Abstract
The extracellular signal-regulated kinases (ERKs) are required for thrombopoietin (TPO) functions on hematopoietic cells, but the ERKs targets involved remain unknown. Here we show that the regulation of the immediate early gene X-1 (IEX-1), identified as an ERK substrate in response to TPO, was mediated by an ERK-dependent phosphorylation of AML1. The addition of TPO to UT7-Mpl cells and primary megakaryocytes induced gene expression of IEX-1. Neither erythropoietin (EPO) nor granulocyte macrophage-colony stimulating factor (GM-CSF) was able to activate IEX-1 gene expression in UT7-Mpl cells. The induced expression was mediated by a transcriptional activation of the IEX-1 promoter and required an AML1-binding site located at –1068. The direct involvement of AML1 in the regulation of IEX-1 gene expression was shown by both the use of AML1 mutants and by shRNA experiments targeting endogenous AML1. Finally, the ability of TPO to induce the IEX-1 gene expression was inhibited by U0126, a specific inhibitor of the ERKs activator MEK and AML1 transcriptional activity was shown to be modulated by TPO through ERK-dependent phosphorylation. Taken together, these data suggest that AML1 plays a role in modulating the IEX-1 expression and that the ERK-dependent AML1 phosphorylation regulates the TPO-mediated activation of IEX-1.
Introduction
Thrombopoietin (TPO) is a cytokine that regulates both megakaryopoiesis and the stem cell compartment.1-3 Binding of TPO to its receptor c-Mpl activates a number of distinct intracellular signaling cascades; that is, the Janus kinase (JAK) signal transducer and activator of transcription (STAT),4,5 PKC,6 PI3-kinase,7,8 Shc/Ras/mitogen-activated protein kinase (MAPK),9-11 and cAMP/PKA pathways.12 These activations result in the proliferation of megakaryocytes progenitors and in the expression of a number of cell-specific genes associated with megakaryocyte differentiation.
MAPK/ERKs are key regulators of cell proliferation, differentiation, and survival. The different responses elicited by this cascade in various cell types are presumably determined by the cell-specific combination of the numerous ERK substrates. The requirement of the ERK pathway in megakaryocytic differentiation was suggested by studies in several leukemic cell lines9,13-15 and was finally shown using primary hematopoietic progenitors.16,17 Prolonged ERK signaling appears to be necessary for megakaryocyte marker induction and cell growth arrest,9,10,13,14 but the ERK substrates that control these 2 effects are not well defined.
Acute myelogenous leukemia 1 (AML1)/Runt-related transcription factor 1 (Runx1) belongs to a family of transcriptional regulators called Runx, which contain a conserved 128-amino acid Runt domain responsible for sequence-specific DNA binding (for a review, see Kurokawa and Hirai18 ). Runx proteins make heterodimeric complexes with a partner protein, core-binding factor/polyomavirus enhancer-binding protein 2 (CBF/PEBP2), and this association is essential for AML1 transcriptional activity. AML1 functions as a transcriptional activator of target gene expression, but it can also repress the transcription of specific genes. AML1 was originally identified on chromosome 21 as the gene that is disrupted in the (8;21)(q22;q22) translocation, which is one of the most frequent chromosome abnormalities associated with human AML (for a review, see Look19 ). Subsequently, AML1 was shown to be one of the most frequent targets of leukemia-associated gene aberrations.18 Gene-targeting studies in mice have demonstrated that, in addition to having a role in leukemic transformation, AML1 is essential for early development of definitive hematopoiesis. AML1-deficient embryos develop through the yolk sac stage but die at around 12 to 13 days of gestation, following complete block of fetal liver hematopoiesis.20 In addition, AML1 haploinsufficiency resulted in a decrease in the number of stem cells and of circulating platelets.21,22 Indeed, AML1 has a specific role in megakaryopoiesis, as shown in familial platelet disorders and by genetic studies in mice.22,23 At the molecular level, AML1-binding sites have been shown in various megakaryocytic genes, including β3 integrin,24 α2 integrin,25 platelet basic protein promoter,26 acetylcholinesterase,27 Mpl,28 and αIIb integrin.29 Apart from the αIIb gene, the question of whether these genes are true AML1 transcriptional targets has not been studied. The regulation of AML1 functions through signal transduction pathways has been investigated, and, by overexpression experiments, AML1 has been shown to be phosphorylated by ERK.30,31 ERK-dependent phosphorylation enhances the transcriptional activity of AML1, and mutations of the phosphorylation sites reduce the transforming activity of AML1 in the fibroblast cell line.30 These results indicate a possible link between TPO, ERK, AML1, and megakaryopoiesis.
IEX-1, a gene previously characterized as an early gene, and also referred to as p22/PRG1, Dif-2, or Gly96 (its mouse homologue), has been identified as an ERK substrate in TPO-stimulated UT7-Mpl cells,32 a cell line that acquires megakaryocytic features in the presence of TPO.33 IEX-1 gene transcription can be rapidly activated by stress factors, growth factors, viral infection, inflammatory cytokines, steroids, and retinoic acids.34 The IEX-1 gene encodes a protein whose function is not well defined. Expression of IEX-1 has been found to be associated with cell cycle progression and proliferation, and IEX-1 can regulate apoptosis as a proapoptotic or an antiapoptotic factor.32,34-37 Thus, the effect of IEX-1 on cell growth and apoptosis seems to be influenced by cell type-specific conditions.
In UT7-Mpl cells, IEX-1 was shown to be involved in TPO-mediated ERK-sustained activation,32 suggesting that it may be an actor of TPO functions. We therefore sought to determine the regulation pathway involved in TPO-mediated IEX-1 expression. Here, we show that IEX-1 is a direct transcriptional target of AML1 that is specifically regulated by TPO in UT7-Mpl cells. In addition, we show that TPO enhances AML1 transcriptional activity in an ERK-dependent fashion.
Materials and methods
Cytokines, reagents, and antibodies
The TPO mimetic peptide GW39505838,39 was synthesized by Genosys Biotechnologies (The Woodlands, TX). Recombinant human EPO and stem cell factor (SCF) were from Boehringer-Mannheim (Mannheim, Germany) and PromoCell (Heidelberg, Germany). The MEK inhibitor U0126 was from Promega (Lyon, France). Antibodies against IEX-1 were generated by immunizing rabbits with recombinant GST-IEX-1 fusion protein purified on glutathione-Sepharose. Specific AML1 polyclonal antibodies were from Active Motif (Rixensart, Belgium). Antiphospho-ERK was from Cell Signaling Technology (Beverly, MA) and anti-ERK (C-14, K-23) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antiactin antibody (clone AC-40) was from Sigma (St Louis, MO).
Cell culture
The human UT7-Mpl cell line33 derived from a patient with megakaryoblastic leukemia was maintained in MEM supplemented with 10% fetal calf serum (FCS) and 2 U EPO/mL. HEK293T cells were maintained in DMEM supplemented with 10% FCS.
Isolation of CD34+ cells and liquid suspension cultures
CD34+ cells were isolated from the umbilical cord blood of healthy infants after obtaining informed consent from their mothers. CD34+ cells were purified by immunomagnetic selection.16 Fluorescence-activated cell sorting (FACS) analysis performed on a Beckman Coulter FC500 (Beckman Coulter, Marseille, France) showed that the CD34+ population had a purity of over 90%. Purified CD34+ cells were cultured in Iscove modified Dulbecco medium (IMDM, Gibco-BRL, Carlsbad, CA) supplemented with 15% serum-free medium for expansion of human cells (RM-B00, Ambio-International, Tourcoing, France), in the presence of 50 ng/mL SCF and 20 nM TPO. At day 12, immunophenotyping was performed as previously described,16 and mature megakaryocytes (CD34–CD41+CD42b+) were found to represent more than 90% of the cellular population.
Plasmid constructs
The IEX-1 promoter40 fused into the luciferase report gene, the pLucIEXp plasmid, was kindly provided by Dr O. Bernard (Institut National de la Santé et de la Recherche Médicale, Paris, France). The pLucIEXp/AML1m, with the mutated AML1-binding site of the human IEX-1 promoter, was obtained by appropriate polymerase chain reaction (PCR) on the pLucIEXp with the following oligonucleotides: 5′-AGCTAAGCTTGCTCAAGTGATCCTCCCACCTTAGCCC-3′; 5′-GAACAGAAAACCAAATGTCGACCATTCTCACTTATAAGTG-3′; 5′-CACTTATAAGTGAGAATGGTCGACATTTGGTTTTCTGTTC-3′; and 5′-AATTCCTCGAGGGTCAGCCGAGCGGAGTGTAAGG-3′. The final PCR fragment was cloned between HindIII and SacI sites of the pLucMCS. pME18S-AML1B, pME18S-AML1S249A, pME18S-AML1S266A, and pME18S-AML1S249/S266A were kindly provided by H. Hirai (Tokyo, Japan). The pCMV-AML1a and pCMV-AML1/ETO plasmids were gifts of G. Mouchiroud (Centre National de la Recherche Scientifique, Lyon, France). pGL2-3xAML1wtTATALuc is composed of 3 tandem AML1 consensus-binding sites downstream the reporter luciferase gene. It was built by cloning 5′-CCCGGGGTATTGTGGTGATTGTGGTGATTGTGGTTGGCGAGCTC-3′ between the Xma-1 and Sac-1 restriction sites of the pGL2-TATALuc vector.
The target sequences for AML1 (5′-CCUCGAAGACAUCGGCAGAAA-3′ and 3′-UUGGAGCUUCUGUAGCCGUCUUU-5′) were described previously for their capacities to inhibit the target protein.41 Vectors that drive the expression of small hairpin RNAs (shRNA) were generated by cloning the above nucleotides, followed by a 6-loop and the corresponding antisense sequence and by 5 thymidines, in a pcDNA vector in which the CMV promoter was replaced by H1 promoter (pcDNA-H1). UT7-Mpl cells were transduced with a self-inactivating lentiviral/HIV vector (TRIPΔU3) carrying expression cassettes for the human shRNA for AML1 or control shRNA under the control of the H1 promoter and GFP under the control of the elongation factor 1α promoter. Briefly, UT7-Mpl cells were incubated with lentiviral over a period of 36 hours. Production and infectious particle titration were done as described.42 Transduction efficiency in UT7-Mpl cells was tested by determining the percentage of GFP+ cells by FACS42 (> 98% positive cells).
Cell transfection
Transient transfection of UT7-Mpl cells were performed by using Lipofectamin 2000 Reagent (Invitrogen, Paisley, United Kingdom). In brief, 400 ng luciferase reporter plasmid, 0.1 ng phRL-null used as a Renilla luciferase reference plasmid, and (1-2 μg) of the different plasmids to be tested were transfected in 1 × 106 cells settled in 12-well plates in IMDM. The total amount of plasmid transfected was adjusted to 1 to 2 μg by adding empty pcDNA vector. Five hours after transfection, 500 μL IMDM complemented with 20% FCS and either 2 U EPO/mL or 10 nM TPO peptide was added. The cells were maintained in culture for 20 hours. Luciferase activities were assayed by using the dual luciferase assay kit (Promega, Lyon, France) following the manufacturer's instructions. Data were normalized with the Renilla luciferase. All transfections were performed at least 3 times in triplicate, using plasmids that were independently prepared at least twice.
Western blotting
Total cell extracts were prepared by lysing the cells directly in Laemmli sample buffer (62.5 nM Tris, pH 6.8, 1% SDS, 5% glycerol, 5% β-mercaptoethanol, 0.001% bromophenol blue) at 100°C. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was carried out with horseradish peroxidase-conjugated antirabbit, antirat, or antimouse antibodies (Amersham Pharmacia Biotech, Piscataway, NJ), and revealed by chemiluminescence.
Nuclear extracts and electrophoretic mobility shift assays
Cell nuclear extracts were prepared by lysing the cells in buffer A (20 mM HEPES, pH 7.9, 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% glycerol, 1 mM PMSF, 1 mM DTT, 1 mM orthovanadate) for 5 minutes at 4°C. After centrifugation for 2 minutes at 12 000g, the pellet was lysed in buffer B (0.4 M NaCl, 20% glycerol, 20 mM HEPES, pH 7.9, 10 mM KCl, 1 mM EDTA, 1 mM PMSF, 1 mM DTT, 1 mM orthovanadate) for 30 minutes at 4°C, then the lysate was centrifuged 5 minutes at 15 000g and the nuclear extract was frozen. Double-stranded oligonucleotide, 5′-GTGAGAATGTGTGGTATTTGGTTT-3′, corresponding to the AML1-binding site of IEX-1 promoter, was labeled with γ32P. The oligonucleotides were mixed with 10 μg nuclear extract and 1 μg oligodIdC in DNA-binding buffer (10 mM Tris HCl, pH 7.5, 75 mM KCl, 1 mM DTT, 1 mM EDTA, 4% Ficoll type 400) for 15 minutes at 4°C. Competition was performed by the addition of 150 ng unlabeled specific oligonucleotides (150-fold excess) during the incubation. The oligonucleotide mutated in the AML1-binding site (AML1m) was 5′-GTGAGAATGTGTTAGATTTGGTTT-3′. To identify whether AML1 was associated with the oligonucleotides, supershift experiments were performed by adding 1 μL anti-AML1 to the mixture and incubating it for 15 minutes at room temperature prior to incubation with the labeled oligonucleotide. Rabbit IgG antibody was used as control for supershift experiments. The complexes were analyzed on 5% nondenaturating acrylamide gels in 0.5 × Tris borate-EDTA buffer (TBE). An overnight prerunning of the acrylamide gel was performed for a best resolution.
Northern blot analysis
Total RNA (10 μg) was extracted with TRIzol, fractionated on a 1% agarose-formaldehyde gel, and transferred onto a Hybond NX membrane (Amersham Biosciences, Piscataway, NJ). Probes were radiolabeled with [α32P]dCTP with random primed DNA labeling kit (Roche Diagnostics, Meylan, France). The membrane was hybridized in Church buffer (360 mM Na2HPO4, 140 mM NaH2PO4, 7% SDS, 1 mM EDTA) overnight at 65°C and washed twice in a mixture containing 2 × SSC-0.1% SDS and twice in 0.5 × SSC-0.1% SDS at 65°C before autoradiography.
Preparation of nuclear extracts and oligonucleotide pull-down
For nuclear extract preparation, cells were incubated for 10 minutes at 4°C in buffer A (10 mM HEPES, pH 7.6, 3 mM MgCl2, 10 mM KCl, 5% glycerol, 0.5% NP-40, 1 mM Na2VO4, 20 mM NaF, 1 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM leupeptin, 1 mM pepstatin). After centrifugation, nuclear pellets were resuspended in buffer A containing 300 mM KCl. For oligonucleotide pull-down, AML1-containing complexes were precipitated from nuclear extracts by addition of 1 μg double-strand biotin-labeled AML1wt oligonucleotide at 4°C for 1 hour. DNA-protein complexes were then pelleted using streptavidinagarose beads (Amersham Biosciences, Piscataway, NJ). The beads were washed 4 times with lysis buffer and resuspended in 1 × Laemmli buffer.
In vivo labeling of cells
UT7Mpl cells (10 × 106) were incubated for 2 hours in 3 mL phosphate-free DMEM with the addition of 1 mCi (37 MBq) [32P]Pi and were either left unstimulated or stimulated with 10 nM TPO for 1 hour at 37°C. Nuclear cells extracts were oligoprecipitated as described. Proteins were resolved by SDS-PAGE and transferred to PVDF membranes. Radioactive proteins were visualized by autoradiography and analyzed with a Typhon 9400 (Amersham Biosciences). AML1 was detected by Western blotting.
Statistical analysis
Results were expressed as means plus or minus standard deviation (SD). All statistical analyses were made with a 2-sided Student t test. A P value below .05 was considered statistically significant.
Results
TPO induces IEX-1 expression in the UT7-Mpl cell line and in primary megakaryocytes
To characterize IEX-1 expression during megakaryopoiesis, a time course of TPO-induced IEX-1 gene expression in the UT7-Mpl cell line was performed. Stimulation of UT7-Mpl cells by TPO mimetic led to a rapid increase of the IEX-1 mRNA expression (Figure 1A, lane 3). This increase peaked at around 2 hours of stimulation and was followed by a decrease in the IEX-1 mRNA level to its initial level after 5 hours (Figure 1A). The induction of IEX-1 expression in UT7-Mpl cells was TPO specific because no detectable mRNA was found in the presence of EPO (Figure 1A) or GM-CSF (data not shown). Different experimental procedures have been explored to verify the TPO specificity. UT7-Mpl cells were starved of EPO and then stimulated with either 10% SCF or different EPO concentrations (2, 5, and 10 U/mL). Only TPO could induce IEX-1 in those conditions (data not shown). TPO is the only cytokine that can induce megakaryocytic differentiation of UT7-Mpl cells, suggesting that IEX-1 expression could be involved in TPO signaling in these cells.
Next, IEX-1 expression in normal megakaryocytes was analyzed. Human CD34+ hematopoietic progenitors were grown in the presence of TPO to induce megakaryocytic differentiation. After 12 days of culture, these progenitors were differentiated into megakaryocytes16 and IEX-1 protein level was studied. As shown in Figure 1B, the IEX-1 protein was detected in mature megakaryocytes, but its expression required TPO, as it disappeared within 3 hours of TPO removal and could be reinduced on TPO stimulation of starved megakaryocytes (Figure 1B). Thus, TPO induces IEX-1 expression in both UT7-Mpl cells and normal cells.
TPO induces IEX-1 transcription in UT7-Mpl cells
To determine whether TPO-mediated IEX-1 induction was regulated at the transcriptional level, a 1.4-kb IEX-1 promoter40 was cloned and the time course of IEX-1 promoter activity was analyzed by luciferase assays. UT7-Mpl cells were transfected with pLucIEXp and incubated in the presence of TPO. As shown in Figure 2A, TPO regulated the transcriptional activity of the IEX-1 promoter. Furthermore—and consistent with the data shown on the induction of IEX-1 mRNA level by TPO—luciferase activity was detectable after 2 hours of TPO treatment and then increased gradually with the incubation time, reaching about 70-fold induction by 20 hours of treatment. Similar results were obtained with 10 ng/mL recombinant human TPO (rHuTPO; data not shown). No significant IEX-1 promoter activity could be detected in cells treated with either EPO (Figure 2A) or GM-CSF (data not shown). These results underline the specificity of TPO on IEX-1 promoter activity.
AML1 is involved in the transcriptional regulation of IEX-1
The transcription factors GATA-1, Ets, and AML1 have been shown to be involved in megakaryocytic-specific gene induction.43 Thus, the TPO-specific induction of IEX-1 promoter activity led us to search for potential GATA-, Ets-, and AML1-binding sites in the 1.4-kb IEX-1 promoter used. As shown in Figure 2B, 3 sites that share homology with binding sites for GATA (CTATCC), Ets (TTCCTT), and AML1 (TGTGGT) were found at positions –1322, –1318, and –1068 in the 1.4-kb promoter region. Whereas point mutations that suppress the GATA- or Ets-binding site had no effect on TPO-induced activity of the IEX-1 promoter, mutation of the AML1-binding site decreased the transcriptional activity of this promoter by 2-fold (Figure 2C).
IEX-1 expression in UT7-Mpl cells. (A) Northern blot analysis. UT7-Mpl cells were stimulated or not with 10 nM of peptide TPO. mRNA (10 μg/lane) was then isolated at the indicated times and analyzed by Northern blot using human IEX-1 as a probe. After stripping, the blot was rehybridized with GAPDH as an mRNA loading control. (B) CD34+ cells were isolated from human cord blood and were grown with 10 nM TPO during 12 days to generate megakaryocytes. The cells were either harvested directly (lane 1) or after starvation for 3 hours in medium without TPO (lane 2) and restimulation with 10 nM of TPO for 3 hours (lane 3). Immunoblotting with anti-IEX1 antibody was performed. Antiactin antibody was used as a loading control.
IEX-1 expression in UT7-Mpl cells. (A) Northern blot analysis. UT7-Mpl cells were stimulated or not with 10 nM of peptide TPO. mRNA (10 μg/lane) was then isolated at the indicated times and analyzed by Northern blot using human IEX-1 as a probe. After stripping, the blot was rehybridized with GAPDH as an mRNA loading control. (B) CD34+ cells were isolated from human cord blood and were grown with 10 nM TPO during 12 days to generate megakaryocytes. The cells were either harvested directly (lane 1) or after starvation for 3 hours in medium without TPO (lane 2) and restimulation with 10 nM of TPO for 3 hours (lane 3). Immunoblotting with anti-IEX1 antibody was performed. Antiactin antibody was used as a loading control.
AML1 is involved in the transcriptional regulation of IEX-1. (A) Activation of the IEX-1 promoter in UT7-Mpl cells. UT7-Mpl were transiently transfected with the pLucIEXp plasmid. The cells were treated with TPO (10 nM, ▪) or with EPO (2 U/mL, □) 5 hours after transfection and were assessed for luciferase activities at the indicated times. Transfection efficiencies were normalized to the Renilla luciferase activity from the cotransfected internal control plasmid (phRL-Null). The results are from one representative experiment. (B) Schematic diagram of the wild-type IEX-1 promoter-luciferase reporter. (C) Site-directed mutagenesis of the binding sites for AML1 (–1068), GATA-1 (–1322), or ETS (–1318) in the human IEX-1 promoter affects its transcriptional activation. The site-directed mutants and wild-type promoter luciferase activities were compared after transient transfection and 20 hours of TPO stimulation of UT7-Mpl cells. (D) Effect of AML1 dominant-negative (DN) mutants on IEX-1 promoter induction. The pLucIEXp was cotransfected with wild-type (AML1wt) or DN (AML1a and AML1/ETO) forms of AML1. Cells were harvested and luciferase assays were performed after 20 hours of TPO treatment. Data represent mean ± SD of at least 3 independent experiments. *P < .01.
AML1 is involved in the transcriptional regulation of IEX-1. (A) Activation of the IEX-1 promoter in UT7-Mpl cells. UT7-Mpl were transiently transfected with the pLucIEXp plasmid. The cells were treated with TPO (10 nM, ▪) or with EPO (2 U/mL, □) 5 hours after transfection and were assessed for luciferase activities at the indicated times. Transfection efficiencies were normalized to the Renilla luciferase activity from the cotransfected internal control plasmid (phRL-Null). The results are from one representative experiment. (B) Schematic diagram of the wild-type IEX-1 promoter-luciferase reporter. (C) Site-directed mutagenesis of the binding sites for AML1 (–1068), GATA-1 (–1322), or ETS (–1318) in the human IEX-1 promoter affects its transcriptional activation. The site-directed mutants and wild-type promoter luciferase activities were compared after transient transfection and 20 hours of TPO stimulation of UT7-Mpl cells. (D) Effect of AML1 dominant-negative (DN) mutants on IEX-1 promoter induction. The pLucIEXp was cotransfected with wild-type (AML1wt) or DN (AML1a and AML1/ETO) forms of AML1. Cells were harvested and luciferase assays were performed after 20 hours of TPO treatment. Data represent mean ± SD of at least 3 independent experiments. *P < .01.
To study the role of AML1 in the regulation of the IEX-1 promoter activity by TPO, 2 dominant-negative forms of AML1, the fusion protein AML1/ETO and AML1a, were transfected together with the IEX-1 promoter-driven luciferase gene in UT7-Mpl cells. As shown in Figure 2D, both AML1/ETO and AML1a inhibited the TPO-mediated IEX-1 promoter activity. By contrast, overexpression of wild-type AML1 induces a 2- to 3-fold induction in the luciferase levels in the presence of TPO. These results suggest that AML1 is involved in the TPO-induced IEX-1 promoter activity.
To show that the AML1-binding site present in the IEX-1 promoter can bind to AML1, this sequence was used as a radiolabeled probe for gel shift assays (Figure 3). Using nuclear extracts from UT7-Mpl cells stimulated or not with TPO, 2 major complexes were evidenced: one nonspecific complex (indicated by the asterisk) and a specific one. Indeed, only the upper complex was efficiently competed by cold consensus-binding sequence for AML1 (lane 4) but not by a mutated competitor that cannot bind AML1 (lane 5) and was shifted by an anti-AML1 antibody (lanes 3 and 7), demonstrating a specific binding of AML1 in the IEX-1 promoter. Interestingly, TPO treatment did not modify the AML1 binding to this sequence, suggesting that the TPO regulation of the IEX-1 promoter activity may result from a regulation of the AML1 transcriptional activity.
We thus constructed an artificial promoter containing 3 AML1-binding sites from the IEX-1 promoter upstream from a TATALuc reporter gene. In the absence of TPO, no luciferase activity could be detected, and TPO induced a 1.5-fold increase in this activity (Figure 4). Overexpression of AML1 led to a 5-fold increase of luciferase activity in absence of TPO, whereas a 20-fold increase of luciferase activity was observed in the presence of TPO.
Together, these results indicate that AML1 transcriptional activity involved in IEX-1 promoter induction is TPO dependent.
IEX-1 expression is affected by AML1 knockdown
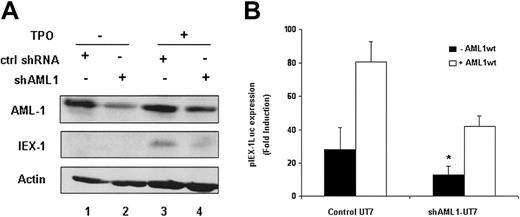
To study the role of endogenous AML1 in TPO-mediated IEX-1 induction, we tested the effects of shRNAs targeting AML1 on both the IEX-1 promoter activity and the endogenous IEX-1 protein level. The AML1-directed shRNA significantly reduced the endogenous AML1 protein level (Figure 5A lanes 2 and 4), as compared to control shRNA (lanes 1 and 3) and led to a significant decrease in TPO-mediated IEX-1 promoter activity (Figure 5B), demonstrating that endogenous AML1 is required for this effect. Moreover, when AML1 shRNA-treated cells were cotransfected with a vector expressing wild-type AML1, the IEX-1 promoter induction was partially restored (Figure 5B). Because the promoter activity of IEX-1 was decreased in AML1-suppressed UT7-Mpl cells treated with TPO, we studied the AML1 dependence of the endogenous IEX-1 level. The IEX-1 protein level induced by TPO in AML1 shRNA-expressing UT7-Mpl cells was greatly decreased as compared to control shRNA (Figure 5A). Together, these results demonstrate that AML1 is involved in TPO-induced IEX-1 endogenous transcription.
TPO-mediated IEX-1 transcription is ERK dependent
TPO treatment of UT7-Mpl cells activates the ERK pathway.9,31 Previous studies have reported that the ERKs phosphorylate and positively regulate AML1 transcriptional activity in COS cells and K562 cells overexpressing either ERK or AML1 or both of them.30 To examine whether this mechanism was involved in TPO-mediated AML1-dependent IEX-1 induction, the effect of U0126, an inhibitor of the ERK activator MEK in TPO-induced activation of the IEX-1 promoter was studied. As shown in Figure 6A, U0126 inhibited about 50% of the TPO-induced IEX-1 promoter activity, suggesting that the ERK pathway is involved in IEX-1 transcription. The effect of U0126 in the regulation of TPO-induced AML1 transcriptional activity was next studied on the artificial AML1 luciferase reporter. As shown in Figure 6B, the MEK inhibitor decreased TPO-mediated activity of this promoter as well. These results indicate that the ERK pathway is involved in TPO-induced AML1 transcriptional activity and suggest that ERK-dependent phosphorylation of AML1 is involved in TPO-induced IEX-1 promoter activity in UT7-Mpl cells.
AML1 DNA binding in UT7-Mpl cells. AML1 DNA binding is independent of TPO stimulation. Nuclear extracts were prepared from 1 × 107 UT7-Mpl cells either nonstimulated (lane 1) or stimulated with 10 nM TPO for 1 hour (lane 2). The γ32P-labeled IEX-1 probe was incubated with 10 μg of the nuclear extracts and subjected to electrophoretic mobility shift analysis. Where indicated, either antibody to AML1 (lane 3), unlabeled wild-type probe (lane 4), or the unlabeled AML1 mutated probe (lane 5) was added to the binding reaction mixtures. The asterisk indicates the nonspecific complex formed. The arrows indicate the complexes supershifted in the presence of an anti-AML1 antibody (αAML1). Rabbit IgG antibody was used as control for the supershift.
AML1 DNA binding in UT7-Mpl cells. AML1 DNA binding is independent of TPO stimulation. Nuclear extracts were prepared from 1 × 107 UT7-Mpl cells either nonstimulated (lane 1) or stimulated with 10 nM TPO for 1 hour (lane 2). The γ32P-labeled IEX-1 probe was incubated with 10 μg of the nuclear extracts and subjected to electrophoretic mobility shift analysis. Where indicated, either antibody to AML1 (lane 3), unlabeled wild-type probe (lane 4), or the unlabeled AML1 mutated probe (lane 5) was added to the binding reaction mixtures. The asterisk indicates the nonspecific complex formed. The arrows indicate the complexes supershifted in the presence of an anti-AML1 antibody (αAML1). Rabbit IgG antibody was used as control for the supershift.
Transcriptional activation of AML1 by TPO. UT7-Mpl cells were transiently cotransfected with 400 ng pGL2-3 × AML1-TATALuc with or without 1μg pME18S-AML1wt. Cells were harvested after 20 hours of culture without (□) or with (▪) TPO, and luciferase assays were performed. Data represent mean ± SD of at least 3 independent experiments. *P < .05.
Transcriptional activation of AML1 by TPO. UT7-Mpl cells were transiently cotransfected with 400 ng pGL2-3 × AML1-TATALuc with or without 1μg pME18S-AML1wt. Cells were harvested after 20 hours of culture without (□) or with (▪) TPO, and luciferase assays were performed. Data represent mean ± SD of at least 3 independent experiments. *P < .05.
IEX-1 expression in UT7-shAML1 cells. (A) Endogenous AML1 and IEX-1 protein expression in the presence of shAML1. UT7-Mpl cells expressing either control shRNA (lanes 1 and 3) or shAML1 (lanes 2 and 4) were stimulated with 10 nM TPO for 1 hour (lanes 3 and 4) or not stimulated (lanes 1 and 2). Immunoblots were performed with IEX-1– or AML1-specific antibodies. The membrane was stripped and reblotted with antiactin antibody as internal loading control. (B) IEX-1 promoter induction in shAML1-expressing cells. Control UT7-Mpl or UT7-shAML cells were transfected with 400 ng pLucIEXp in the presence or not of 1 μg pME18S-AML1wt. Five hours after transfection, the cells were stimulated with 10 nM TPO and 20 hours later lysed and assessed for luciferase activities. Data represent mean ± SD of 4 independent experiments. *P < .05.
IEX-1 expression in UT7-shAML1 cells. (A) Endogenous AML1 and IEX-1 protein expression in the presence of shAML1. UT7-Mpl cells expressing either control shRNA (lanes 1 and 3) or shAML1 (lanes 2 and 4) were stimulated with 10 nM TPO for 1 hour (lanes 3 and 4) or not stimulated (lanes 1 and 2). Immunoblots were performed with IEX-1– or AML1-specific antibodies. The membrane was stripped and reblotted with antiactin antibody as internal loading control. (B) IEX-1 promoter induction in shAML1-expressing cells. Control UT7-Mpl or UT7-shAML cells were transfected with 400 ng pLucIEXp in the presence or not of 1 μg pME18S-AML1wt. Five hours after transfection, the cells were stimulated with 10 nM TPO and 20 hours later lysed and assessed for luciferase activities. Data represent mean ± SD of 4 independent experiments. *P < .05.
TPO induces ERK-dependent phosphorylation of AML1
As shown in Figure 7A-B, AML1 was detected as various electrophoretic mobility species in UT7-Mpl cells. In addition to the main band of AML1, a retarded band appeared on TPO stimulation as shown by Western blot (Figure 7A-B). This effect was inhibited by U0126, indicating that TPO triggers ERK-dependent phosphorylation of AML1 (Figure 7A lane 3). Consistent with this result, oligonucleotide pull-down of AML1 from UT7-Mpl cells labeled with [32P] phosphate showed that TPO induced the appearance of a new slower migrating AML1 species that was oligo-precipitated AML1 (Figure 7B). Autoradiography shows that a constitutive phosphorylation of AML1 occurs in the presence or the absence of TPO. This result is consistent with previous observations by Tanaka et al30 and Zhang et al31 that TPO induced 32P incorporation in the slower migrating band specifically. This result demonstrates that the upper band of AML1 represents a phosphorylated form of AML1 and that TPO induces AML1 phosphorylation.
The Ser249 and Ser266 residues in the AML1 protein have been shown to be phosphorylated by ERK.30 To determine whether these serine sites account for the increase in the level of AML1 phosphorylation, oligonucleotide pull-down experiments were performed in HEK-293T cells transfected with AML1 phosphorylation mutant (S249A/S2466A) compared with AML1 wild-type. As shown in Figure 7C, whereas the wild-type AML1 migrated as 2 bands, only the lower species was detected in cells expressing the S249A/S266A mutant. Thus, ERK-mediated phosphorylation at these 2 residues is sufficient to induce a shift in AML1 mobility. This reinforces the results described showing that the slower migrating form of AML1 observed in response to TPO is AML1 phosphorylated at Ser249/266.
ERKs are involved in the transcriptional regulation of IEX-1. UT7-Mpl cells were transiently transfected with 400 ng pLucIEXp (A) or with 400 ng pGL2-3 × AML1 and AML1 expression plasmid, as indicated (B). Cells were stimulated with 10 nM TPO in the presence or absence of 10 μM U0126 and harvested 20 hours later for luciferase assays. The results shown represent the mean ± SD of at least 3 separate experiments. *P < .05.
ERKs are involved in the transcriptional regulation of IEX-1. UT7-Mpl cells were transiently transfected with 400 ng pLucIEXp (A) or with 400 ng pGL2-3 × AML1 and AML1 expression plasmid, as indicated (B). Cells were stimulated with 10 nM TPO in the presence or absence of 10 μM U0126 and harvested 20 hours later for luciferase assays. The results shown represent the mean ± SD of at least 3 separate experiments. *P < .05.
ERK activation is involved in AML1 transcriptional activation of IEX-1. (A) TPO induces an ERK-dependent increased phosphorylation of AML1. UT7-Mpl cells were stimulated for 1 hour with 10 nM TPO (lanes 2 and 3) in the absence (lane 2) or the presence of 10 μM U0126 (lane 3). Total cell lysates were analyzed by immunoblotting with anti-AML1–, antiphospho-ERK–, and anti-ERK–specific antibodies. (B) TPO induces specific AML1 phosphorylation. UT7-Mpl cells were in vivo labeled with [32P]Pi and stimulated (lane 2) or not (lane 1) with 10 nM TPO. Endogenous AML1 was oligo-precipitated and detected by Western blot analysis. Radioactive proteins were analyzed with a Typhoon 9400. (C) The ERK-dependent phosphorylation of Ser249 and Ser266 is sufficient to induce a shift in AML1 mobility. HEK293T cells were transfected with mock vector (lane 1), pME18SAMLwt (lane 2), or pME18SAML1S249/266A (lane 3) and oligo-precipitation was performed. Results were analyzed by immunoblot with anti-AML1–specific antibody. (D) IEX-1 promoter induction by AML1 phospho sites mutants. UT7-Mpl cells were cotransfected with 400 ng pLucIEXp and 400 ng either wild-type or mutated AML1 as indicated and stimulated with 10 nM TPO. The cells were harvested 20 hours after transfection for luciferase assays. Data represent means ± SD of at least 3 independent experiments. *P < .05.
ERK activation is involved in AML1 transcriptional activation of IEX-1. (A) TPO induces an ERK-dependent increased phosphorylation of AML1. UT7-Mpl cells were stimulated for 1 hour with 10 nM TPO (lanes 2 and 3) in the absence (lane 2) or the presence of 10 μM U0126 (lane 3). Total cell lysates were analyzed by immunoblotting with anti-AML1–, antiphospho-ERK–, and anti-ERK–specific antibodies. (B) TPO induces specific AML1 phosphorylation. UT7-Mpl cells were in vivo labeled with [32P]Pi and stimulated (lane 2) or not (lane 1) with 10 nM TPO. Endogenous AML1 was oligo-precipitated and detected by Western blot analysis. Radioactive proteins were analyzed with a Typhoon 9400. (C) The ERK-dependent phosphorylation of Ser249 and Ser266 is sufficient to induce a shift in AML1 mobility. HEK293T cells were transfected with mock vector (lane 1), pME18SAMLwt (lane 2), or pME18SAML1S249/266A (lane 3) and oligo-precipitation was performed. Results were analyzed by immunoblot with anti-AML1–specific antibody. (D) IEX-1 promoter induction by AML1 phospho sites mutants. UT7-Mpl cells were cotransfected with 400 ng pLucIEXp and 400 ng either wild-type or mutated AML1 as indicated and stimulated with 10 nM TPO. The cells were harvested 20 hours after transfection for luciferase assays. Data represent means ± SD of at least 3 independent experiments. *P < .05.
To investigate whether ERK-dependent phosphorylation at these sites alters the transactivation activity of AML1, the abilities of AML1 phosphorylation mutants (S249A, S266A, S249A/S266A) to induce IEX-1 promoter activity in TPO-stimulated UT7-Mpl cells were analyzed. As shown in Figure 7D, AML1 mutated at either Ser249 or Ser266 exhibited a reduced ability to increase TPO-mediated IEX-1 promoter activity, as compared to wild-type AML1. Mutation of both Ser249 and Ser266 to alanine resulted in an almost complete loss of AML1 transcriptional activity (Figure 7D). However, because the levels of luciferase activity were similar to those of nontransfected cells, this AML1 double mutant did not appear to act as a dominant-negative on the function of endogenous AML1. These results show that Ser249 and Ser266 are phosphorylated by ERK in response to TPO and suggest the involvement of ERK phosphorylation in the regulation of AML1 activity in UT7-Mpl cells.
Discussion
IEX-1 transcription can be rapidly and transiently activated by a broad range of mediators. In accordance with this, the IEX-1 promoter is rather complex, consisting of multiple consensus sequences for transcription factor.34,40,44-46 IEX-1 has been identified as an ERK substrate involved in the prolongation of ERK signal in response to TPO.32 We show here that TPO, but not EPO or GM-CSF, induces IEX-1 expression in UT7-Mpl cells.
IEX-1 expression is also induced in megakaryocytes derived from TPO-stimulated CD34+ cord blood cells, suggesting that IEX-1 may play a role in the functions of TPO. Supporting this possibility, although IEX-1 has been shown to be induced by numerous ubiquitous transcription factors in different cell context, we found that IEX-1 expression in response to TPO is controlled at least in part by a transcription factor with known functions in megakaryocyte development. We also show that ERK-dependent phosphorylation of AML1 is involved in TPO-induced IEX-1 expression in UT7-Mpl cells. The ERK-mediated increase in AML1 phosphorylation and transcriptional activity in response to TPO could be one of the mechanisms underlying the function of the ERK pathway in megakaryopoiesis. Interestingly, although it has been previously shown that EPO and GM-CSF can also activate the ERK pathway in UT7-Mpl cells,9 TPO only induces IEX-1. This indicates that ERK activation by TPO but, not by EPO nor GM-SCF, leads to AML-1 phosphorylation and activates its transcriptional activity. The reason for that may be the fact that ERK activation by TPO and the other cytokines is qualitatively different, differing in both kinetics and magnitude.9,11 A likely explanation for the difference between EPO, GM-CSF, and TPO in their ability to induce IEX-1 promoter expression and increase AML-1 transcriptional activity is the capacity of phospho-ERK to reach the nucleus where AML1 is located, only in response to TPO.
The involvement of AML1 in IEX-1 gene expression was demonstrated by mutation of the AML1-binding site in the IEX-1 promoter and by the regulation of its activity by wild-type or dominant-negative mutants of AML1, because the dominant-negative AML1 proteins, AML1a and the fusion protein AML1/ETO, greatly decreased the TPO-induced IEX-1 expression. More importantly, shAML1, which has been shown to down-regulate AML1 expression, prevented the induction of endogenous IEX-1 by TPO, identifying IEX-1 as a direct transcriptional target of AML1 in UT7-Mpl cells. Consensus AML1-binding sites for AML1 are present in many megakaryocytic promoters and enhancers. However, only the αIIb gene has been shown to be an AML1 direct target gene, and Elagib et al29 have recently demonstrated that a functional and physical interaction among AML1 and GATA-1 contributes to the activation of the αIIb gene promoter in K562 cells. Most megakaryocytic promoters possess Ets-binding sites adjacent to GATA-binding sites.43 One function of AML1 could be to assemble a megakaryocytic-specific transcriptional complex through the recruitment of GATA-1 and Ets-1 factors, as has been suggested for the αIIb gene promoter.29 However, although GATA-1 and Ets-1 putative binding sites have also been found in the IEX-1 promoter, our study indicates that they are not involved in its induction by TPO. These results suggest a different requirement of transcription factors for TPO-specific induction of IEX-1 gene expression. Furthermore, in the IEX-1 murine gene promoter, we found 3 consensus-binding sites for AML1, but none for GATA-1 or Ets-1.
This is the first report showing that the activity of AML1 is regulated by TPO. Although TPO did not induce any difference in the DNA-binding affinity of AML1, TPO increases the transactivation ability of AML1 on the IEX-1 promoter. Furthermore, using an artificial promoter containing multimerized AML1 IEX-1–binding sites, we demonstrate a positive regulatory effect of TPO on the transcriptional function of AML1. Several AML1-binding motifs were characterized in the Mpl promoter. Moreover, a new AML1 mutation in a pedigree of familial platelet disorder with predisposition to acute myelogenous leukemia (FPD/AML) is associated with a reduced Mpl expression,28 suggesting Mpl is an AML1 target. Together with the results shown here, these data suggest that AML1 and Mpl could regulate each other. Our results also show that the MEK inhibitor U0126 has an inhibitory effect on TPO-induced both IEX-1 and 3 × AML1 promoter activities, indicating that the ERK pathway contributes to AML1 regulation in response to TPO. Although the importance of the ERK pathway in TPO functions and megakaryocyte differentiation has long been recognized,9,10,16,17 AML1 is, to our knowledge, the first transcription factor implicated in megakaryopoiesis directly regulated by ERK phosphorylation in response to TPO. Recently, Sevinsky et al47 have shown that the regulation of αIIb expression in K562 cells is mediated through ERK-dependent up-regulation of the transcription factor MafB/Kreisler, which then promotes αIIb transcription, but MafB has not been found as a substrate of ERK. We demonstrate in this study that AML1 is phosphorylated by ERK in vivo in TPO-stimulated UT7-Mpl cells.
The regulatory mechanisms by which ERK phosphorylation control AML1 activity remain elusive so far. Previous studies have identified several ERK phosphorylation sites on AML1.30,31 Phosphorylation at Ser249 and Ser266 has been shown to be induced in COS cells treated with EGF and PMA-treated K562 cells.30,31 Mutation of Ser249 and Ser266 to alanine results in the loss of AML1-induced IEX-1 promoter activity in TPO-stimulated UT7-Mpl cells. This result suggests that Ser249 and Ser266 are phosphorylated in response to TPO and strengthens the involvement of ERK in the regulation of AML1 activity, consistent with previous data.30 Surprisingly, although the S/A double mutant has almost completely loss its activity, this mutant does not act as a dominant negative. Although TPO did not modify AML1-binding affinity on the IEX-1 promoter site, we cannot rule out the possibility that a slight alteration in DNA-binding affinity of AML1 was induced by ERK-dependent phosphorylation, as was previously suggested.30 On the other hand, we cannot preclude that other phosphorylation sites could be involved. This would be consistent with the results of Zhang et al.31 In fact, the AML1 protein contains 11 ERK consensus phosphorylation sites. Based on the results obtained for various promoters, Zhang et al31 suggest that specific phosphorylation sites on AML1 may be important for its activity on some promoters but not on others and that mutation of the 11 phosphorylated sites is required to generate a dominant-negative form of AML1. ERK-mediated phosphorylation in response to TPO may either favor the recruitment of coactivators (p300/CBP) or prevent its binding to corepressors mSin3A, HDAC, and TLE.18,48 The precise mechanisms by which the ERK-dependent phosphorylation of AML1 is involved in IEX-1 gene expression are under investigation.
In addition, to be a master factor for definitive hematopoiesis, AML1 has been shown to be required for megakaryocyte differentiation. In primary human hematopoietic progenitors cultures, AML1 up-regulation preceded megakaryocytic differentiation, and down-regulation of this factor preceded erythroid differentiation.23,49 Also, haploinsufficiency of AML1 resulted in a decrease in the number of circulating platelets.21,22 Point mutations that impair AML1 function are found in patients with AML,50 FPD with predisposition to AML,22 and myelodysplastic syndrome (MDS).51,52 It could be assumed that a deregulation of AML1 activity could have major effects on IEX-1 expression and IEX-1 inhibition by the fusion protein AML1/ETO supports this hypothesis. Interestingly, IEX-1 expression has been shown to be decreased in CD34+ cells from patients with MDS.53
The ERK pathway, although not required for megakaryocytic formation, is involved in the transition from proliferation to maturation in this lineage.16,17 It has been previously shown that IEX-1 has a dual role in ERK signaling, acting both as a specific substrate and a positive regulator of ERK activation.32 Indeed, IEX-1 maintains the ERK signal in response to TPO, suggesting that it may play a role in megakaryocyte differentiation. Supporting this possibility, our preliminary results indicate an inhibition of the megakaryocytic differentiation in UT7-Mpl cells in which TPO-mediated IEX-1 induction was inhibited by IEX-1-shRNA (M.G., unpublished data, July 2005). Further studies on human and murine normal progenitors cells are, however, required to determine the role of IEX-1 in hematopoiesis and in megakaryopoiesis.
Prepublished online as Blood First Edition Paper, December 20, 2005; DOI 10.1182/blood-2005-07-2953.
Supported by INSERM and grants from the Ligue Nationale Contre le Cancer and the Association pour la Recherche sur le Cancer. V.H. is supported by a fellowship from the Ligue Nationale Contre le Cancer and by the Societé Française d'Hématologie.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






![Figure 7. ERK activation is involved in AML1 transcriptional activation of IEX-1. (A) TPO induces an ERK-dependent increased phosphorylation of AML1. UT7-Mpl cells were stimulated for 1 hour with 10 nM TPO (lanes 2 and 3) in the absence (lane 2) or the presence of 10 μM U0126 (lane 3). Total cell lysates were analyzed by immunoblotting with anti-AML1–, antiphospho-ERK–, and anti-ERK–specific antibodies. (B) TPO induces specific AML1 phosphorylation. UT7-Mpl cells were in vivo labeled with [32P]Pi and stimulated (lane 2) or not (lane 1) with 10 nM TPO. Endogenous AML1 was oligo-precipitated and detected by Western blot analysis. Radioactive proteins were analyzed with a Typhoon 9400. (C) The ERK-dependent phosphorylation of Ser249 and Ser266 is sufficient to induce a shift in AML1 mobility. HEK293T cells were transfected with mock vector (lane 1), pME18SAMLwt (lane 2), or pME18SAML1S249/266A (lane 3) and oligo-precipitation was performed. Results were analyzed by immunoblot with anti-AML1–specific antibody. (D) IEX-1 promoter induction by AML1 phospho sites mutants. UT7-Mpl cells were cotransfected with 400 ng pLucIEXp and 400 ng either wild-type or mutated AML1 as indicated and stimulated with 10 nM TPO. The cells were harvested 20 hours after transfection for luciferase assays. Data represent means ± SD of at least 3 independent experiments. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/8/10.1182_blood-2005-07-2953/4/m_zh80080694380007.jpeg?Expires=1767719414&Signature=eehcqs-A9pHRSaC-1xGVIoWvx9fTdmtH~Y7bJXUMG44aYeTUBkkuSDixn8cJ6jzoWkiqhFH2Eeea4Ha1~VZ60mtUq~nrJd4mj4A6BXjdXBVIe3sg-xYRkHtGBzo8tCA-aLN~~C5Z6F7wdswZ-8wRNZXtCk~CB9s65D4~E3QhB65~mXH1UZlh8kbMg7sY2TDLUjeA2Bt6-4lJRhG9CFq~0DB89vQtCyH8yai36sshdZGhKzhyG0T1nXJV31VGeEXOaVr9swmcqpPGdtRmEFL1uc7ITO0csWfK7mnYmicGn6bjJhoWB-WGWZtcp9PlwrN26CF8ns63ZHu~hf44WK2rRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal