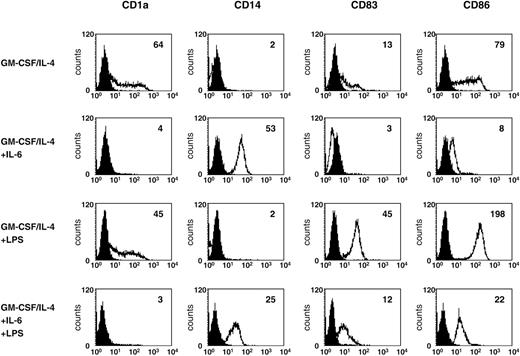
To further analyze the possible physiologic significance of increased IL-6 production, we examined the phenotype of DCs generated in the presence of IL-6. As demonstrated in Figure 7, preincubation of monocytes for 2 days with IL-6 and further cultivation with GM-CSF, IL-4, and IL-6 resulted in reduced expression of CD1a, CD83, and CD86. Additionally, these cells remain CD14+ even after stimulation with LPS (Figure 7).
Down-regulation of ESE-3 during DC development leads to an impaired DC phenotype. CD14+ peripheral-blood monocytes were isolated from buffy coat preparations with magnetic beads and transfected with 5 nM siRNA specific for ESE-3 (siRNA ESE3) or firefly luciferase GL2 (siRNA GL2). Mock transfection was carried out with the equivalent amount of water. (A) After 48 hours of culturing the cells in RP10 medium supplemented with GM-CSF and IL-4, the phenotype of the cells was analyzed by FACS staining. Shaded histograms represent staining with the indicated antibody; open histograms, the isotype control. The level of surface expression is indicated as mean fluorescence intensity. (B) LPS was added as a maturation stimulus 24 hours before harvesting the cells. The data shown are representative of 4 independent experiments. (C) Real-time quantitative RT-PCR to analyze the amount of ESE-3 mRNA after siRNA treatment. The amount of mRNA was determined in relation to GAPDH.
Down-regulation of ESE-3 during DC development leads to an impaired DC phenotype. CD14+ peripheral-blood monocytes were isolated from buffy coat preparations with magnetic beads and transfected with 5 nM siRNA specific for ESE-3 (siRNA ESE3) or firefly luciferase GL2 (siRNA GL2). Mock transfection was carried out with the equivalent amount of water. (A) After 48 hours of culturing the cells in RP10 medium supplemented with GM-CSF and IL-4, the phenotype of the cells was analyzed by FACS staining. Shaded histograms represent staining with the indicated antibody; open histograms, the isotype control. The level of surface expression is indicated as mean fluorescence intensity. (B) LPS was added as a maturation stimulus 24 hours before harvesting the cells. The data shown are representative of 4 independent experiments. (C) Real-time quantitative RT-PCR to analyze the amount of ESE-3 mRNA after siRNA treatment. The amount of mRNA was determined in relation to GAPDH.
ESE-3 down-regulation leads to an increase in IL-6 secretion. CD14+ peripheral-blood monocytes were isolated from buffy coat preparations with magnetic beads and transfected with 5 nM siRNA specific for ESE-3 (siRNA ESE3) or firefly luciferase GL2 (siRNA GL2). Mock transfection was carried out with the equivalent amount of water. After 48 hours of culturing the cells in RP10 medium supplemented with GM-CSF and IL-4, supernatants were collected. The data shown are representative of 4 independent experiments.
ESE-3 down-regulation leads to an increase in IL-6 secretion. CD14+ peripheral-blood monocytes were isolated from buffy coat preparations with magnetic beads and transfected with 5 nM siRNA specific for ESE-3 (siRNA ESE3) or firefly luciferase GL2 (siRNA GL2). Mock transfection was carried out with the equivalent amount of water. After 48 hours of culturing the cells in RP10 medium supplemented with GM-CSF and IL-4, supernatants were collected. The data shown are representative of 4 independent experiments.
Discussion
Monocyte-derived DCs (MDCs) are used as antigen-presenting cells (APCs) in many immunotherapeutic approaches and in vitro models. Despite this broad application, molecules determining the fate during the development and function of these cells have not yet been fully defined. In this study, we demonstrate that the epithelial-specific Ets transcription factor ESE-3 is expressed and plays an important role during DC development. So far, the function of ESE-3 has been elusive. It has been shown to be exclusively expressed in epithelial cells,23 and due to its binding to the c-Met promoter, a role in morphogenesis was suggested.23 Moreover, ESE-3 seems to be an important regulator in the control of epithelial differentiation and carcinogenesis.14 Recently, Galang et al24 reported elevated expression of ESE-3 in mammary tumors.
Using RT-PCR and Western blot analyses, we found that ESE-3 expression is induced at an early stage during DC development from peripheral-blood monocytes. Moreover, depending on the stimulus, maturation of MDCs leads to distinct up-regulation of ESE-3. According to our results, DCs are the only hematopoietic cells that express this transcription factor.
Addition of IL-6 during DC generation leads to an impaired DC phenotype. DCs generated in the presence of IL-6 were analyzed for surface expression of CD1a, CD14, CD83, and CD86. LPS was added 24 hours before harvesting of the cells. Open histograms represent staining with the indicated antibody; shaded histograms, the isotype control. The level of surface expression is indicated as mean fluorescence intensity.
Addition of IL-6 during DC generation leads to an impaired DC phenotype. DCs generated in the presence of IL-6 were analyzed for surface expression of CD1a, CD14, CD83, and CD86. LPS was added 24 hours before harvesting of the cells. Open histograms represent staining with the indicated antibody; shaded histograms, the isotype control. The level of surface expression is indicated as mean fluorescence intensity.
To further analyze the role of ESE-3 during DC development and maturation, we used siRNA technology to silence ESE-3 transcription in DCs. Down-regulation of ESE-3 led to an impaired DC phenotype characterized by a decreased expression of DC marker CD1a. Moreover, the responsiveness of the generated cells to a maturation stimulus was reduced, characterized by low surface expression of MHC class II molecules compared with control cells transfected with siRNA against firefly luciferase GL2. Further analysis revealed that IL-6 secretion was elevated in these cells. This is one possible explanation for the inhibitory effects of ESE-3 down-regulation, as, in line with our results, IL-6 was shown to inhibit DC development from CD14+ monocytes and CD34+ progenitor cells25-27 and maturation of DCs.28,29 Moreover, it was reported that elevated IL-6 level switches the differentiation from DCs to macrophages,25 which might be the reason for the higher CD14 expression on the cells with down-regulated ESE-3.
Taken together, our study demonstrates that ESE-3 expression is induced in DCs during their differentiation and up-regulated upon activation by maturation stimuli. ESE-3 transcripts and protein can be found in different DC subpopulations including peripheral-blood DCs and Langerhans cells. According to our results, this transcription factor is critically involved in DC development from peripheral-blood monocytes.
Prepublished online as Blood First Edition Paper, December 27, 2005; DOI 10.1182/blood-2005-06-2480.
Supported by a grant from Deutsche Forschungsgemeinschaft (SFB510).
S.A. and A.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank A. Tugores (University of California, La Jolla, CA) for the rat monoclonal ESE-3 antibody 5A5.5 as well as Sylvia Stephan, Bruni Schuster, Tina Hörnle, and Birgit Fehrenbacher for excellent technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal