Abstract
Monozygotic twin sisters, with nonhematologic symptoms of Fanconi anemia (FA), were discovered to be somatic mosaics for mutations in the FANCA gene. Skin fibroblasts, but not lymphocytes or committed hematopoietic progenitors, were sensitive to DNA cross-linking agents. Molecular analysis revealed, in skin cells of both twins, a frameshift causing deletion in exon 27 (2555ΔT) and an exon 28 missense mutation (2670G>A/R880Q). The latter resulted in primarily cytoplasmic expression and reduced function of the mutant FANCA (R880Q) protein. Surprisingly, the same acquired exon 30 missense change (2927G>A/E966K) was detected in the hematopoietic cells of both sisters, but not in their fibroblasts, nor in either parent. This compensatory mutation existed in cis with the maternal exon 28 mutation, and it restored function and nuclear localization of the resulting protein. Both sisters have been free of hematologic symptoms for more than 2 decades, suggesting that this de novo mutation occurred prenatally in a single hematopoietic stem cell (HSC) in one twin and that descendants of this functionally corrected HSC, via intra-uterine circulation, repopulated the blood lineages of both sisters. This finding suggests that treating FA patients with gene therapy might require transduction of only a few hematopoietic stem cells.
Introduction
Fanconi anemia (FA; OMIM no. 227650)1 is a heterogeneous recessive genetic disorder with clinical symptoms including bone marrow failure, multiple birth defects, and cancer predisposition.2-4 Currently, there are 12 known complementation groups (A, B, C, D1, D2, E, F, G, I, J, L, and M).5-8 The diagnostic hallmark of Fanconi anemia is hypersensitivity to DNA cross-linking agents such as diepoxybutane (DEB) or mitomycin C (MMC).9,10 About 25% of Fanconi anemia patients11 have somatic mosaicism in their hematopoietic system, which contains clastogen-sensitive mutant cells as well as cross-linker–resistant revertant cells. Multiple molecular mechanisms for restoration of normal function in mutant FA alleles have been demonstrated, including compensatory mutations in cis, intragenic crossovers, and gene conversion.11-14 Reversion is thought to occur in a single hematopoietic progenitor, followed by selection and growth expansion of the cells with a functional FA allele. Clinically, somatic mosaicism for functionally corrected hematopoietic cells could prevent the onset of anemia, at least temporarily. Indeed, in murine models of FA, selection of functionally corrected hematopoietic cells has been clearly demonstrated.15-17
Clinical case reports of somatic mosaicism have involved reversions in the FANCA, FANCC, and FANCD2 genes and have demonstrated correction of multiple blood lineages, suggesting growth selection of corrected progenitors.11-14,18 Some of the patients had milder than expected hematologic symptoms. However, to date, somatic mosaicism caused by functional reversion at the level of the hematopoietic stem cell (HSC) and oligoclonal repopulation by unassisted natural selection has yet to be demonstrated. Here we provide evidence that molecular reversion can occur at the level of the HSC and result in long-term restoration of hematopoiesis. We describe the molecular events in monozygotic twin sisters19 who have mutations in the FANCA gene but, at age 28, are free of any hematologic signs or symptoms.
Patients, materials, and methods
Cell lines and patient samples
The FANCA functionally null GM6914 cell line has been previously described,20 as has wild-type lymphoblast cell line PD7.21 Primary fibroblasts (.F) were obtained from skin biopsy, and lymphoblast cell lines (.L) were obtained by immortalization of peripheral blood lymphocytes. Patient cells were designated as follows: PD845 and PD839 cell lines were derived from the affected twins, PD846 from the mother, and PD852 from the father. Skin and blood samples were obtained as per international review board (IRB) protocol no. 3582, Oregon Health and Science University (OHSU). All cells were grown in a humidified 5% CO2-containing atmosphere at 37°C in standard accepted media. Bone marrow was obtained for Wright-Giemsa–stained smears and for in vitro assays from the patients and volunteers according to OHSU IRB protocol no. ise823. Samples were obtained with informed consent, provided according to the Declaration of Helsinki.
Cytogenetic analysis
Cytogenetic analysis of mitomycin C (MMC; Sigma-Aldrich, St Louis, MO) and diepoxybutane (DEB; Sigma-Aldrich) sensitivity, by chromosomal breakage (DEB) and radial formation (MMC) assays, was performed as previously reported.22-25 At least 35 G-banded metaphase cells from each culture were scored for DNA breaks and radial formations by microscopic analysis. For complementation analysis, primary PD839 fibroblasts (PD839.F) were infected with retroviral supernatant representing various FA genes (A, C, D2, F, and G) as previously described.26 Karyotyping of bone marrow was done according to standard procedures, similar to those previously described.24
Mutation confirmation in genomic DNA
Mutations discovered by sequencing were confirmed in genomic DNA, obtained as previously described,27 using allele-specific restriction enzyme digests of polymerase chain reaction (PCR) products generated using the following cycling conditions and standard buffers: 95°C for 5 minutes, followed by 37 cycles of 95°C for 30 seconds, X°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 1 minute.
X = 51°C for exon 27 assay, 54°C for exon 28 assay, and 52°C for exon 30 assay.
Exon 27 assay for the 2555ΔT mutation: forward primer: 5′CCATCCAGTTCGGAATGC3′, reverse primer: 5′AAGAAAACTTGCAGAGAGAGCAA3′.
155-bp product; mutant allele-specific cleavage to 131- and 23-bp products with BstAPI.
Exon 28 assay for the 2670G>A mutation: forward primer: 5′GGTCTGTTTCCACCTGAGCATTTGC3′, reverse primer: 5′CTGGCTACGTCCTCCTCACCAAGAG3′.
153-bp product; mutant allele-specific cleavage to 130- and 23-bp products with XcmI.
Exon 30 assay for the 2927G>A mutation; 10% DMSO was required in the PCR mix: forward primer: 5′AGTGCTGTGTGTCCCTTACTATG3′, reverse primer 5′CTGTCCCTCCAGAGAACCC3′.
269-bp product; wild-type allele-specific cleavage to 143- and 126-bp products with HinfI.
Constitutive expression constructs
The retroviral expression vectors pMMP-puro26 and pMMPpuroFANCA28 were described previously and include an internal ribosomal entry site (IRES)–driven puromycin-resistance cassette.28
To obtain retroviral vectors for the constitutive expression of patient-derived, variant FANCA alleles, RNA was extracted from cells as previously described.29 Reverse transcriptase–polymerase chain reaction (RT-PCR) products, produced with primers 5′AATGAGGATGACAGCAGCG3′ and 5′GACAGACGAAGGCAGGCG3′, were used to replace the Bpu1102I to BamHI fragments in pMMPpuroFANCA. The PCR cycling program for this was 95°C for 5 minutes, followed by 40 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 2 minutes, followed by 72°C for 10 minutes. In addition, site-directed mutagenesis was performed on the pMMPpuroFANCA construct to produce the R880Q, E966K, and R880Q-E966K (double) mutant FANCA cDNAs with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The cDNA inserts were verified by direct DNA sequencing. Production of retroviral supernatants from pMMP-based constructs and infection of fibroblasts were performed as previously described.30,31 The resulting cell lines were cultured under 1 μg/mL puromycin selection.
Immunoblotting for FANCA in patient cell lines
Immunofluorescence microscopy
Cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes, followed by permeabilization with 0.2% Triton X-100 in PBS (3 minutes). They were then incubated in anti-FANCA (N-terminal) antibody,32 diluted 1:200 in 3% bovine serum albumin/0.05% Triton X-100/0.04% sodium azide/PBS for 1 hour at room temperature. Cells were subsequently washed 3 times in PBS, then incubated in fluorescein-conjugated anti–rabbit IgG secondary antibody (Jackson Immunoresearch, West Grove, PA) diluted 1:500 in 3% bovine serum albumin/0.05% Triton X-100/0.04% sodium azide/PBS for 30 minutes at room temperature. Three more washes were applied, and the nuclei were counterstained with DAPI (4,6 diamidino-2-phenylindole) diluted in PBS at 1 μg/mL for 5 minutes. Three more washes were applied, and the slides were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were captured on a Nikon Zeiss Axioplan 2 microscope (Tokyo, Japan) using a Zeiss Neofluar Infinite 0.17 40 ×/1.3 numeric aperture objective oil-immersion lens, a mounted Zeiss AxioCam HRc CCD camera, and OpenLab (Improvision) software. Images were trimmed, and the overall brightness and contrast adjusted, using Adobe Photoshop software (Adobe Systems, San Jose, CA).
Conditional expression constructs
The FA-A cell line GM6914 was transfected with plasmid pREV-Tet-Off (BD Biosciences, San Jose, CA, catalog no. 6140-1), and stable integrants were selected with G418. Lysates from individual clones were analyzed for expression of tetracycline repressible transcription activator (tTA) by polyvinylidenefluoride (PVDF) membrane Western blot using the anti-VP16 (1-21) monoclonal IgG1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA; catalog no. sc-7545), diluted 1:1000 in 1% milk, in PBS overnight at 4°C. An anti–mouse IgG1 horseradish peroxidase (HRP)–conjugated secondary antibody (Santa Cruz; catalog no. sc-2969) was used at 1:3000 dilution in 1% milk in PBS. The 3 pREV-Tet-Off transfected GM6914 clones demonstrating the highest levels of tTA expression were retained. Next, the patient-derived variant FANCA cDNAs were inserted into plasmid pREV-TRE (BD-Biosciences, catalog no. 6137-1). Retroviral supernatants were produced from the various resulting constructs using the packaging cell line PA317.33 The recombinant vectors containing the wild-type, paternal, maternal, or lymphoblast allele of FANCA were individually used to infect the 3 selected clones, as previously described,30,31 to produce a set of 4 cell lines for each clone. Resulting cell lines were placed under 250 μg/mL hygromycin B and 400 μg/mL G418 double selection.
Doxycyclin-dependent FANCA expression was tested for each population of double-resistant cells of a representative set of cell lines by culturing cells for 3 days under 0, 7.8, 31, 125, 500, or 2000 ng/mL Dox and performing Western blot on cell lysates using PVDF membrane and the monoclonal anti–FANCA 5G9 antibody,34 diluted 1:500 in 5% milk in PBS, followed by Santa Cruz goat anti–mouse IgG HRP-conjugated secondary antibody (catalog no. sc-2005), diluted 1:3000 in 5% milk in PBS.
MMC resistance assays
Cell survival assay for constitutively expressing cells. 1000 cells per well were seeded, in triplicate, to 96-well plates. Media was changed the next day to add 0, 10, 25, 50, 75, or 100 nM MMC plus 1 μg/mL puromycin, and cells were cultured for 7 days. Cell number per well was quantitated using the CyQUANT system (Molecular Probes, Eugene, OR) and FLUOstar 403 fluorescence plate reader (BMG LabTechnologies, Durham, NC) set to 485-nm excitation and 535-nm emission as previously described.35 Gain was set based on cells expressing wild-type allele in 0 nM MMC wells.
Clonal growth of cell line. GM6914 cell lines expressing pMMPpuro-FANCA constructs with wild-type allele, maternal allele, and lymphoblast allele, or vector only (pMMPpuro), were seeded to 100-mm plates, 500 cells per plate. Puromycin selection was removed, and 0, 5, 10, 20, 40, or 80 nM MMC was added, in triplicate, per cell line. Media was changed about 2.5 days later, without puromycin or MMC. Plates of cells were stained 8 days later with methylene blue stain (Fisher Scientific; 1%-2%, dissolved in 50% methanol). Colonies were counted and averaged for each cell line for each concentration of MMC, and the percent colony-forming abilities (percent CFAs) relative to 0 nM MMC were graphed.
Growth assay with doxycyclin-regulated FANCA expression. Cells containing doxycyclin-repressible FANCA constructs were seeded into 96-well plates, 1000 cells per well. G418 and hygromycin B selection was removed 12 hours later and replaced with media containing 0, 7.8, 31, 125, 500, or 2000 ng/mL Dox. Twenty-four hours later, media was replaced again, this time containing both appropriate Dox levels and 0, 20, 40, or 80 nM MMC. Thus, for each cell line, cells were exposed, in triplicate, to each of the 4 MMC concentrations at each of the 6 Dox concentrations, in a Dox by MMC matrix. Cells were cultured for between 5 and 7 days and then were analyzed using the CyQUANT assay and FLUOstar reader as described in “Cell survival assay for constitutively expressing cells.”
CFU-GMs and BFU-Es. Low-density bone marrow cells were obtained as previously described,36 except for the use of the EasySep kit (Stem Cell Technologies, Vancouver, BC, Canada). These cells were exposed to various doses of mitomycin C in suspension culture for only 24 hours, after which they were washed in complete medium and plated in methylcellulose with complete medium, Steel factor (50 ng/mL), interleukin-3 (IL-3) (10 ng/mL), and erythropoietin (2 U/mL). Granulocyte-macrophage colony-forming units (CFU-GMs) and erythrocyte burst-forming units (BFU-Es) were counted 14 days after plating.
Multilineage progenitor assays
Multilineage progenitor assays were undertaken as previously described14 ; DNA obtained from individually plucked CFU-GMs and BFU-Es from both twins was analyzed via the PCR digest assay for the acquired exon 30 sequence change (2927G>A) described in “Mutation confirmation in genomic DNA.”
X-inactivation assay
An X chromosome inactivation assay was applied to genomic DNA from patients' whole blood and fibroblasts, as well as to DNA from the mother, from one male control, and from one female control. The assay was performed essentially as previously described.37 DNA samples were digested with HpaII prior to PCR.
Pedigree and haplotypes of immediate family members. Parents carried one mutation each; daughters were compound heterozygotes. An acquired mutation downstream of the maternally inherited mutation was found in the whole blood of both patients. Inset: Western blot showing FANCA levels for cell lines derived from sisters versus controls. Fibroblasts and lymphoblasts were analyzed separately.
Pedigree and haplotypes of immediate family members. Parents carried one mutation each; daughters were compound heterozygotes. An acquired mutation downstream of the maternally inherited mutation was found in the whole blood of both patients. Inset: Western blot showing FANCA levels for cell lines derived from sisters versus controls. Fibroblasts and lymphoblasts were analyzed separately.
Results
Clinical description of patients
The patients were previously described monozygotic twin sisters,19 with normal complete blood counts and differential counts from initial diagnosis at age 6 months through follow-up at age 13. One twin had initially elevated chromosome breakage in DEB-treated peripheral blood lymphocytes (PBLs), which resolved by age 13, while the other twin demonstrated normal breakage even at 6 months. Both twins exhibit short stature and thumb defects, and one has abdominal skin hyperpigmentation and had surgically corrected malformed kidneys. The twins are currently 28 years old and remain free of hematologic signs and symptoms. Their complete blood counts are normal, the bone marrows are not hypocellular and show no evidence of myeloid leukemia or myelodysplasia, and neither twin has developed a solid tumor.
MMC survival curves of cells highly expressing differentFANCAalleles. (A) Relative percent cell growth, as measured by percent fluorescence relative to 0 nM MMC. (B) Percent colony-forming ability. Maternal and lymphoblast alleles both generated intermediate phenotypes in both assays. Error bars indicate standard error of the mean (SEM).
MMC survival curves of cells highly expressing differentFANCAalleles. (A) Relative percent cell growth, as measured by percent fluorescence relative to 0 nM MMC. (B) Percent colony-forming ability. Maternal and lymphoblast alleles both generated intermediate phenotypes in both assays. Error bars indicate standard error of the mean (SEM).
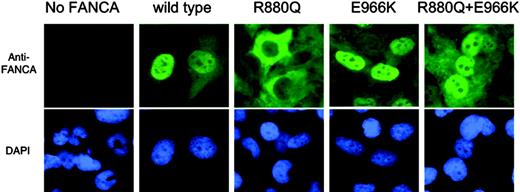
Subcellular localization of FANCA. Immunofluorescence in GM6914 cells retrovirally infected with constitutively expressed site-directed mutagenesis constructs. The R880Q mutation, but not the E966K mutation, prevents nuclear localization; both mutations in cis restore nuclear localization.
Subcellular localization of FANCA. Immunofluorescence in GM6914 cells retrovirally infected with constitutively expressed site-directed mutagenesis constructs. The R880Q mutation, but not the E966K mutation, prevents nuclear localization; both mutations in cis restore nuclear localization.
Determination of complementation group and mutation detection
Fibroblasts from one twin (proband) were cytogenetically analyzed for chromosome radial formation with mitomycin C (MMC) and diepoxybutane (DEB) treatment and were found to be fully sensitive to both agents (Table 1). However, lymphocytes from the same twin were found to be MMC resistant (Table 1). The MMC sensitivity of fibroblasts was corrected with a FANCA retrovirus (Table 2).
Cytogenetic analyses of PD839 cells
PD839 cells and clastogen concentration . | Radials, no. (%) . | FA range, % . | Normal range, % . |
|---|---|---|---|
| Fibroblasts | |||
| None | 0 (0) | 0-8 | 0 |
| MMC, 12 ng/mL | 11 (22) | 16-100 | 0 |
| MMC, 20 ng/mL | 25 (50) | 47-67 | 0 |
| DEB, 75 ng/mL | 18 (35) | 20-36 | 0 |
| Lymphocytes | |||
| MMC, 40 ng/mL | 3 (6) | 30-100 | 0-4 |
| DEB, 100 ng/mL | 0 (0) | 0-32 | 0 |
PD839 cells and clastogen concentration . | Radials, no. (%) . | FA range, % . | Normal range, % . |
|---|---|---|---|
| Fibroblasts | |||
| None | 0 (0) | 0-8 | 0 |
| MMC, 12 ng/mL | 11 (22) | 16-100 | 0 |
| MMC, 20 ng/mL | 25 (50) | 47-67 | 0 |
| DEB, 75 ng/mL | 18 (35) | 20-36 | 0 |
| Lymphocytes | |||
| MMC, 40 ng/mL | 3 (6) | 30-100 | 0-4 |
| DEB, 100 ng/mL | 0 (0) | 0-32 | 0 |
MMC/DEB-treated fibroblasts and lymphocytes were scored for formation of chromosomal radials.
Cytogenetic analyses of PD839 fibroblasts
PD839 fibroblasts and clastogen concentration . | Radials, no. (%) . | 1 Rad/cell, no. . | More than 1 Rad/cell, no. . |
|---|---|---|---|
| No vector | |||
| None | 0 (0) | 0 | 0 |
| MMC, 15 ng/mL | 23 (46) | 9 | 14 |
| DEB, 150 ng/mL | 17 (47) | 7 | 10 |
| RV:FANCA | |||
| None | 0 (0) | 0 | 0 |
| MMC, 15 ng/mL | 3 (6) | 2 | 1 |
| DEB, 150 ng/mL | 0 (0) | 0 | 0 |
PD839 fibroblasts and clastogen concentration . | Radials, no. (%) . | 1 Rad/cell, no. . | More than 1 Rad/cell, no. . |
|---|---|---|---|
| No vector | |||
| None | 0 (0) | 0 | 0 |
| MMC, 15 ng/mL | 23 (46) | 9 | 14 |
| DEB, 150 ng/mL | 17 (47) | 7 | 10 |
| RV:FANCA | |||
| None | 0 (0) | 0 | 0 |
| MMC, 15 ng/mL | 3 (6) | 2 | 1 |
| DEB, 150 ng/mL | 0 (0) | 0 | 0 |
Exon-by-exon sequencing of FANCA from genomic DNA revealed sequence changes as follows, with numbering referring to National Center for Biotechnology Information (NCBI) nucleotide database entry no. NM_000135; the start codon is at position 32. A single thymidine was deleted at position 2555 (2555ΔT) in exon 27, leading to a frameshift and resulting premature termination codon in exon 28. In exon 28, a nucleotide change of 2670G>A, resulting in an arginine-to-glutamine missense change at amino acid 880 (R880Q), was found. Both the previously described38 paternal 2555ΔT mutation and the novel maternal 2670G>A mutation were found in fibroblasts from the parent of origin as well as in the twins' fibroblasts, lymphoblasts, and whole blood DNA. In contrast, a third sequence change was detected distinctively in lymphoblast and peripheral blood DNA from both twins. This secondary alteration, in exon 30, was a 2927G>A nucleotide change, resulting in a glutamate-to-lysine change at amino acid 966 (E966K). By sequencing of RT-PCR products, this mutation was found to be in cis with the maternal exon 28 mutation (data not shown). The summary of these findings, confirmed by allele-specific restriction digests, can be found in Figure 1.
Constitutive expression of patient-derived alleles
To test the functional properties of the alleles found, the alleles were engineered into the pMMPpuroFANCA retroviral vector, and the corresponding retroviral supernatants were used to infect FANCA-null GM6914 cells. Transduced GM6914 cell lines were tested both by growth assay and by colony-forming ability assay for mitomycin C sensitivity. This demonstrated a clearly mutant phenotype for the paternal allele (2555ΔT), whereas the maternal allele, containing the exon 28 mutation (2670G>A), and the mosaic lymphoblast allele, containing both the exon 28 mutation and the acquired exon 30 mutation (2927G>A), demonstrated only intermediate phenotypes (Figure 2A). The latter outcome was confirmed with site-directed mutagenesis-derived constructs (data not shown). In colony formation assays, the mitomycin C sensitivity of the cell lines expressing either the maternal allele or lymphoblast allele construct was intermediate between that of wild-type and vector-only expressing cell lines (Figure 2B).
Cellular localization by immunofluorescence
As nuclear localization of FANCA protein is known to be required for function,31,39 the cellular localization of each of the mutants was studied. GM6914 cell lines infected with site-directed mutagenesis pMMPpuroFANCA-derived constructs were assayed for protein localization by immunofluorescence (Figure 3). As expected, wild-type FANCA was largely nuclear, but FANCA protein with the R880Q mutation localized mostly to the cytoplasm, indicating that not much FANCA was required to enter the nucleus for functional correction to occur. FANCA protein carrying either an E966K mutation or the R880Q + E966K double mutation localized mostly to the nucleus. Hence, the initial maternal mutation negatively affected subcellular localization of FANCA protein, and the acquired downstream mutation corrected this mislocalization. As these findings were observed in cell lines with overexpression-prone constructs, the erroneous localization of the mutant may not be the only reason for the mutant phenotype; protein and mRNA stability also may play a role at endogenous expression levels.
Doxycyclin-regulated expression of patient-derived constructs
It was hypothesized that the intermediate phenotype of the maternal allele displayed in Figure 2 might be due to constitutive, high-level expression, common to retroviral vectors, which would mask a mutation that resulted in reduced function (hypomorph). This possibility was supported by the immunofluorescence data, which clearly indicated a mutant phenotype for the maternal allele.
To test this hypothesis, the patient-derived alleles of FANCA were transferred into the tetracycline-repressible pREV-TRE vector (BD Biosciences), and the pREV-TRE-FANCA constructs, containing wild-type, maternal, paternal, or lymphoblast FANCA allele, were used to make retroviruses. GM6914 cells were made to express tTA via transfection with pREV-Tet-Off (BD Biosciences), and the 3 transfected clones that expressed the highest levels of tTA (clones D3, E1, and I15) were infected with the 4 pREV-TRE-FANCA retroviruses, each bearing a different FANCA allele. FANCA expression was determined to be Dox regulateable in the 4 cell lines derived from clone D3 by Western blot; regulated expression of the maternal allele is shown in Figure 4. Expression of other alleles also was repressed in the presence of Dox (data not shown).
Doxycyclin-regulated FANCA expression. Western blot demonstrating repression of expression of FANCA in Dox-responsive clone D3–derived cell lines, with addition of 0 to 2000 ng/mL Dox to cells. Lanes: norm indicates endogenous level control, wild-type cell line; neg indicates clone D3 expressing empty vector as negative control; maternal indicates D3 cell line expressing FANCA maternal allele, at 0, 31, and 500 ng/mL Dox; and wt indicates D3 cell line expressing wild-type FANCA allele, no Dox. Arrow indicates FANCA in endogenous level lane.
Doxycyclin-regulated FANCA expression. Western blot demonstrating repression of expression of FANCA in Dox-responsive clone D3–derived cell lines, with addition of 0 to 2000 ng/mL Dox to cells. Lanes: norm indicates endogenous level control, wild-type cell line; neg indicates clone D3 expressing empty vector as negative control; maternal indicates D3 cell line expressing FANCA maternal allele, at 0, 31, and 500 ng/mL Dox; and wt indicates D3 cell line expressing wild-type FANCA allele, no Dox. Arrow indicates FANCA in endogenous level lane.
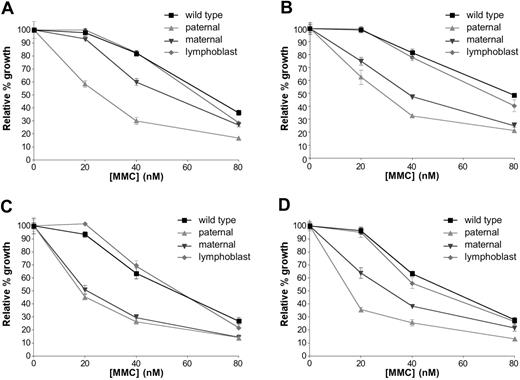
MMC survival curves of Dox-responsive clone-derived cell lines. (A) Clone D3–derived cell lines, without Dox, (B) D3-derived cell lines at 500 ng/mL Dox, (C) clone E1–derived cell lines at 31 ng/mL Dox, and (D) clone I15–derived cell lines at 125 ng/mL Dox. Error bars indicate SEM.
MMC survival curves of Dox-responsive clone-derived cell lines. (A) Clone D3–derived cell lines, without Dox, (B) D3-derived cell lines at 500 ng/mL Dox, (C) clone E1–derived cell lines at 31 ng/mL Dox, and (D) clone I15–derived cell lines at 125 ng/mL Dox. Error bars indicate SEM.
Cell growth was compared, in response to MMC, under different Dox concentrations. Without Dox, the maternal allele exhibited a phenotype intermediate between mutant and wild type (Figure 5A). With increasing Dox concentrations, resulting in reduced FANCA protein levels, the maternal form (R880Q) displayed reduced function, whereas the phenotypically reverted hematopoietic form (R880Q + E966K) demonstrated near wild-type function (Figure 5B-D). These findings demonstrated that the secondary change found in hematopoietic cells represents a functional reversion of the maternal allele, and that the 2670G>A (R880Q) change is a hypomorphic mutation that also can be compensated for by overexpression.
Secondary mutation acquired in HSC, which repopulated the blood
Since the acquired mutation was found in peripheral blood from both twins, and since these twins were more than 2 decades of age, it seemed likely that the secondary mutation occurred in a hematopoietic stem cell, leading to repopulation of the blood. This hypothesis was tested by confirming multilineage clonal hematopoiesis using X-inactivation assays and committed progenitor assays.
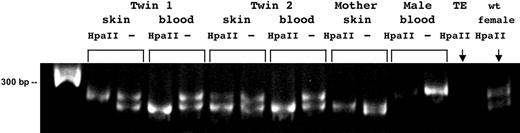
First, an assay for X-inactivation skewing was undertaken using genomic DNA extracted from fibroblasts and whole blood of both twins. One twin exhibited a greater degree of skewing of X-inactivation in fibroblasts than the other twin, a phenomenon that previously has been reported in monozygotic twins.40-47 More importantly, the assay demonstrated concordant extreme skewing in the whole blood (Figure 6). Specifically, one twin showed skewing of X-inactivation in opposite directions in her blood versus her skin, and X-inactivation in her blood was skewed in the same direction as it was for her sister's blood. This finding suggests that she was repopulated by a hematopoietic precursor cell from her twin sister,44 presumably with the hematopoietic progenitor or stem cell containing the acquired mutation.
Skewing of X chromosome inactivation. Products from PCR with Hpa II digested and mock-digested templates. Order of lanes: 100-bp ladder; then, paired, PCRs of digested and mock-digested DNA from: fibroblasts from one twin, whole blood from that twin, fibroblasts from second twin, whole blood from second twin, and fibroblasts from mother; then, as controls, Hpa II-digested and mock-digested male DNA as digestion controls, Hpa II-“digested” TE and Hpa II-digested female DNA as nonskewed control.
Skewing of X chromosome inactivation. Products from PCR with Hpa II digested and mock-digested templates. Order of lanes: 100-bp ladder; then, paired, PCRs of digested and mock-digested DNA from: fibroblasts from one twin, whole blood from that twin, fibroblasts from second twin, whole blood from second twin, and fibroblasts from mother; then, as controls, Hpa II-digested and mock-digested male DNA as digestion controls, Hpa II-“digested” TE and Hpa II-digested female DNA as nonskewed control.
Next, bone marrow was obtained from both sisters, and CFU-GMs and BFU-Es were plucked and analyzed for presence of the acquired mutation. The acquired mutation was present in 100% of colonies evaluated (180 CFU-GMs and 211 BFU-Es from one twin, 99 CFU-GMs and 96 BFU-Es from the other twin). Along with the finding of the acquired mutation in whole blood and in cultured lymphoblasts, this result validates the notion that a multilineage bone marrow–derived hematopoietic progenitor, probably an HSC, acquired the 2927G>A compensatory mutation.
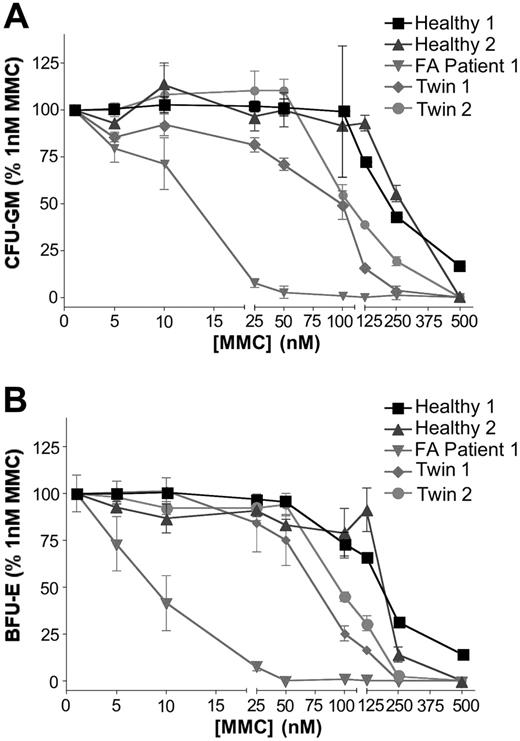
When CFU-GMs and BFU-Es from both twins were tested for MMC sensitivity, both cell types from both twins demonstrated dose-response curves similar to those for wild-type CFU-GMs and BFU-Es, respectively (Figure 7). This indicated normal MMC resistance and, therefore, functional correction of bone marrow–derived committed hematopoietic progenitor cells in both patients, implying correction of bone marrow HSCs. In addition, the karyotype for bone marrow from both twins was normal (46 X,X) in 100% of metaphases examined (data not shown).
Taken together, these data support the hypothesis that a single HSC in one twin acquired the 2927G>A compensatory FANCA mutation and, via self renewal, gave rise to more hematopoietic stem cells, which repopulated the bone marrow and peripheral blood of both twins.
Clonal growth of hematopoietic progenitor cells. The MMC dose-survival response of bone marrow–derived clonal progenitors from one Fanconi anemia patient (FA patient 1), 2 healthy volunteers (Healthy 1 and 2), and the twins (Twin 1 and 2). are shown. (A) CFU-GM, (B) BFU-E.
Clonal growth of hematopoietic progenitor cells. The MMC dose-survival response of bone marrow–derived clonal progenitors from one Fanconi anemia patient (FA patient 1), 2 healthy volunteers (Healthy 1 and 2), and the twins (Twin 1 and 2). are shown. (A) CFU-GM, (B) BFU-E.
Discussion
The case of somatic reversion reported here has important implications regarding the biology of human hematopoietic stem cells and the prospects for successful gene therapy in FA. The identical twins described herein were originally reported as examples of an FA-like syndrome because, while they had some classic features of FA, they were hematologically normal and did not display abnormal cross-linking agent–induced chromosome breakage in peripheral blood lymphocytes.19 Our results demonstrate conclusively that the patients in fact have FA, complementation group A. The severity of their nonhematologic findings and the in vitro expression studies of the mutant alleles demonstrate the expected loss of FANCA function, which, while partially due to mislocalization, also may result from reduced mRNA or protein stability. The fact that hematopoiesis is normal is due to an acquired compensatory mutation in hematopoietic cells. Importantly, both twins have the same single-base change in lymphoid, myeloid, and erythroid DNA. No additional blood-specific DNA alterations were found. Repopulation by maternal cells heterozygous for FA was ruled out. These observations can be explained in 2 ways. First, the reversion could have occurred independently in both twins early in life, followed by subsequent strong selective pressure at the stem cell level that resulted in normal hematopoiesis for more than 28 years. The second and more likely explanation is that the reversion occurred prenatally in a single HSC, resulting in engraftment of both patients via the shared circulation known to exist in monozygotic twins. The presence of multiple independent reversions rarely has been seen in other cases of mosaicism in FA. Furthermore, unassisted normalization of hematopoiesis for more than 2 decades also has not been formally reported. It therefore appears very unlikely that reversion at the stem cell level is common enough to occur multiple times, independently, in different stem cells of an individual patient. Regardless of whether the reversion occurred once in a prenatal HSC or independently in both twins postnatally, clearly very few HSCs were responsible for extremely long-term hematopoiesis, spanning decades of postnatal life. To our knowledge, this is the first formal report of curative, unassisted oligoclonal hematopoiesis over such a long time period in humans. Our findings powerfully confirm the substantial capacity of human HSCs to self renew.
Regarding the potential for gene therapy in FA, this case of “natural gene therapy” suggests that genetically corrected HSCs can prevent/correct the hematologic abnormalities characteristic of this disease. Here, the functionally normal HSC was spontaneously selected to become the dominant clone without the use of any exogenous myeloablative stress. Therefore, correction of only very few long-term repopulating HSCs could be sufficient to have significant therapeutic benefit, given sufficient time. The time required for successful in vivo selection of normal HSCs is not known, but it is noteworthy that one of the twins was reported to have some cells with abnormal chromosome breakage at 6 months of age. Spontaneous selection may therefore be a slow process, taking years. However, gentle, nonmyeloablative selection of corrected HSCs has been reported in murine gene therapy for Fancc16 and could potentially be used to safely accelerate the selection process, so long as the corrected cells are introduced prior to onset of hematologic symptoms, including anemia, pancytopenia, or clonal karyotypic abnormality.14 Based on the data presented in this paper, once the appropriate conditions for safe selection are established, very limited numbers of HSCs should be capable of repopulating the bone marrow and blood of an affected FA patient, as well as any histocompatible siblings, via serial stem cell transplantation.
Prepublished online as Blood First Edition Paper, January 5, 2006; DOI 10.1182/blood-2005-07-2638.
Supported by the National Heart, Lung, and Blood Institute, National Institutes of Health Program Project grant 1P01 HL48546; and an American Society of Hematology fellowship (T.T., A.D'A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal