Abstract
Protein inhibitor of activated STAT3 (PIAS3) functions in vivo as a key molecule in suppressing the transcriptional activity of both microphthalmia transcription factor (MITF) and signal transducer and activator of transcription 3 (STAT3), 2 transcription factors that play a major role in the regulation of growth and function in mast cells and melanocytes. Previously, we have demonstrated binding of PIAS3 to MITF leading to the inhibition of MITF transcriptional activity. Following cellular activation, PIAS3 is released from MITF and binds to STAT3. Now we have localized a common binding motif in PIAS3 for MITF and STAT3. This motif (PIAS82-132), which contains 50 amino acids, is sufficient for the inhibition of both MITF and STAT3. Three-dimensional protein modeling demonstrated that this motif contains 2 alpha helices. Disruption of one of the helices led to the loss of PIAS3 inhibitory activity. In addition to contributing to our understanding of the mechanisms of PIAS3 activity, these results could pave the way toward the formulation of an antioncogenic agent for the inhibition of both STAT3 and MITF.
Introduction
Microphthalmia transcription factor (MITF) is a basic helix-loop-helix leucine zipper (bHLH-Zip) DNA-binding protein.1 Its gene resides at the mi locus in mice,2 and mutation of this gene results in deafness, bone loss, small eyes, and poorly pigmented eyes and skin.3 The primary cell types affected in MITF-deficient mice are mast cells, osteoclasts, and melanocytes.3 In humans, mutation in this gene causes Waardenburg syndrome type II.4 MITF regulates the expression of mouse mast-cell protease 6 (mMCP-6),5 mMCP-5,6 c-kit,7 p75 nerve growth factor,6 granzyme B,8 tryptophan hydroxylase,9 and others. MITF has been described as a master regulatory gene for melanocytes10 and has been shown to play important roles in the regulation of Bcl-2 and CDK2.11
Recently, it has been demonstrated that MITF is amplified in a significant number of malignant melanomas and can act as an important lineage survival oncogene in these tumors.12 It has been proposed that, similar to the androgen receptor, MITF is important both for differentiation of melanocytes and for tumor transformation.12
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor involved in signal transduction pathways that are activated by several extracellular stimuli including the IL-6 family of cytokines. It is tyrosine phosphorylated by Janus kinase (JAK) and translocates as a dimer into the nucleus where it activates specific genes.13 STAT3 signaling has been shown to prevent programmed cell death and enhance cellular proliferation through the regulation of genes such as Bcl-XL, Mcl-l, c-Myc, and cyclin D1.14-17 Activated STAT3 is involved in the regulation of cell growth, differentiation, and survival.18 In many cancers, overactivation of STAT3 has been identified and was described to have a pivotal role in cancer progression through direct effects on the tumor-cell proliferation.19
Protein inhibitor of activated STAT3 (PIAS3)20 has been identified as an inhibitor of both activated STAT320 and MITF.21,22 In our previous work, we reported that the MITF-PIAS3-STAT3 network of interactions is set in motion when mast cells are activated by interleukin-6 (IL-6) or stem-cell factor (SCF). This leads to the phosphorylation and transcriptional activation of both MITF and STAT3 transcription factors.23 Moreover, PIAS3 stimulates MITF sumoylation at 2 sumoylation sites, MITF K182 and K316.24
The PIAS family of proteins contains several conserved domains: the SAP domain, which is required for repression of STAT1 by PIASy25 ; the Miz-Zn finger/RING domain, which is essential for SUMO (small ubiquitin-like modifier) ligase activity26 ; and the PINIT motif, which is required for nuclear retention of PIAS3.27
In the present work, we identified a short stretch of 50 amino acids in the PINIT27 domain of PIAS3 (PIAS382-132) that can directly interact with MITF and STAT3. Transfection of constructs containing PIAS382-132 resulted in down-regulation of MITF- and STAT3-dependent transcriptional activity. Furthermore, as a result of cross-species sequence analysis of PIAS3 and 3-dimensional peptide modeling, we identified an amino acid in this sequence, Y94, that is critical for PIAS3 binding to its target proteins. We explored the role played by this conserved Y94 in the structure of PIAS3 and in its function as an inhibitor of both MITF and STAT3.
Materials and methods
Cell culture and treatments
Melanoma BL6-B16 (ATCC no. CRL-6322; Manassas, VA) and RBL (ATCC no. CRL-2256) cells were cultured in a growth medium containing RPMI-1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, 2 mM nonessential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco Invitrogen, Grand Island, NY), and 50 μM β-mercaptoethanol (Fisher Scientific, Medford, MA). NIH 3T3 (ATCC no. CRL-6361) cells were cultured and maintained in a growth medium containing DMEM, supplemented as for RPMI-1640. All the cells were grown in a humidified incubator at 37°C with 5% CO2
NIH 3T3 cells were stimulated with a chimeric molecule of IL-6/IL-6R kindly provided by Dr Michel Revel (Weizmann Institute, Rehovot, Israel).
Plasmid construction
Mouse MITF (1129 bp) was subcloned into the XbaI and HindIII sites of the pcDNA3.1 (-) vector (Gibco Invitrogen). Mouse MITF (1129 bp) was inserted into the pGEX-4T-3 vector (Stratagene, La Jolla, CA). The MITF responsive reporter plasmid pTRPM128 was generously provided by Dr David E. Fisher (Harvard Medical School, Boston, MA). The luciferase reporter plasmid pSP72,29 containing the MITF binding region of the promoter and the first exon of the mMCP-6 gene (-191 to +26), was generously provided by Y. Kitamura (Osaka, Japan).
pGEX230 -mouse STAT3 plasmid was kindly provided by Dr Dov Zipori (Weizmann Institute). pcDNA-STAT3 flag tagged and the M67 pTATA Tk-Luc reporter gene31 were kindly provided Dr James E. Darnell (The Rockefeller University, New York, NY).
The cDNA encoding the open reading frame of mouse PIAS3 was subcloned into the XbaI and HindIII sites of the pcDNA3.1 (-) vector (Gibco Invitrogen). PIAS382-132 (150 bp) was subcloned into the XbaI and BamHI sites of the pcDNA3.1 (-) vector (Gibco Invitrogen). Fidelity of all constructs was verified by direct sequencing.
Transient cotransfection and luciferase assay
NIH 3T3 cells (5 × 105) were used in various luciferase assay experiments. NIH 3T3 cells were cotransfected with liposomes (Promega, San Luis Obispo, CA) with 0.1 μg luciferase reporter gene (of MITF or STAT3), 0.1 μg pcDNA-MITF, or 0.1 μg pcDNA-PIAS3, or with 0.1 μg pcDNA-PIAS382-132 (wild type or mutant), 0.1 μg pcDNA-STAT3, or 0.1 μg pcDNA (no insert was used as a nonspecific control). The cells were incubated in 24-well plates for 48 hours, treated or not with IL-6/IL-6R, lysed, and assayed for luciferase activity.
RBL cells (5 × 106) were also used in luciferase assay experiments. Cells were transfected using nucleofector technology (solution R, program T-20; Amaxa Biosystems, Gaithersburg, MD). Cells were transfected with 5 μg luciferase reporter gene (MITF or STAT3). pcDNA (5 μg) with no insert was used as a nonspecific control. The cells were incubated in plates for 24 hours, lysed, and assayed for luciferase activity. All of the luciferase activity was normalized to the total protein concentration. The normalized value was then divided by the luciferase activity obtained by cotransfection of the reporter with pcDNA alone. The ratio was expressed as the relative luciferase activity.
Real-time quantitative polymerase chain reaction (PCR)
MITF and STAT3 responsive genes were measured by using real-time quantitative PCR. Total RNA was extracted from transfected melanoma BL6-B16 and RBL cell line. The cells were transfected with 0.1 μg pcDNA-PIAS31-628 or with 0.1 μg PIAS382-132, and mRNA levels of various genes were quantified by SYBR-green incorporation (SYBR Green PCR Master Mix; Applied Biosystems, Foster City, CA). SYBR-green incorporation to double-strand DNA permits the direct detection of PCR product after each amplification cycle (ABI Prism 7000 sequence detection system; Applied Biosystems). The genes whose mRNA levels were quantified by real-time PCR are as follows: β-actin; STAT3 target genes: CyclinD1 and VEGF; MITF target genes: Tbx2, c-kit, Granzyme B, and tryptophan hydroxylase; and the MITF and STAT3 target gene: c-Fos.
Primers
Bacteriophage T7 transcription regulatory element and ribosome binding site were fused with the PIAS3 primers. The pcDNA3.1-mouse PIAS3 was used as a template. Primers for the PIAS382-132 and PIAS3443-585 were as follows: PIAS382-132: sense, 5′-CTAATACGACTCACTATAGGGAAGGAGATATACATATGGTCACCATG and antisense, 5′ TTACCTGGACGTGAGAATCTGCTGCAGCTGCTG; PIAS3443-585: sense, 5′ CTAATACGACTCACTATAGGGAAGGAGATATACATATGGGTCACCAGCCATCTTCGGTG and antisense, 5′ TCAGTCCAAGGAAATGACGTCTGACCG.
In vitro GST pull-down assay
MITF or STAT3 GST fusion proteins were expressed in protease-deficient E coli strain B12 and purified on glutathione-sepharose beads (Amersham Biosciences, Freiburg, Germany) essentially as described before.21 Pull-down assays32 were performed with GST-MITF or GST-STAT3 fusion protein (1 μg-5 μg) bound to sepharose beads and preincubated for 1 hour at 4°C in 1 mL binding buffer (PBS, 1 mM DTT, 1 mM EDTA, 5% glycerol, 0.1% NP40). 35S-labeled PIAS31-628 (1-10 μL) or its fragments were synthesized using the TNT-coupled rabbit reticulocyte lysate system (Promega) and then added to each preincubation mix, and the binding reaction was carried out overnight at 4°C. Beads were washed 4 times in 1 mL PBS/290 mM NaCl and boiled for 7 minutes in sample buffer, and aliquots were examined by electrophoresis. Integrity and quantity of GST fusions were confirmed by Blue stain reagent (Pierce Biotechnology, Rockford, IL), and autoradiography detected the amount of retained full-length PIAS3 and its fragments.
Computational structure prediction
Secondary structure prediction of both PIAS31-628 and PIAS382-132 was made using the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/33 ). Models of the tertiary structure of PIAS382-132 were created by the de novo structure prediction method ROBETTA. Initial models were obtained from the ROBETTA server (http://www.robetta.org34 ) and further refined by a standard full atom relax protocol.35 The final models were selected from the inspection of the 10 lowest scoring and structurally distant models.
Multiple sequence alignments
Protein sequences were retrieved from GenBank,36 and alignments were performed with ClustalW (EMBL, Heidelberg, Germany; http://www.ebi.ac.uk/clustalw/37 ).
Results
Characterization of the 50-amino acid domain in PIAS3 that is associated with both MITF and STAT3
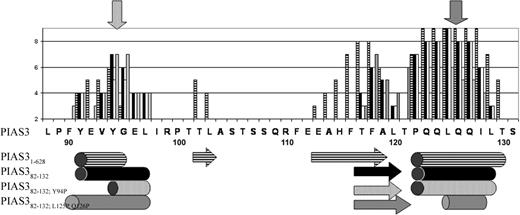
We have previously shown that PIAS3 associated with both MITF and STAT3 through the same region, which is a major part of the PINIT domain. Using ClustalW software, we observed that a 50-amino acid motif of the N-terminus of PIAS3 (PIAS582-132) is well conserved among PIAS family members (Figure 1).
In order to identify the minimal motif in PINIT that is responsible for the interaction with both MITF and STAT3, PCR fragments coding for 2 domains of PIAS3 were constructed (Figure 2A), PIAS382-132 and PIAS3443-585. MITF and STAT3 were expressed in bacteria as glutathione S-transferase (GST) fusion proteins, immobilized on glutathione-sepharose beads, and assayed for their ability to retain the in vitro-translated PIAS3 domains labeled with [35S]methionine.
Alignment of PIAS family members. Fifty amino acids of the PIAS proteins (with their indicated GenBank accession numbers) have been aligned using ClustalW software. Mus indicates Mus musculus; Rat, Rattus norvegicus; and Homo, Homo sapiens. Arrows point to the 3 amino acids that were mutated.
Alignment of PIAS family members. Fifty amino acids of the PIAS proteins (with their indicated GenBank accession numbers) have been aligned using ClustalW software. Mus indicates Mus musculus; Rat, Rattus norvegicus; and Homo, Homo sapiens. Arrows point to the 3 amino acids that were mutated.
The results clearly show that PIAS382-132 is responsible for the association with both MITF and STAT3 (Figure 2B). Both PIAS382-132 and full-length PIAS3 (PIAS31-628) were associated only with GST-MITF or GST-STAT3, but not with the control GST (Figure 2B) or with PIAS3443-585 (data not shown). The unlabeled PIAS382-132 and PIAS3443-585 that were synthesized in vitro with similar efficiency (data not shown) were used as competitors for PIAS31-628 that was labeled with [35S]methionine (Figure 2C). These results clearly show that PIAS382-132 associated strongly and specifically with both MITF and STAT3 in contrast to PIAS3443-585, and thus that MITF and STAT3 interact specifically with the same domain (PIAS382-132) in PIAS3.
A domain of 50 amino acids in PIAS3 (PIAS382-132) is associated with both MITF and STAT3. (A) Schematic representation of the full-length mouse PIAS3 (PIAS31-628), including the SAP domain, PINIT motif, the zinc finger motifs, and the 2 domains of PIAS3 investigated in this work. (B) Fragments PIAS382-132 and full-length PIAS3 (PIAS31-628) labeled with [35S]methionine in vitro were added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS31-628 or PIAS382-132 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Input and GST alone are shown as control. One representative of 3 experiments is shown. (C) PIAS31-628 labeled with [35S]methionine in vitro was added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Increasing concentrations of in vitro-translated unlabeled PIAS382-132 or PIAS3443-585 was added to the mixtures. Retained [35S]-labeled PIAS31-628 was determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown.
A domain of 50 amino acids in PIAS3 (PIAS382-132) is associated with both MITF and STAT3. (A) Schematic representation of the full-length mouse PIAS3 (PIAS31-628), including the SAP domain, PINIT motif, the zinc finger motifs, and the 2 domains of PIAS3 investigated in this work. (B) Fragments PIAS382-132 and full-length PIAS3 (PIAS31-628) labeled with [35S]methionine in vitro were added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS31-628 or PIAS382-132 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Input and GST alone are shown as control. One representative of 3 experiments is shown. (C) PIAS31-628 labeled with [35S]methionine in vitro was added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Increasing concentrations of in vitro-translated unlabeled PIAS382-132 or PIAS3443-585 was added to the mixtures. Retained [35S]-labeled PIAS31-628 was determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown.
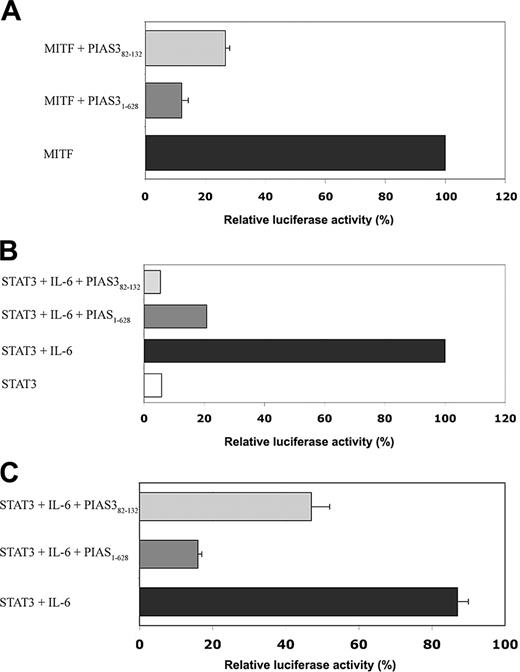
The inhibitory effect of PIAS382-132 on MITF and STAT3 transcriptional activity. (A) NIH-3T3 cells were cotransfected with an expression plasmid containing MITF, with a MITF luciferase reporter gene containing the mMCP-6 promoter, and with an expression plasmid containing either PIAS31-628 or PIAS382-132. Mean ± SE of 4 experiments is shown. (B) NIH-3T3 cells were cotransfected with the STAT3 luciferase reporter gene (M67), with an expression plasmid containing STAT3, and with expression plasmids containing either PIAS31-628 or PIAS382-132. Cells were then treated with IL-6/IL-6R for 6 hours, with untreated cells as the control. One representative of 3 experiments is shown. (C) RBL cells were nuclear transfected with the STAT3 luciferase reporter gene (M67), and with expression plasmids containing either PIAS31-628 or PIAS382-132. Mean ± SE of 4 experiments is shown. In all cases, luciferase activity of lysed cells was measured and normalized against protein concentration. For each transfection, the total DNA concentration was kept constant by complementing with the empty vector pcDNA.
The inhibitory effect of PIAS382-132 on MITF and STAT3 transcriptional activity. (A) NIH-3T3 cells were cotransfected with an expression plasmid containing MITF, with a MITF luciferase reporter gene containing the mMCP-6 promoter, and with an expression plasmid containing either PIAS31-628 or PIAS382-132. Mean ± SE of 4 experiments is shown. (B) NIH-3T3 cells were cotransfected with the STAT3 luciferase reporter gene (M67), with an expression plasmid containing STAT3, and with expression plasmids containing either PIAS31-628 or PIAS382-132. Cells were then treated with IL-6/IL-6R for 6 hours, with untreated cells as the control. One representative of 3 experiments is shown. (C) RBL cells were nuclear transfected with the STAT3 luciferase reporter gene (M67), and with expression plasmids containing either PIAS31-628 or PIAS382-132. Mean ± SE of 4 experiments is shown. In all cases, luciferase activity of lysed cells was measured and normalized against protein concentration. For each transfection, the total DNA concentration was kept constant by complementing with the empty vector pcDNA.
Effect of PIAS382-132 on MITF and STAT3 transcriptional activity
To examine the effect of PIAS382-132 on the transcriptional activity of MITF, we used the transient cotransfection assay. The luciferase reporter plasmid containing the mMCP-6 (MITF target gene) promoter was used. The luciferase reporter construct was cotransfected into 3T3-NIH (mouse embryo fibroblast) cells with an expression plasmid (pcDNA) containing MITF, and with expression plasmids containing PIAS31-628, PIAS382-132, or no insert (control). The expression of PIAS382-132 resulted in about 73% inhibition of MITF transcriptional activity (Figure 3A), compared with about 88% inhibition seen with PIAS31-628.
In a complementary set of experiments, we examined the effect of the PIAS382-132 on the transcriptional activity of STAT3. The STAT3 reporter gene (M67) was cotransfected in 3T3-NIH cells with an expression plasmid containing STAT3, and with expression plasmids containing PIAS31-628, PIAS382-132, or no insert. After incubation of 48 hours, the cotransfected cells were treated for 6 hours with IL-6/IL-6R, a known STAT3 activator,38 and then luciferase activity was measured. As shown in Figure 3B, IL-6/IL-6 receptor (IL-6/IL-6R) induced STAT3 luciferase activity, whereas the expression of PIAS31-628 or PIAS382-132 resulted in a substantial reduction of the luciferase activity. In addition, rat basophilic leukemia (RBL) cells were nuclear transfected with STAT3 reporter gene (M67) and expression plasmid containing PIAS31-628, PIAS382-132, or no insert. Similar reductions in luciferase activity were obtained in these cells when transfected with PIAS31-628 and PIAS382-132 (Figure 3C).
These results indicate the ability of PIAS382-132 to inhibit the transcriptional activity of both MITF and STAT3 in transiently transfected cell lines.
The effect of PIAS382-132 on STAT3- and MITF-mediated gene expression
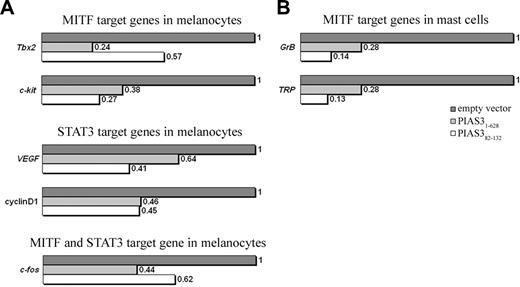
The inhibitory effect of PIAS382-132 on the endogenous transcriptional activity of MITF and STAT3 in melanocytes and mast cells was assessed using B16 (mouse melanoma cell line) or RBL cells, respectively, transfected with expression plasmids containing either PIAS31-628 or PIAS382-132. Quantitative real-time PCR was performed on the RNA samples for the following genes: (1) MITF target genes: c-kit,7 T-box transcription factor 2 (Tbx2),17,39 Granzyme B8 (GrB), and tryptophan hydroxylase40 (TRP); (2) STAT3 target genes: CyclinD126 and vascular endothelial growth factor (VEGF)41 ; (3) MITF and STAT3 target gene: c-fos.42
The effect of PIAS382-132 on STAT3- and MITF-mediated gene expression. (A) Quantitative real-time PCR analysis of MITF target genes (c-Kit and Tbx2), STAT3 target genes (CyclinD1 and VEGF), and MITF and STAT3 target gene (c-fos) from transfected melanocytes. (B) Quantitative RT-PCR analysis of MITF mast cell target genes (GrB and TRP) from transfected mast cells. In both cases, cells were transfected with expression plasmids containing PIAS31-628, PIAS382-132, or an empty vector. Total RNA (1.7 μg) was used from each transfection experiment. Transcript levels of MITF and STAT3 target genes were normalized to actin and performed in triplicate.
The effect of PIAS382-132 on STAT3- and MITF-mediated gene expression. (A) Quantitative real-time PCR analysis of MITF target genes (c-Kit and Tbx2), STAT3 target genes (CyclinD1 and VEGF), and MITF and STAT3 target gene (c-fos) from transfected melanocytes. (B) Quantitative RT-PCR analysis of MITF mast cell target genes (GrB and TRP) from transfected mast cells. In both cases, cells were transfected with expression plasmids containing PIAS31-628, PIAS382-132, or an empty vector. Total RNA (1.7 μg) was used from each transfection experiment. Transcript levels of MITF and STAT3 target genes were normalized to actin and performed in triplicate.
The real-time PCR analysis on the melanoma cells clearly shows that the transcript levels of MITF target genes such as c-kit and Tbx2 and of STAT3 target genes such as CyclinD1 and VEGF were significantly suppressed when cells were transfected with the expression plasmid containing PIAS382-132 (Figure 4A). No such suppression of transcript levels of MITF and STAT3 target genes in these cells was observed with the empty vector only (pcDNA).
Recently, it was demonstrated by promoter analysis and chromatin immunoprecipitation assay that both STAT3 and MITF cooperatively up-regulate the expression of c-fos.42 In accordance with this observation, we demonstrate here that c-fos expression is also suppressed in melanocytes transfected with expression vector containing PIAS382-132 (Figure 4A).
The transcript levels of MITF target genes such as GrB and TRP in mast cells were measured by real-time PCR analysis on transfected RBL cells. Both the expression of PIAS382-132 and of PIAS31-628 in RBL cells caused a decrease in the transcript levels of MITF target genes (Figure 4B).
Secondary structure prediction of PIAS382-132
The function of PIAS382-132 was further evaluated based on computational structure prediction. Secondary structure prediction programs such as PSIPRED33 suggested several segments of secondary structure for PIAS31-628 and PIAS382-132, of which the most confident predictions include a short N-terminal helix (positions F90-L97) and a C-terminal helix (positions P122-L129) (Figure 5). An additional strand in the central region (positions F116-L120) was predicted with less confidence, due to the absence of a pairing strand and low hairpin probability.43
Predicted effects of the Y94P and L125P, Q126P mutations on the secondary structure of the PIAS382-132 segment. The secondary structure was predicted with PSIPRED. Confidence values (ranging from 0-9; 9 indicates highest confidence) are shown in bars along the PIAS3 sequence for the PIAS31-628 protein ( ), the segment PIAS382-132 (▪), and the mutants PIAS382-132; Y94P (□) and PIAS382-132; L125P, Q126P (
), the segment PIAS382-132 (▪), and the mutants PIAS382-132; Y94P (□) and PIAS382-132; L125P, Q126P ( ). Predicted α helices and β strands are shown below the graph as tubes and arrows, respectively. Predictions are similar for PIAS31-628 and PIAS382-132, except for more confident location of a central β strand in the full sequence secondary structure prediction. Mutation(s) to proline reduce the confidence level of the helix prediction, both for the N- and C-terminal helix. This effect is somewhat more expressed at the N-terminus due to its initially weaker helical signal. This figure indicates that the N-terminal helix might be abolished by the Y94P mutation, while the C-terminal helix is significantly shortened by the L125P, Q126P double mutation.
). Predicted α helices and β strands are shown below the graph as tubes and arrows, respectively. Predictions are similar for PIAS31-628 and PIAS382-132, except for more confident location of a central β strand in the full sequence secondary structure prediction. Mutation(s) to proline reduce the confidence level of the helix prediction, both for the N- and C-terminal helix. This effect is somewhat more expressed at the N-terminus due to its initially weaker helical signal. This figure indicates that the N-terminal helix might be abolished by the Y94P mutation, while the C-terminal helix is significantly shortened by the L125P, Q126P double mutation.
Predicted effects of the Y94P and L125P, Q126P mutations on the secondary structure of the PIAS382-132 segment. The secondary structure was predicted with PSIPRED. Confidence values (ranging from 0-9; 9 indicates highest confidence) are shown in bars along the PIAS3 sequence for the PIAS31-628 protein ( ), the segment PIAS382-132 (▪), and the mutants PIAS382-132; Y94P (□) and PIAS382-132; L125P, Q126P (
), the segment PIAS382-132 (▪), and the mutants PIAS382-132; Y94P (□) and PIAS382-132; L125P, Q126P ( ). Predicted α helices and β strands are shown below the graph as tubes and arrows, respectively. Predictions are similar for PIAS31-628 and PIAS382-132, except for more confident location of a central β strand in the full sequence secondary structure prediction. Mutation(s) to proline reduce the confidence level of the helix prediction, both for the N- and C-terminal helix. This effect is somewhat more expressed at the N-terminus due to its initially weaker helical signal. This figure indicates that the N-terminal helix might be abolished by the Y94P mutation, while the C-terminal helix is significantly shortened by the L125P, Q126P double mutation.
). Predicted α helices and β strands are shown below the graph as tubes and arrows, respectively. Predictions are similar for PIAS31-628 and PIAS382-132, except for more confident location of a central β strand in the full sequence secondary structure prediction. Mutation(s) to proline reduce the confidence level of the helix prediction, both for the N- and C-terminal helix. This effect is somewhat more expressed at the N-terminus due to its initially weaker helical signal. This figure indicates that the N-terminal helix might be abolished by the Y94P mutation, while the C-terminal helix is significantly shortened by the L125P, Q126P double mutation.
The importance of Y94 at the N-terminal helix of PIAS382-132 for binding
In order to assess the importance of the N- and C-terminal-predicted helices to the structure and binding function of PIAS382-132, we mutated residues in those helices to proline, a helix breaker.44 The importance of the N-terminal helix was assessed by the Y94P mutation, while the C-terminal helix was assessed by a double mutation of L125P and Q126P. Secondary structure prediction for the mutants indicates that the helices will be significantly shortened—and thus probably removed (Figure 5). In addition, sequence alignment of PIAS sequences showed that Y94 is conserved in the subset of PIAS3 family members (Figure 1, arrows), suggesting a possible functional role of this residue.
The PIAS382-132; Y94P mutation has lost its ability both to interact with MITF and STAT3 and to inhibit their transcriptional activity. (A) In vitro-translated and [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were incubated with GST-MITF, GST-STAT3, or GST immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown. (B) Luciferase reporter plasmids, containing a MITF promoter region of the mMCP-6, or a STAT3 reporter gene, M67, were cotransfected into NIH-3T3 cells, with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, or PIAS382-132; Q125P, L126P. Luciferase activity of lysed cells was measured and normalized against protein concentration. For each transfection, the total DNA concentration was constant by complementing with the empty vector pcDNA. The mean ± SE of 4 experiments is shown. (C) Quantitative RT-PCR analysis of MITF target gene (TRP) from transfected RBL cells. Cells were nuclear transfected with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, PIAS382-132; Q126P, L125P, or no insert. Transcript levels were normalized to actin and performed in triplicates. One representative of 3 experiments is shown.
The PIAS382-132; Y94P mutation has lost its ability both to interact with MITF and STAT3 and to inhibit their transcriptional activity. (A) In vitro-translated and [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were incubated with GST-MITF, GST-STAT3, or GST immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown. (B) Luciferase reporter plasmids, containing a MITF promoter region of the mMCP-6, or a STAT3 reporter gene, M67, were cotransfected into NIH-3T3 cells, with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, or PIAS382-132; Q125P, L126P. Luciferase activity of lysed cells was measured and normalized against protein concentration. For each transfection, the total DNA concentration was constant by complementing with the empty vector pcDNA. The mean ± SE of 4 experiments is shown. (C) Quantitative RT-PCR analysis of MITF target gene (TRP) from transfected RBL cells. Cells were nuclear transfected with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, PIAS382-132; Q126P, L125P, or no insert. Transcript levels were normalized to actin and performed in triplicates. One representative of 3 experiments is shown.
MITF and STAT3 were expressed in bacteria as GST fusion proteins, immobilized on glutathione-sepharose beads, and assayed for their ability to retain the in vitro-translated PIAS382-132 or its mutants (PIAS382-132; Y94P, PIAS382-132; L125P, Q126P) labeled with [35S]methionine. The results clearly show that the Y94P mutation abolished the ability of PIAS3 to associate with either MITF or STAT3, either due to disruption of the N-terminal helix by the mutation to proline or to the direct involvement of Y94 in binding (Figure 6A).
We then determined the effect of PIAS382-132 and its mutants on MITF or STAT3 transcriptional activity. NIH 3T3 fibroblasts were cotransfected either with an MITF reporter gene (mMCP-6) or with a STAT3 reporter gene (M67), and expression plasmids containing MITF, STAT3, PIAS31-628, PIAS382-132, PIAS382-132; Y94P, PIAS382-132; L125P, Q126P, or no insert. Almost no inhibitory effect was observed when cells were transfected with the PIAS382-132; Y94P mutant (Figure 6B). In contrast, the PIAS382-132; L125P, Q126P mutant caused about 50% inhibition of both MITF and STAT3 transcriptional activity. In the same experiment, PIAS31-628 and PIAS382-132 wild type showed about 60% inhibition of both MITF and STAT3 transcriptional activity (Figure 6B).
The inhibitory effect of different mutations of PIAS382-132 on the endogenous transcriptional activity of MITF in mast cells was assessed using RBL cells transfected with expression plasmid containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, PIAS382-132; L125P, Q126P, or no insert. Quantitative real-time PCR was performed to analyze the transcript level of the MITF target gene TRP.40 Real-time PCR analysis clearly shows a significantly suppressed expression of the MITF target gene upon transfection with the expression plasmid containing PIAS382-132; L125P, Q126P (Figure 6C). No such suppression was observed for the MITF target gene in cells transfected with the expression plasmid containing either PIAS382-132; Y94P or empty vector (pcDNA).
These experimental results indicate that the N-terminal helix is responsible for the binding of PIAS3 to STAT3 and MITF, while shortening the C-terminal helix does not abolish these interactions. Furthermore, these data indicate that Y94 of the N-terminal helix plays a critical role in this interaction and in suppression of MITF and STAT3 transcriptional activity.
Discussion
PIAS3 is a regulator protein of several key transcription factors, including NFκB,45 estrogen receptor,46 STAT3,20 and MITF.23 Several functions have been described for this protein—as an E3 SUMO ligase,46 as a transcriptional coactivator/repressor,45 and as an inhibitor of the binding of the transcription factor to DNA.23 The mechanisms involved in the binding of PIAS3 to its target and the modulation of their transcription have just begun to be explored. For example, it was found that the N-terminal part of PIAS3 is sufficient for binding to NFκB and that the transcriptional inactivation of this factor depends on a short sequence in the N-terminus of PIAS3.45 Others have found that the protein domain PINIT allows nuclear translocation of PIAS3.27
In the current study, we defined the specific sequence that allows PIAS3 to bind to 2 different transcription factors—STAT3 and MITF. Using the GST pull-down assay, we defined a short domain of PIAS3 (PIAS382-132), which can bind both to MITF and STAT3. These 50 amino acids are required for binding and inhibition of both MITF and STAT3 transcriptional activity in melanocytes and mast cells (Figures 3, 4). To verify the potential inhibition activity of the PIAS382-132 peptide on the endogenous transcriptional activities of MITF and STAT3, we used real-time PCR to demonstrate that introduction and expression of this peptide into mast cells or melanocytes inhibits the expression of some of the known target genes of these transcription factors (Figure 4).
It has previously been described that PIAS3, but not PIAS1, promotes sumoylation of MITF.47 Since this short amino acid sequence does not contain the sumoylation domain of PIAS3 and also does not contain the short sequence important for NFκB inactivation,45 our current findings imply that PIAS3 can inhibit both MITF and STAT3 by a mechanism independent of sumoylation and of NFκB inactivation. Furthermore, whether PIAS3 works in vivo under most circumstances mainly as a sumo ligase modulating the gene-specific transcriptional activity of MITF, or via stoichiometric binding and inhibition of MITF transcriptional activity, is not clear at this point. It seems that PIAS3 behaves in a similar way to an enzyme, so that lower levels of PIAS3 compared with MITF might still be sufficient to allow the sumoylation of many MITF molecules. Thus the mode of action of PIAS3 as a transcriptional repressor might depend on the specific stoichiometric relationship between itself and its target proteins in the cell at a specific time point.
As described in “Introduction,” overactivation of both MITF and STAT3 has an important pro-oncogenic role in certain cancers.12,19 These observations provide substantial support to the notion that PIAS382-132 contains amino acids that are essential for the inhibitory activity of PIAS3 on MITF and STAT3. Small peptides constructed on the basis of PIAS382-132 could be the starting point for the formulation of potent inhibitors for both of these transcription factors. Such an approach might prove to be especially effective for tumors such as melanoma where both MITF and STAT3 transcription factors play an important role in tumor development and growth.
Using computational models of the secondary and tertiary structure of PIAS382-132, we predicted that this region contains 2 peptide helices. Cross-species analysis showed that Y94 is conserved in PIAS3 sequences only, suggesting its involvement in PIAS3-specific interactions to STAT3 and MITF. When we replaced this tyrosine with proline, a helix breaker, PIAS3 lost its ability to bind and inhibit MITF and STAT3 activity both in vitro and in vivo (Figure 6). The loss of binding could be due either to the disruption of the N-terminal helix or to the elimination of important contributions of the tyrosine side chain at the interface of PIAS3-STAT3 and PIAS3-MITF complexes. In order to evaluate these possibilities further, models of the tertiary structure of PIAS382-132 were created with the de novo structure prediction protocol of ROBETTA.34,35 The models were clustered based on structural similarity, and lowest energy models of large clusters were further inspected for the location and importance of the N- and C-terminal helices for the protein fold, with specific emphasis on potential contributions of Y94.
Figure 7A shows the lowest energy model encountered in this prediction, containing a tightly packed core. According to this model, either the relatively exposed Y94 side chain itself or the N-terminal helix in general could contribute directly to binding. Figure 7B shows a low-energy model taken from a largely populated cluster (large clusters indicate that a particular solution is found frequently, increasing the confidence in a correct prediction). Here, Y94 points more into the protein center, suggesting possible contributions to protein stability in addition to binding. Finally, Figure 7C shows another low-energy model from a highly populated cluster, including a beta hairpin to satisfy the hydrogen bond pairing in beta sheets. In this model, Y94 points into the protein center, and L125 and Q126 are in a distinct region of the protein, suggesting a structural role for Y94. These models imply the importance of Y94 in stability of the protein structure. In all models, the importance of the N-terminal helix to binding and stability cannot be precisely defined, and additional experiments are needed to evaluate the precise role of this helix in general, and of Y94 specifically.
Possible structures for the PIAS382-132 segment created by ROBETTA de novo modeling. Three representative models are shown: (A) the lowest energy model of the prediction runs; (B) low-energy model of largest cluster that contains lowest energy models; and (C) a model that includes a beta hairpin. The predicted elements of secondary structure (according to PSIPRED, Figure 5) are colored in red (helix) and yellow (strand). The positions that were mutated to proline (eg, Y94, L125, and Q126) are shown in sticks. Y94 points mostly into the core but is also accessible to interactions with protein partners. In contrast, Q126 is mostly exposed.
Possible structures for the PIAS382-132 segment created by ROBETTA de novo modeling. Three representative models are shown: (A) the lowest energy model of the prediction runs; (B) low-energy model of largest cluster that contains lowest energy models; and (C) a model that includes a beta hairpin. The predicted elements of secondary structure (according to PSIPRED, Figure 5) are colored in red (helix) and yellow (strand). The positions that were mutated to proline (eg, Y94, L125, and Q126) are shown in sticks. Y94 points mostly into the core but is also accessible to interactions with protein partners. In contrast, Q126 is mostly exposed.
To summarize, our results demonstrate that a short amino acid sequence from the N-terminus of PIAS3, PIAS382-132, is able to bind and inhibit the transcription factors MITF and STAT3. Furthermore, replacement of Y94 by proline leads to loss of the transcriptional repression of STAT3 and MITF by PIAS3. These results shed new light on possible regulatory mechanisms of PIAS3 activity on the pro-oncogenic transcription factors MITF and STAT3. In addition, these findings open unique experimental approaches for the formulation of effective inhibitors based on the interaction between PIAS3 and these transcription factors.
Prepublished online as Blood First Edition Paper, December 20, 2005; DOI 10.1182/blood-2005-08-3325.
Supported by United States Binational Science Foundation (grant 2003009; E.R.), the Israeli Academy of Science (grant 144/04; E.R.), and the German-Israeli Foundation for Scientific Research and Development (grant I-726-10.2/2002; E.R.).
C.L. and Y.-N.L. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr G. Kay for article preparation.


![Figure 2. A domain of 50 amino acids in PIAS3 (PIAS382-132) is associated with both MITF and STAT3. (A) Schematic representation of the full-length mouse PIAS3 (PIAS31-628), including the SAP domain, PINIT motif, the zinc finger motifs, and the 2 domains of PIAS3 investigated in this work. (B) Fragments PIAS382-132 and full-length PIAS3 (PIAS31-628) labeled with [35S]methionine in vitro were added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS31-628 or PIAS382-132 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Input and GST alone are shown as control. One representative of 3 experiments is shown. (C) PIAS31-628 labeled with [35S]methionine in vitro was added to GST-MITF or GST-STAT3 that was immobilized on glutathione-sepharose beads. Increasing concentrations of in vitro-translated unlabeled PIAS382-132 or PIAS3443-585 was added to the mixtures. Retained [35S]-labeled PIAS31-628 was determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/7/10.1182_blood-2005-08-3325/5/m_zh80070693320002.jpeg?Expires=1769119629&Signature=CJXHbr3jHk48JOpFXfNDrrQ9xWOlfX52LG24zXmZ3h2xvgieY7t-GKrAJ789Cwth9hHFY2zwLp5YCjt0V8iBl-dP5edqzPb38dQkrdBjXNfrPgZJ4SpDZNc5Ldg~a1UGou-Y5C9UVVevvmUIw-clcwvUppqmzKGNKY6r393U8iGk86V1ql3X5RQK2WBZ9IYS4nssNgbIMKAjzeAJiJ8Brvry4AJWanwhIibxLMw1huf~ZCENhfF3NlC7GvnqXV1VEyqd44GrNiSxnez3yZUehkM9PmtqJ2Ah7hkJZpfrxl7i4aZnbbalC96p4xVDZrW61wV8p4-UZ-xWuVoxDE06ug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. The PIAS382-132; Y94P mutation has lost its ability both to interact with MITF and STAT3 and to inhibit their transcriptional activity. (A) In vitro-translated and [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were incubated with GST-MITF, GST-STAT3, or GST immobilized on glutathione-sepharose beads. Retained [35S]-labeled PIAS382-132, PIAS382-132; Y94P, and PIAS382-132; Q125P, L126P were determined by SDS-PAGE and autoradiography. One representative of 3 experiments is shown. (B) Luciferase reporter plasmids, containing a MITF promoter region of the mMCP-6, or a STAT3 reporter gene, M67, were cotransfected into NIH-3T3 cells, with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, or PIAS382-132; Q125P, L126P. Luciferase activity of lysed cells was measured and normalized against protein concentration. For each transfection, the total DNA concentration was constant by complementing with the empty vector pcDNA. The mean ± SE of 4 experiments is shown. (C) Quantitative RT-PCR analysis of MITF target gene (TRP) from transfected RBL cells. Cells were nuclear transfected with expression plasmids containing PIAS31-628, PIAS382-132, PIAS382-132; Y94P, PIAS382-132; Q126P, L125P, or no insert. Transcript levels were normalized to actin and performed in triplicates. One representative of 3 experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/7/10.1182_blood-2005-08-3325/5/m_zh80070693320006.jpeg?Expires=1769119629&Signature=QVf0oUL7bAOWPG~jF4v09gecD84R3f1CF7rHT0ifsYl2UWrvbTfEZcfHvtqG9zaFPkaJME1l0gdy8ASN7WE9nlZ7lew5nUGvu~AfZ~54pm~JNPW~42MrtqY9RrVWlAZJFUIjXm8alQ1cwM5r6f30MTiDi3Ijpm5MWACR69plBFABXMheMWFoBcd933LQfgIbtDU8Go6kpqtMvtd3MWXNH8zKVFR~zAwUJo8le6fPOa566jmGCKNvrhnFX5uiA6lfZwgZKNWZCQmRZ6sV3csYR30X9OMdg~ib5fTxk5rVPbGl~xJEhBdN9FDJgHPMveg9psaW5bVzPHXNlNprj67mqA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal