Abstract
We previously identified a small number of genes using cDNA arrays that accurately diagnosed patients with Sézary Syndrome (SS), the erythrodermic and leukemic form of cutaneous T-cell lymphoma (CTCL). We now report the development of a quantitative real-time polymerase chain reaction (qRT-PCR) assay that uses expression values for just 5 of those genes: STAT4, GATA-3, PLS3, CD1D, and TRAIL. qRT-PCR data from peripheral blood mononuclear cells (PBMCs) accurately classified 88% of 17 patients with high blood tumor burden and 100% of 12 healthy controls in the training set using Fisher linear discriminant analysis (FLDA). The same 5 genes were then assayed on 56 new samples from 49 SS patients with blood tumor burdens of 5% to 99% and 69 samples from 65 new healthy controls. The average accuracy over 1000 resamplings was 90% using FLDA and 88% using support vector machine (SVM). We also tested the classifier on 14 samples from patients with CTCL with no detectable peripheral involvement and 3 patients with atopic dermatitis with severe erythroderma. The accuracy was 100% in identifying these samples as non-SS patients. These results are the first to demonstrate that gene expression profiling by quantitative PCR on a selected number of critical genes can be employed to molecularly diagnosis SS.
Introduction
Cutaneous T-cell lymphoma (CTCL) refers to a heterogeneous group of non-Hodgkin lymphomas of “skin-homing” T lymphocytes. The most common forms of CTCL are mycosis fungoides (MF) and Sézary syndrome (SS), an erythrodermic and leukemic variant of CTCL.1-3 Early detection and treatment are directly correlated with favorable outcome for both MF and SS,4-6 but diagnosis is frequently difficult in early stages. In this study we have focused on the more aggressive SS. The identification of the malignant Sézary cell is based primarily on characteristic cytologic features (“cerebriform” nucleus) and the presence of 20% or more Sézary cells among peripheral blood lymphocytes. Otherwise, 5% or more Sézary cells plus evidence of a T-cell clone is the currently recognized threshold level for blood involvement in CTCL.7 However, the detection of early blood involvement using cytologic criteria can be problematic, because Sézary-like cells may be found in healthy individuals and unrelated diseases. Quantitative measurement of lymphocytes with reduced expression of surface CD26 8-11 and CD71,12 have also been reported to be informative for subclasses of patients, but a more sensitive method for diagnosis based on quantitative polymerase chain reaction (qPCR) technique for the early detection of neoplastic cells in the blood would be rapid and less costly.
In a previous study, we analyzed gene expression profiles of peripheral blood mononuclear cells (PBMCs) from patients with bona fide SS using cDNA arrays.13 Using penalized discriminant analysis (PDA),14,15 a machine learning algorithm that is an extension of Fisher linear discriminant analysis (FLDA), to analyze these data, we identified several groups of genes whose expression profiles accurately discriminated patients with high proportions of circulating neoplastic cells (60% to 99% of lymphocytes) from healthy controls with 100% accuracy. By using the information obtained from the analysis of samples from patients with high blood tumor burden, we found that several nonoverlapping subsets of 8 to 20 genes could identify patients with as few as 5% circulating neoplastic lymphocytes with 100% accuracy.13 Other gene expression studies using microarrays have demonstrated that patterns of gene expression can be identified that distinguish cancer cells from their normal counterparts, and one cancer from another,16-19 but they have for the most part not been tested on blood samples containing low numbers of neoplastic cells. It has also been possible to identify groups of genes whose expression levels correlate with prognosis and predict responsiveness to therapy17,20-23 using gene expression arrays, further supporting the power of this approach.
In our previous study,13 we reported qPCR-validated expression patterns for 35 genes that included 26 of the most significantly differentially expressed genes identified by the array studies on RNA derived from PBMC samples from 18 patients with SS and high blood tumor burden and 9 T helper-2 (Th2)–skewed healthy controls. The remaining 9 genes were specifically selected to confirm by real-time (RT)–PCR the consistent levels of their expression across these samples as measured by microarrays. These top 26 genes were chosen for their low P value by t test and high fold change. However, while these data were used to successfully validate the microarray results, they were not used to develop a qPCR classifier for CTCL.
We now report qPCR measurements of the expression values for 5 of the informative genes identified in our microarray studies: STAT4, GATA-3, CD1D, TRAIL, and Plastin-T (PLS3)13 from 125 new samples. FLDA applied to the qPCR data correctly separates patient samples from controls with 90% accuracy, including patients with as few as 5% circulating neoplastic T cells.
Patients, materials and methods
Preparation of patient and control sample RNA
All RNA samples were prepared from Ficoll (Amersham Biosciences, Uppsala, Sweden)–purified PBMCs, and RNA was isolated using Tri reagent (Sigma-Aldrich, St. Louis, MO) as previously described.13 RNA integrity was assayed by agarose gel or Bioanalyzer (Agilent, Palo Alto, CA). A total of 125 samples from patients and controls were analyzed by PCR. The 69 samples from healthy individuals included 8 samples that were skewed to a Th1 phenotype and 2 samples that were skewed to a Th2 phenotype.13 Controls were selected to mimic the demographics of the patient sample group as best as possible and were selected from volunteers ranging in age from 35 to 67 years. Fifty-six samples were from patients with SS that had tumor burdens ranging from 5% to 99%. Both patient and control samples were collected and processed with the approval of The Wistar Institute Internal Review Board, and informed consent was provided in accordance with the Declaration of Helsinki. The proportion of neoplastic cells in the lymphocyte population was estimated by counting the number of atypical lymphocytes with cerebriform nuclei (Sézary cells) in a buffy coat preparation or blood smears as previously described.13 To supplement these findings, either gene rearrangement studies or flow cytometry studies focusing on CD4+/CD7– or CD26– cells was performed. Such studies provided the clear determination of blood involvement. Patients were selected for this study based on percent atypical lymphocytes detected regardless of whether erythroderma was present.
Quantitative real-time PCR
Gene-specific primers (IDT, Coralville, IA) were designed with the Light Cycler Probe Design Software, Version 1.0 (Idaho Technology, Salt Lake City, UT). Primers were selected from the 3′ half of the message and usually from the PCR sequence that was spotted. PCR was performed in 20 μL in a Light Cycler Instrument (Roche Diagnostics, Mannheim, Germany) as previously described.13 All primers were designed to have a melting temperature of 60°C. The PCR cycle parameters were 94°C for 3 minutes, hot start; and 40 cycles of 94°C for 10 seconds, 56°C or 60°C for 10 seconds, and 72°C for 25 seconds. SYBR Green I fluorescence intensity was measured at the end of each 72°C extension as previously described.13 Product specificity was assessed by melting curve analysis, and selected samples were run on 1% agarose gels for size assessment. The cDNA for PCR amplification was prepared from approximately 0.5 to 1 μg total RNA using Superscript II as previously described. Some samples were also assayed on the Opticon IV (MJ Research, Waltham, MA) with similar results.
Primer sequences
Gene-specific primers (IDT) were designed with the Light Cycler Probe Design Software, Version 1.0 from the sequence of spotted cDNA clones. Primer sequences are as follows: GATA-3 (forward: 5′TATCCATCGCGTTTAGGC3′; reverse: 5′CCCAAGAACAGCTCGTTTA3′); PLS3 (forward: 5′GCTTGACAAAGCAAGAGT3′; reverse: 5′GCATCTTCCCTCTCATACC3′); STAT4 (forward: 5′TCCTAGAACCTGGTATTTACAAAG3′; reverse: 5′GTGTATGCCGGTGTTGA3′); CD1D (forward: 5′TGAGACGCCTCTGTTTC3′; reverse: 5′ACACCTCAAATACATACCTACT3′); TRAIL (forward: 5′ACGTGTACTTTACCAACGA3′; reverse: 5′ATGCCCACTCCTTGAT3′); and MBD4 (forward: 5′CACATCTCTCCAGTCTGC3′; reverse: 5′CGACGTAAAGCCTTTAAGAA3′).
qPCR normalization
Values for fluorescence intensity of each gene for each sample were reported as the ratio of its determined value compared with a standard expression curve determined using the human Universal Standard RNA (Stratagene, La Jolla, CA). The expression levels for each gene (relative to that of the reference sample, in our case, the Stratagene Universal Standard RNA) were derived from the fluorescence intensity measurements determined using the Light Cycler Analysis Software, Version 3.5. The housekeeping gene methyl-CpG binding domain protein 4 (MBD4) was used as an internal control for the amount of cDNA in each assay based on its constant expression observed in our previous microarray and PCR studies.13 The calculated gene expression measurements were then (natural) log-transformed for analysis by FLDA and support vector machine (SVM).
Discriminant models
Two linear discriminant analysis models have been applied for classification of qRT-PCR data: classical FLDA24 and a relatively recent machine learning technique: SVM.25 The formula for both techniques can be expressed as follows:
where f(x) expresses the classification score as a function of the measured expression levels for our 5 genes, x = (xCD1D,xGATA3,xPLS3,xSTAT4,xTRAIL), a = (aCD1D,aGATA3,aPLS3,aSTAT4,aTRAIL) is a set of 5 discriminant coefficients associated with each of our 5 selected genes, and a0 is a constant term allowing for adjustment of the sensitivity versus specificity of the model. These coefficients, of course, depend on the discriminant model chosen to differentiate 2 groups of samples as well as on the training set used for fitting the model. Table 1 gives computed average values for these coefficients along with their estimated standard deviation.
Computed average values for the discriminant coefficients along with the corresponding standard deviation for PCR classifiers
. | FLDA . | SVM . |
|---|---|---|
| a0 | 7.548 ± 0.123 | 1.298 ± 0.141 |
| aCD1D | 0.077 ± 0.015 | -0.064 ± 0.017 |
| aGATA-3 | 1.534 ± 0.027 | 0.177 ± 0.031 |
| aPLS3 | -0.425 ± 0.012 | 0.068 ± 0.014 |
| aSTAT4 | -3.817 ± 0.027 | -0.482 ± 0.031 |
| aTRAIL | 2.219 ± 0.027 | 0.435 ± 0.031 |
. | FLDA . | SVM . |
|---|---|---|
| a0 | 7.548 ± 0.123 | 1.298 ± 0.141 |
| aCD1D | 0.077 ± 0.015 | -0.064 ± 0.017 |
| aGATA-3 | 1.534 ± 0.027 | 0.177 ± 0.031 |
| aPLS3 | -0.425 ± 0.012 | 0.068 ± 0.014 |
| aSTAT4 | -3.817 ± 0.027 | -0.482 ± 0.031 |
| aTRAIL | 2.219 ± 0.027 | 0.435 ± 0.031 |
Cluster analysis
The clustering was performed using the Pearson correlation–based distance metric and Ward linkage. The expression measurements of each gene were converted to z scores by subtracting the mean value of the given gene (computed across all samples that are being clustered) and dividing by the corresponding standard deviation, thus bringing the measurements of every gene to a common scale.
Results
Selection of the genes used for classification
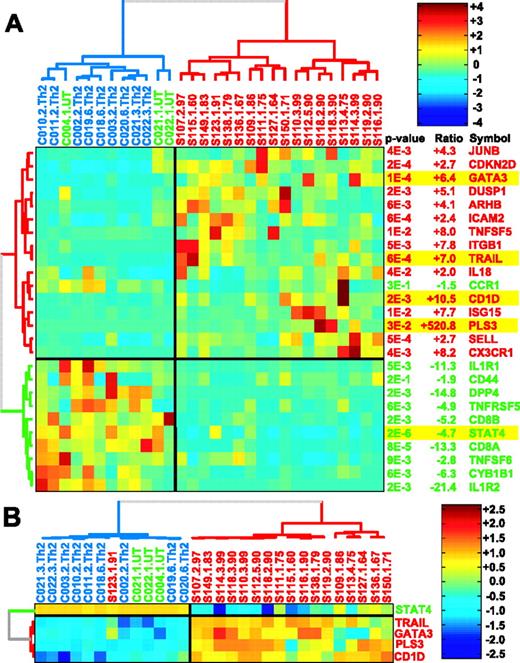
The genes used in the present analysis were in the top 100 genes selected at a P value below .01 that could distinguish 18 patients with SS and high proportions (60% to 90%) of circulating neoplastic lymphocytes from Th2-skewed healthy controls.13 They also appear in the list of the top 10 up-regulated and 10 down-regulated genes that could accurately distinguish the same 18 patients from 12 control PBMCs that were identified using PDA, a machine learning algorithm that is used to carry out supervised sample classification.13 PDA is an extension of FLDA,24 applied to the cases in which the features (in this case genes) outnumber the samples trained on. Additional results on selection of top genes by PDA using recursive feature elimination (RFE) along with an estimated classification accuracy (corrected for selection bias, following Ambroise and McLachlan26 ) as a function of the number of genes is shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The Treeview in Figure 1A shows the results of qRT-PCR studies on 26 of the top genes on RNA from the 18 Sézary patients and 12 healthy controls. The 5 genes examined in this study, STAT4, CD1D, GATA-3, TRAIL, and PLS3, are highlighted in the figure. Selection of these genes was based on their relative changes in gene expression levels measured across the 30 samples used for gene selection and their relevance to the Th2 phenotype of CTCL. Selection of the 5 genes was based on the following criteria: (1) They are consistently expressed across both untreated and Th2-skewed controls (this eliminated ARHB, DUSP1, ICAM2, CD-KND2, and JUNB, as is evident from the heatmap in Figure 1A), and (2) expression levels should be high enough so that variations in expression could be reliably measured by qRT-PCR (this eliminated, for example, MGAM; data not shown). All of the selected 5 genes had median changes in gene expression levels that were more than 4-fold (–4.7-fold to +520-fold), and all except PLS3 (P = .03) had P values below .001. In addition, each gene is a member of a different gene cluster. The Treeview in Figure 1B shows the relative expression levels of the 5 genes on the same 30 patient and control samples.
Dendrograms showing gene expression measured by qRT-PCR on RNA from 18 Sézary patients with high blood tumor burden and 12 healthy controls, including 3 untreated and 9 skewed to Th2 phenotype. Hierarchical clustering was applied to both the genes and the samples and was performed using Pearson correlation–based distance metric and Ward linkage. Samples from SS patients, untreated controls, and Th2-skewed controls are colored in red, green, and blue, respectively. Genes up- and down-regulated in the patients as compared with Th2-skewed controls are colored red and green, respectively. For visual enhancement, expression levels for each gene are converted to z scores. (A) Dendrogram of qRT-PCR on 26 of the genes with P < .01 measured by qRT-PCR on amplified RNA. (B) Dendrogram of 5 genes selected for our discriminant model on the same set of 30 samples assayed by qRT-PCR on total RNA derived from the same samples.
Dendrograms showing gene expression measured by qRT-PCR on RNA from 18 Sézary patients with high blood tumor burden and 12 healthy controls, including 3 untreated and 9 skewed to Th2 phenotype. Hierarchical clustering was applied to both the genes and the samples and was performed using Pearson correlation–based distance metric and Ward linkage. Samples from SS patients, untreated controls, and Th2-skewed controls are colored in red, green, and blue, respectively. Genes up- and down-regulated in the patients as compared with Th2-skewed controls are colored red and green, respectively. For visual enhancement, expression levels for each gene are converted to z scores. (A) Dendrogram of qRT-PCR on 26 of the genes with P < .01 measured by qRT-PCR on amplified RNA. (B) Dendrogram of 5 genes selected for our discriminant model on the same set of 30 samples assayed by qRT-PCR on total RNA derived from the same samples.
Classification of 125 new samples using qRT-PCR data for 5 differentially expressed genes
Although the original array studies were carried out using amplified RNA (aRNA), all of the PCR data on the new samples were derived from total RNA (tRNA) to simplify the assay and to avoid any biases that might be introduced by amplifications carried out with different procedures in different laboratories. We had shown in 2 previous studies on approximately 40 genes, including the 5 assayed in this study, that the qPCR results were similar whether aRNA or tRNA was used.13,27 We used FLDA to analyze the qPCR data in this study because cases (125 samples) now outnumber features (5 genes). Fifty-six samples were from 49 new patients with widely diverse tumor burdens ranging from very low (about 5%) to very high (99%) and included 69 new control samples (65 individuals). Tumor burden was assessed by determining the number of circulating cells with cerebriform nuclei. Patients with fewer than 20% circulating lymphocytes with cerebriform nuclei had either an identified clonal expansion or other confounding factors, including evidence of lymph node involvement. To supplement gene rearrangement studies, flow cytometry focusing on CD4+/CD7– or CD26– cells was also performed. Such studies provided the clear determination of blood involvement. Almost all patients had erythrodermic disease. A few cases had been initially diagnosed with CTCL skin disease and later with low numbers of circulating cerebriform cells (S151, S156). In most cases, diagnosis was reaffirmed over time as almost all patients were seen over a period of 2 years.
The qPCR data for STAT4, CD1D, GATA-3, TRAIL, and PLS3 were used to train the FLDA algorithm to identify patterns of gene expression that were best at distinguishing the 2 classes of samples (in this case, patients and controls) from one another. The accuracy of the 5-gene discriminator was measured by the percent correct sample classification obtained. To eliminate bias associated with the gene selection,26 none of the samples assayed on the microarrays that were used for gene selection were included in these studies. The accuracy of the PCR classifier was determined only using the independent set of 56 new patient samples and 69 new control samples. Multiple samples from the same patient were used only when there was some evidence of a change in disease status or samples were taken several years apart.
To obtain estimates of the accuracy of our PCR classifier and the statistical significance of the classification prediction for each sample, we performed 10-fold cross-validation with 1000 random resamplings of our dataset. In each resampling, we withheld a random 10% of the patient and the control samples (test set) to be used for the subsequent validation step. The remaining 90% of the samples were used to train the discriminant model, which was subsequently applied to the classification of the 10% withheld samples in the independent test set. Thus, each sample in the dataset gets, on average, 100 classification scores corresponding to how it performs with the different discriminant models generated on each of the training sets. The 100 classification scores derived from the resampling studies were then used to estimate average error rates.
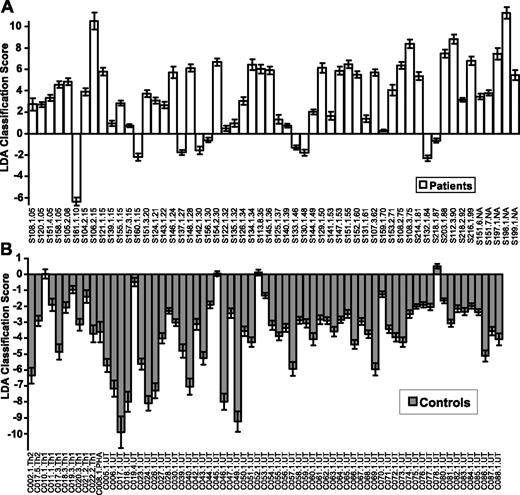
Classification of Sézary patients using qPCR data. Data shown are for analysis using FLDA. Scores for patient samples are shown in panel A, and controls are shown in panel B. The dataset consists of 125 samples: 56 from 49 patients with erythrodermic CTCL and 69 from 65 controls. The controls are untreated (UT), Th1 skewed (Th1), Th2 skewed (Th2), and PHA treated (PHA). The patient samples are sorted according to the blood tumor burden (5% to 99%) indicated by the last 2 digits of the label. Patients in whom percent Sézary was not available are indicated as “NA.” A positive score indicates the sample is classified as a patient; a negative score indicates the sample is classified as a control. The height of each bar represents the average score that a given sample received, when tested approximately 100 times, during 1000 random resamplings. The error bars indicate the standard deviation in the generated set of scores for the sample. Sézary samples (S) are followed by a 3-digit patient donor code. Serial samples taken at different times from the same patient are indicated by an additional number (ie, S151.1, S151.4) and followed by the percent Sézary cells present. None of the controls was sampled more than once, but samples from donors C017, C018, and C019 were also skewed to the Th1 (C017, C018, C019) and Th2 (C017) phenotype. The classifier has a sensitivity of 86% and a specificity of 95% on this dataset.
Classification of Sézary patients using qPCR data. Data shown are for analysis using FLDA. Scores for patient samples are shown in panel A, and controls are shown in panel B. The dataset consists of 125 samples: 56 from 49 patients with erythrodermic CTCL and 69 from 65 controls. The controls are untreated (UT), Th1 skewed (Th1), Th2 skewed (Th2), and PHA treated (PHA). The patient samples are sorted according to the blood tumor burden (5% to 99%) indicated by the last 2 digits of the label. Patients in whom percent Sézary was not available are indicated as “NA.” A positive score indicates the sample is classified as a patient; a negative score indicates the sample is classified as a control. The height of each bar represents the average score that a given sample received, when tested approximately 100 times, during 1000 random resamplings. The error bars indicate the standard deviation in the generated set of scores for the sample. Sézary samples (S) are followed by a 3-digit patient donor code. Serial samples taken at different times from the same patient are indicated by an additional number (ie, S151.1, S151.4) and followed by the percent Sézary cells present. None of the controls was sampled more than once, but samples from donors C017, C018, and C019 were also skewed to the Th1 (C017, C018, C019) and Th2 (C017) phenotype. The classifier has a sensitivity of 86% and a specificity of 95% on this dataset.
The results of classification are shown as bar plots in Figure 2. Patient classifications are shown in panel A and controls in panel B. The false-positive error rate is the percentage of healthy control samples classified as patients (4 of 69; 6%); the false-negative error rate is the percentage of patient samples classified as healthy controls (9 of 56; 16%). The average overall error rate is computed as the percentage of misclassified samples (13 of 125; 10%), thus leading to the classification accuracy with FLDA of 90%. Similar results were obtained when the data were analyzed by a different machine learning algorithm: linear SVM. The overall accuracy by SVM (data not shown) is slightly less (88%). In both the FLDA and the SVM analysis, the misclassified control samples are borderline, and only one control sample has a significant, but low, positive score. Table 2 is a comparison of the samples misclassified by the 2 approaches. Nine of the 13 samples misclassified by FLDA were also misclassified by SVM, which misclassified a total of 15 samples. Notably, only 2 of the patients misclassified by FLDA are in the low blood tumor burden class (5% to 20% Sézary cells), the group we expected to be most difficult to classify. A Treeview of the expression patterns of the 5 genes on the 125 samples is shown in Figure 3.
Samples misclassified by the 5-gene model using FLDA and/or SVM
Sample . | FLDA . | SVM . | PLS3 . |
|---|---|---|---|
| C010.1.Th1* | 0.02 ± 0.29† | 0.12 ± 0.04† | 0.007 |
| C021.2.Th1‡ | -1.40 ± 0.40 | -0.03 ± 0.07 | 0.009 |
| C019.4.UT‡ | -0.49 ± 0.27 | -0.02 ± 0.05 | 0.007 |
| C052.1.UT | 0.12 ± 0.16† | -0.10 ± 0.03 | 0.007 |
| C078.1.UT | 0.50 ± 0.15† | -0.15 ± 0.03 | 0.006 |
| C045.1.UT | 0.05 ± 0.15† | -0.22 ± 0.03 | 0.006 |
| S108.1.05 | 2.73 ± 0.56 | -0.04 ± 0.09† | 0.033 |
| S161.1.10* | -6.35 ± 0.40† | -0.56 ± 0.05† | 0.001 |
| S157.3.15 | 0.74 ± 0.14 | -0.12 ± 0.03† | 0.004 |
| S160.1.15* | -2.19 ± 0.33† | -0.25 ± 0.05† | 0.075§ |
| S137.1.27* | -1.77 ± 0.25† | -0.16 ± 0.03† | 0.001 |
| S142.1.30* | -1.55 ± 0.36† | -0.08 ± 0.08† | 0.700§ |
| S156.1.30* | -0.61 ± 0.21† | -0.12 ± 0.04† | 0.001 |
| S140.1.39 | 0.75 ± 0.17 | -0.04 ± 0.04† | 0.004 |
| S133.1.46* | -1.30 ± 0.21† | -0.15 ± 0.03† | 0.002 |
| S130.1.48 | -1.78 ± 0.30† | 0.10 ± 0.07 | 0.939§ |
| S159.1.70 | 0.28 ± 0.10 | -0.15 ± 0.03† | 0.035 |
| S132.1.84* | -2.31 ± 0.25† | -0.24 ± 0.03† | 0.001 |
| S218.1.87* | -0.66 ± 0.20† | -0.44 ± 0.07† | 0.218§ |
Sample . | FLDA . | SVM . | PLS3 . |
|---|---|---|---|
| C010.1.Th1* | 0.02 ± 0.29† | 0.12 ± 0.04† | 0.007 |
| C021.2.Th1‡ | -1.40 ± 0.40 | -0.03 ± 0.07 | 0.009 |
| C019.4.UT‡ | -0.49 ± 0.27 | -0.02 ± 0.05 | 0.007 |
| C052.1.UT | 0.12 ± 0.16† | -0.10 ± 0.03 | 0.007 |
| C078.1.UT | 0.50 ± 0.15† | -0.15 ± 0.03 | 0.006 |
| C045.1.UT | 0.05 ± 0.15† | -0.22 ± 0.03 | 0.006 |
| S108.1.05 | 2.73 ± 0.56 | -0.04 ± 0.09† | 0.033 |
| S161.1.10* | -6.35 ± 0.40† | -0.56 ± 0.05† | 0.001 |
| S157.3.15 | 0.74 ± 0.14 | -0.12 ± 0.03† | 0.004 |
| S160.1.15* | -2.19 ± 0.33† | -0.25 ± 0.05† | 0.075§ |
| S137.1.27* | -1.77 ± 0.25† | -0.16 ± 0.03† | 0.001 |
| S142.1.30* | -1.55 ± 0.36† | -0.08 ± 0.08† | 0.700§ |
| S156.1.30* | -0.61 ± 0.21† | -0.12 ± 0.04† | 0.001 |
| S140.1.39 | 0.75 ± 0.17 | -0.04 ± 0.04† | 0.004 |
| S133.1.46* | -1.30 ± 0.21† | -0.15 ± 0.03† | 0.002 |
| S130.1.48 | -1.78 ± 0.30† | 0.10 ± 0.07 | 0.939§ |
| S159.1.70 | 0.28 ± 0.10 | -0.15 ± 0.03† | 0.035 |
| S132.1.84* | -2.31 ± 0.25† | -0.24 ± 0.03† | 0.001 |
| S218.1.87* | -0.66 ± 0.20† | -0.44 ± 0.07† | 0.218§ |
Patient samples are sorted by their tumor burden. Columns LDA and SVM contain average scores obtained by these samples ± standard deviation over the approximately 100 times that these samples were tested during 1000 random resamplings. The column labeled PLS3 displays the expression level of PLS3 (in terms of its ratio to the housekeeping gene relative to that in the Stratagene reference RNA sample).
Samples that were misclassified by both methods.
Scores assigned these samples to incorrect class (positive average scores for controls and negative average scores for patients).
Two control samples, C021.2.Th1 and C019.4.UT, were marginally classified by SVM, even though the average score turned out to be negative, and were considered to be misclassified.
Misclassified patients with high levels of PLS3 (with z score more than 5, relative to controls). These high-PLS3 patients may deserve special treatment, because high levels of PLS3 alone are very unusual for healthy individuals.
Because PLS3 expression is unique to patients, we examined PLS3 expression in the misclassified samples. Relative PCR expression levels of PLS3 are shown in the last column of Table 2. Because 4 of the 9 patients misclassified by FLDA had very high levels of PLS3, these patients would have been recommended for further analysis, as PLS3 is not expressed in normal PBMCs.28,29
Dendrogram of 125 samples on 5 genes selected for our discriminant model. Hierarchical clustering was carried out using Pearson correlation–based distance metric with Ward linkage on both genes and the samples. For visual enhancement, the gene expression values for each gene shown are converted to z scores. Patient samples are shown in red, untreated controls are shown in green, and Th1- and Th2-skewed controls are shown in light and dark blue, respectively. Yellow highlights indicate the samples that were misclassified by FLDA.
Dendrogram of 125 samples on 5 genes selected for our discriminant model. Hierarchical clustering was carried out using Pearson correlation–based distance metric with Ward linkage on both genes and the samples. For visual enhancement, the gene expression values for each gene shown are converted to z scores. Patient samples are shown in red, untreated controls are shown in green, and Th1- and Th2-skewed controls are shown in light and dark blue, respectively. Yellow highlights indicate the samples that were misclassified by FLDA.
We also applied hierarchical clustering to the data for the 5 genes on the 125 samples. Figure 3 is a dendogram that shows the results of the analysis. Hierarchical clustering, unlike FLDA, is an unsupervised technique and therefore does not incorporate the information regarding the sample phenotype. The only input data are the gene expression values that were converted to z scores to ensure that each gene would contribute equally to the classification rather than having it dominated by the most abundant gene. The results, shown in Figure 3, further support the significance of the selected genes for differentiating Sézary patients from various controls. It can be seen that the expression patterns of the 5 selected genes separated the samples into 2 distinct clusters based on their phenotype. The left cluster overwhelmingly consists of the control samples (with only 6 patient samples, which were misclassified by both FLDA and SVM). The right cluster is composed predominantly of patient samples (with only 6 control samples; all but one sample were correctly classified by SVM, and 4 were also misclassified by FLDA). The average error rate of 12 samples out of 125 is 10%. While the overall accuracy is comparable to that achieved by FLDA and SVM, both FLDA and SVM had significantly fewer false positives than clustering. Clustering provides a good tool for illustrative purposes but does not provide a direct measure of confidence in classification for each sample, as is the case for both FLDA and SVM. In addition, clustering results are sensitive to systematic bias caused by other experimental factors unrelated to sample phenotype.
Classification of samples with skin disease and no peripheral involvement
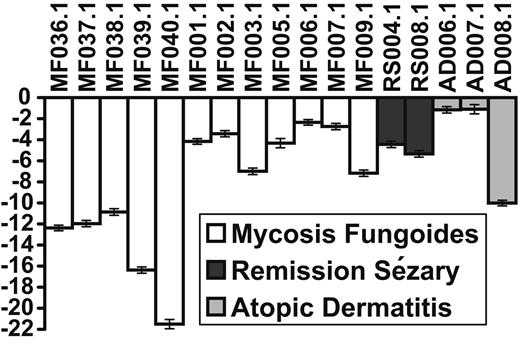
To further test the specificity of the 5 genes for leukemic CTCL, we tested an additional 17 patients. The results are shown in Figure 4. The RNA was derived from PBMCs from 12 patients with MF with no evidence of blood involvement; 2 patients were originally classified as SS but were in remission after treatment with no evidence of a circulating malignant clone as demonstrated by flow cytometry and V gene rearrangement. The 3 atopic dermatitis (AD) patients had severe erythroderma with possible diagnosis of CTCL, but at the time the samples were taken, none of the AD patients had evidence of a malignant clone. One of the patients, AD007, was diagnosed as MF/CTCL with no blood involvement a few months later. Another patient (AD006) was suspected to have CTCL based on lymph node histology. No clonal expansion could be detected by V gene amplification or by flow cytometry. These patients were all correctly classified as not having leukemic CTCL by the 5-gene assay. Samples RS004 and RS008 both had originally presented with a circulating malignant clone. At the time the samples were taken, no evidence of a malignant clone could be detected by flow cytometry or V gene amplification. These patients were also found by our qRT-PCR assay to be free of peripheral disease.
Discussion
From cDNA arrays to qPCR
We have demonstrated in these studies that gene expression profiling can be employed to molecularly diagnose leukemic CTCL and that this can be accomplished by qPCR assays carried out on a selected number of critical genes. The goal of our studies was to develop a method that would reliably detect early involvement of the blood in CTCL. Consequently, the assay had to be able to accurately diagnose patients with low numbers of circulating neoplastic cells as well as those with high tumor burden and be adaptable to a clinical laboratory. Gene expression assays using qPCR are more suitable for this purpose than microarray assays, because they use a robust and well-established technology. The patient and control samples used in these studies were collected in 4 different laboratories. The RNA was prepared and the reactions were carried out by 3 different technicians. A subset of the samples was assayed on 2 different PCR machines (Roche's “Light Cycler” and MJ Research's “Opticon 4”) without any effect on the classification results.
Previous studies that also attempt to make the transfer from the microarray to qPCR platform include that of Gordon et al,30 which described a “gene expression ratio” method to differentiate patients with mesotheliomas from those with adenocarcinomas of the lung by using simple ratios of pairs of expressed genes as determined by qPCR. This method is dependent on being able to identify gene pairs with extreme differences in expression levels in the 2 classes of samples being compared. These great differences may be expected when comparing different cell types or different tumors. For SS, only PLS3 showed striking differences between the patients, and diversified control classes we tested. However, PLS3 was not informative for 30% of the samples tested in our previous array studies13 and is not informative for 50% of the more diverse samples tested in this study. While single-gene diagnostics would-simplify studies, in reality, at least for our CTCL patients who vary considerably, a more robust classification can be made using several genes. Whether PLS3 expression is indicative of a specific subclass of patients is under investigation.
Classification of samples from MF/CTCL, atopic dermatitis, and SS/CTCL in remission. A positive score indicates the sample is classified as an SS; a negative score indicates the sample is classified as a non-SS. The length of each bar represents the average score over 1000 random resamplings, and the error bars indicate the standard deviation in the scores generated by the resamplings. MF indicates mycosis fungoides; RS, SS patient in remission; AD, atopic dermatitis.
Classification of samples from MF/CTCL, atopic dermatitis, and SS/CTCL in remission. A positive score indicates the sample is classified as an SS; a negative score indicates the sample is classified as a non-SS. The length of each bar represents the average score over 1000 random resamplings, and the error bars indicate the standard deviation in the scores generated by the resamplings. MF indicates mycosis fungoides; RS, SS patient in remission; AD, atopic dermatitis.
The 5-gene classifier we have tested successfully identified patient samples with blood tumor burdens ranging from as little as 5% to 99%. In the case of at least one patient originally diagnosed with MF who had a very low blood tumor burden (S151), flow cytometry failed to identify an expanded T-cell clone using loss of CD7 to identify a T-cell clonal population. The loss of CD7 on the neoplastic cells has been used as a marker for CTCL in both skin and peripheral blood,1,12 but neither loss of CD7 nor loss of CD26, also used as a marker for the neoplastic T cells, can be used exclusively as a parameter for early detection of blood disease. Our 5 genes assayed on a concurrent sample easily classified the Sézary signature in this individual.
More recently Lossos et al31 were able to use expression profiles of 6 genes to predict survival in large B-cell lymphoma. Although we previously showed that a relatively small number of genes predicted survival in a class of CTCL patients with survival of less than 6 months from the time of sampling,13 the genes in this study were not those that were informative for survival. This is an area we are pursuing, but because CTCL is such a rare cancer, finding sufficient samples of this class to make sound predictions has been difficult.
Significance of the selected genes
The identification of transcription factors STAT4 and GATA-3 as diagnostic genes is consistent with the most striking biologic aspect of CTCL, the skewing of the patient immune system to a T helper-2 state. GATA-3, whose expression is consistently increased in patient samples, not only induces Th2 T-cell differentiation32 but also suppresses a Th1 T-cell response important for tumor suppression and for protection against the infections that plague these patients. Similarly, expression of STAT4, which is required for Th1 differentiation,33-35 is consistently decreased in patient samples. We previously demonstrated that purified CD4+ cells from patients with SS had little or no STAT4 protein as measured by Western blotting.36 Although most of the patients analyzed in the STAT4 study had tumor burdens above 90%, we found that even in patients with tumor cells present at 15% to 30% of lymphocytes, the reduction of STAT4 protein was characteristic of the entire CD4+ population. We see similar reductions of STAT4 message in patients with levels of circulating tumor cells ranging from 5% to 90% both by arrays and PCR, suggesting STAT4 expression is being actively repressed in the normal CD4+ population as well as in the tumor cells.36 This global suppression of STAT4 in the CD4+ cells is not found when normal CD4 cells are skewed to the Th2 phenotype by culturing with IL-4 and anti–IL-12 in vitro14,27 and is likely a tumor effect. By contrast, the CD8+ cells in the patients tested by Western blotting were found to be protected from this suppression, because they appear to express normal levels of STAT4 protein, at least during early disease when there are sufficient numbers of CD8+ cells remaining to assay.36
PLS3, located on the X chromosome, is expressed in a variety of tissues but is normally never expressed in T cells, and there is also no correlation between expression levels in the CTCL cells and sex (data not shown). PLS3 message levels frequently do not correlate with tumor burden, and we do not know if this is due to lower levels of expression in some tumor cells or to expression in only a subclass of tumor cells. Its expression is regulated by CpG methylation,37,38 suggesting chromatin remodeling is required for the dysregulated expression in the Sézary T cell. In addition, the neoplastic cells of CTCL have unchanged levels of message for the lymphoid plastin, LCP1.13,39 The presence of both LCP1 and PLS3 proteins in CTCL cells has also been confirmed by Western blotting.39 Additional studies on the coexpression of the 2 proteins in transfected cells suggest that the cellular associations of the 2 proteins are not identical, because they must be extracted using different conditions.40,41 LCP1's actin-bundling function has been shown to be important in signaling pathways associated with activation and migration of T cells.42 The presence of PLS3 could possibly interfere with that function. Conversely, aberrant expression of LCP1 has been reported in many cancers, including breast, prostate,43 and colon cancer,44 tissues that normally express PLS3, suggesting that coexpression may not be an uncommon feature of malignant cells.
The possible roles of the overexpressed TRAIL and CD1D genes in CTCL are less clear. TRAIL is a member of the TNF receptor/ligand family and a powerful inducer of apoptosis. Altered expression of several members of this gene family have been described in both SS and MF.13,45 TRAIL preferentially induces apoptosis in tumor cells, where its receptors are more abundantly expressed.46-48 Resistance to TRAIL-induced apoptosis has been suggested to be due to the overexpression of nonsignaling “decoy receptors” by the tumor cells,38,49 but more recent studies have uncovered alternative mechanisms of TRAIL resistance. At the heart of these observations is the constitutive activation of the AKT kinase, which can inactivate several different apoptotic pathways, and the loss of the AKT regulator and tumor suppressor, PTEN.50-53 Based on our array studies, PTEN message levels are not significantly reduced in patients as compared with controls (data not shown), although we have not determined whether protein levels are also unchanged.
The misclassified patient samples have no common difference among them, but the most frequent differences appear to be associated with reduced CD1D and increased STAT4. CD1D is a nonclassical major histocompatibility complex (MHC) class I–like molecule that can present glycolipid or phospholipid bacterial or self antigens to a restricted class of T-cell receptors on natural killer (NK) T cells.54-56 Most functional studies have used presentation of a sponge glycolipid (α-galactosylceramide), but recent studies have identified endogenous antigens that are presented by CD1D in mice and humans.57 CD1D message was not detected in purified CD4+ cells from 7 patients with more than 90% tumor cells (data not shown). Its overexpression appears to be induced in the normal cells by the malignant environment. The importance of NK T cells in tumor surveillance has recently been demonstrated for multiple myeloma58 and acute lymphoblastic leukemia (ALL).59 However, activation of these cells requires stimulation by IL-12 as well as receptor activation through CD1D antigen presentation.60 Our previous studies have demonstrated that CTCL patients are profoundly deficient in IL-12 production.61,62
The overexpression of TRAIL and CD1D in progressive CTCL suggests the cell death pathways controlled by the products of these 2 genes are not functional and that therapies focused on the activation of these pathways may provide new avenues for treatment. Our studies in vitro36,61 and in phase 1 and 2 clinical trials63,64 suggest that IL-12 treatment may be beneficial for certain CTCL patients and that this effect may be due, at least in part, to the activation of the CD1D/NK T-cell pathway.
Despite all the similarities described for MF/CTCL and SS/CTCL when assayed on peripheral blood, these 5 genes accurately diagnosed the 12 MF/CTCL samples we have tested and, although the number of AD samples tested was small, these patients had severe erythrodermic involvement but were still properly classified. One area that we are now pursuing based on the studies on the 2 SS/CTCL patients in remission is whether this PCR test can provide an assessment of response to therapy based on changes in the CTCL predictive score.
Prepublished online as Blood First Edition Paper, January 10, 2006; DOI 10.1182/blood-2005-07-2813.
Supported by U01 CA85060, NSF RCN 0090286 (M.K.S.), NCI T32 CA09171 (A.L., L.K.), R01 CA 106553-02, P30 CA10815-34S3, and the Pennsylvania Department of Health (PA DOH Commonwealth Universal Research Enhancement Program: Tobacco Settlement grant ME01-740).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal