Abstract
Tumor necrosis factor (TNF)–related activation-induced cytokine (TRANCE) induces osteoclast formation from monocyte/macrophage lineage cells via various transcription factors, including the Mi transcription factor (Mitf). Here, we show that inhibitors of differentiation/DNA binding (Ids), helix-loop-helix (HLH) transcription factors, negatively regulate TRANCE-induced osteoclast differentiation. Expression levels of Id1, Id2, and Id3 genes are significantly reduced by TRANCE during osteoclastogenesis. Interestingly, overexpression of the 3 Id genes in bone marrow–derived monocyte/macrophage lineage cells (BMMs) inhibits the formation of tartrate-resistant acid phosphatase (TRAP)–positive multinuclear osteoclasts, but it does not alter the ability of BMMs to either phagocytose or differentiate into dendritic cells (DCs). Overexpression of Id2 in BMMs attenuates the gene induction of nuclear factor of activated T cells c1 (NFATc1) and osteoclast-associated receptor (OSCAR) during TRANCE-mediated osteoclastogenesis. Furthermore, Id proteins interact with Mitf, a basic HLH (bHLH) transcription factor, and inhibit its transactivation of OSCAR, which is a costimulatory receptor expressed by osteoclast precursors, by attenuating the DNA binding ability of Mitf to the E-box site of the OSCAR promoter. Taken together, our results reveal both a new facet of negative regulation, mediated by Id proteins, as well as the mechanism whereby TRANCE signaling overcomes it, allowing osteoclastogenesis to proceed.
Introduction
Osteoclasts play an important role in bone metabolism by resorbing the bone matrix. These cells originate from hematopoietic precursors and share a common progenitor with macrophages and dendritic cells (DCs). Two essential cytokines, macrophage colony-stimulating factor (M-CSF) and tumor necrosis factor (TNF)–related activation-induced cytokine (TRANCE; also called RANKL, OPGL, and ODF), enable osteoclast differentiation from their monocyte/macrophage lineage precursors.1-3
It has been shown that TRANCE induces activation and/or induction of transcription factors such as Mi transcription factor (Mitf), PU.1, and nuclear factor of activated T cells c1 (NFATc1).4-6 Mitf is known to be important for osteoclastogenesis in vitro and in vivo.7-9 TRANCE activates Mitf via the MKK6/p38 signaling cascade. Subsequently, activated Mitf induces the expression of target genes, including tartrate-resistant acid phosphatase (TRAP), cathepsin K, and osteoclast-associated receptor (OSCAR), which are important for osteoclast differentiation or function,9-13 by binding to the canonical E-box sequence in the promoter region of those genes.
The helix-loop-helix (HLH) family of transcriptional regulatory proteins have important roles in developmental processes including neurogenesis, myogenesis, and hematopoiesis.14 Tissue-specific basic HLH (bHLH) proteins form dimers with E proteins, ubiquitously expressed bHLH transcription factors, which bind to the E-box sequence CANNTG. HLH proteins can bind to bHLH transcription factors via their HLH domain; the resulting heterodimers are unable to bind to DNA because HLH proteins lack the necessary basic motif.15 Thus, HLH proteins act as dominant-negative regulators of bHLH transcription factors. One of the HLH subfamilies is composed of Id genes 1 to 4. Id genes are thought to affect the balance between cell growth and differentiation in many cell types, including osteoblasts, keratinocytes, myoblasts, and mammary epithelial cells. Differentiation of various cell lineages is shown to be accompanied by the down-regulation of Id expression, while overexpression of Id genes in cultures of mammalian cells is shown to inhibit their ability to differentiate under appropriate conditions.16 Although Id genes regulate cell proliferation and differentiation in many cell types, the role of Id genes in osteoclast differentiation has not been determined.
We report here that TRANCE down-regulates the expression of Id genes. In addition, the overexpression of Id genes in bone marrow–derived monocyte/macrophage lineage cells (BMMs) inhibits TRANCE-mediated osteoclastogenesis, but does not affect the phagocytic activity or differentiation of common precursors into DCs. Furthermore, Id proteins can associate with Mitf and can inhibit Mitf binding to the promoter of OSCAR, a costimulatory molecule in osteoclast formation. Thus, this study shows that the Id family, down-modulated by TRANCE, is the first example of repressors that may play an important role in TRANCE-mediated osteoclastogenesis.
Materials and methods
Reagents and mice
Antibodies specific for c-fms and FcRγ were from Upstate Biotechnology (Lake Placid, NY); TREM2 was from R & D Systems (Minneapolis, MN); CD86, CD11b, I-Ab, CD11c, and NFATc1 were from BD Biosciences (San Jose, CA); Id2 was from Santa Cruz Biotechnology (Santa Cruz, CA); FLAG and actin were from Sigma-Aldrich (St Louis, MO); and F4/80 was from Serotec (Raleigh, NC). A polyclonal antibody for OSCAR was prepared as previously described.17 Anti-DAP12 antibody was kindly provided by M. Colonna (Washington University, St Louis, MO).18 The breeding and genotyping of Id2-deficient mice was performed as previously described.19
Constructs
Id1, Id2, and Id3 genes were prepared by reverse transcriptase–polymerase chain reaction (RT-PCR) using RNA from BMMs. The primers sequences are as follows: Id1, 5′-CGG GAT CCA CCA TGA AGG TCG CCA GTG GCA GTG-3′ (5′-Id1-BamHI) and 5′-GGA ATT CTC AGC GAC ACA AGA TGC GAT C-3′ (3′-Id1-EcoRI); Id2, 5′-CGG GAT CCA CCA TGA AAG CCT TCA GTC CGG TG-3′ (5′-Id2-BamHI) and 5′-ATA AGA ATG CGG CCG CTT AGC CAC AGA GTA CTT TGC T-3′ (3′-Id2-NotI); Id3, 5′-CGG GAT CCA CCA TGA AGG CGC TGA GCC CGG T-3′ (5′-Id3-BamHI) and 5′-GGA ATT CTC AGT GGC AAA AGC TCC TCT TG-3′ (3′-Id3-EcoRI). These amplified PCR fragments were digested with BamHI and EcoRI or BamHI and NotI and cloned into pMX-IRES-EGFP vector (courtesy of T. Kitamura, University of Tokyo, Tokyo, Japan). All constructs were confirmed by DNA sequencing (Korea Basic Science Institute, Gwangju, Korea). To generate expression constructs, inserts were digested with BamHI and NotI and subcloned into pGEX-6p-1 (Amersham Biosciences, Freiburg, Germany), pEBG, or HA-pCDNA3.1 vectors. Flag-tagged Mitf and pGL2 OSCAR 1.7, which contains 1.7-Kb promoter region of OSCAR, were generated by PCR.12
Osteoclast formation
Murine osteoclasts were prepared from bone marrow cells as previously described.12,20 In brief, bone marrow cells were cultured in α–minimal essential medium (MEM) containing 10% fetal bovine serum (FBS) with M-CSF (5 ng/mL) for 16 hours. Nonadherent cells were harvested and cultured for 3 days with M-CSF (30 ng/mL). Floating cells were removed and attached cells were used as osteoclast precursors (BMMs). To generate osteoclasts, BMMs were cultured with M-CSF (30 ng/mL) and TRANCE (100 ng/mL) for 3 days. To generate osteoclasts from coculture with osteoblasts and bone marrow cells, primary osteoblasts were prepared from calvariae of newborn mice as previously described.21 Bone marrow cells and primary osteoblasts were cocultured for 7 days in the presence of 1,25(OH)2D3 (1 × 10–8 M) and PGE2 (1 × 10–6 M). Cultured cells were fixed and stained for TRAP as previously described.12 TRAP-positive multinuclear cells (TRAP+ MNCs), which contain more than 3 nuclei, were counted.
Retroviral infection
To generate retrovirus stock, retroviral vectors were transfected into packaging cell line Plat E (a gift from T. Kitamura) using FuGENE 6 (Roche Applied Sciences, Indianapolis, IN). Virus supernatant was collected from cultured media 24 to 48 hours after transfection. BMMs were incubated with virus supernatant for 8 hours in the presence of polybrene (10 μg/mL). After removing the virus supernatant, BMMs were used for generating macrophages (30 ng/mL M-CSF), osteoclasts (30 ng/mL M-CSF and 100 ng/mL TRANCE), and DCs (10 ng/mL GM-CSF).
TRAP solution assay and MTT assay
BMMs were infected with virus supernatant for 8 hours. Cells were cultured for 3 days in the presence of M-CSF (30 ng/mL) and various concentrations of TRANCE as indicated in Figure 2. To determine the TRAP activity of osteoclasts, cultured cells were washed with phosphate-buffered saline (PBS) and lysed in TRAP buffer (120 mM sodium acetate [pH5.2], 0.012% sodium tartrate, and 1% Triton X-100). Cell lysates were incubated with P-nitrophenyl phosphate (pNPP) solution (Sigma-Aldrich) at 37°C for 20 minutes. The reactions were stopped with 1 M NaOH and optical density (OD) values were determined at 405 nm. To determine cell proliferation, an MTT assay was performed. The cultured cells were incubated with 25 μL/well of MTT solution (5 mg/mL) for 2 hours at 37°C. After incubation for 24 hours, formazan crystals were dissolved in 100 μL extraction buffer (20% SDS/50% DMF) and the absorbance was measured at 570 nm using a microplate reader.
Phagocytosis assay
BMMs were infected with retroviral supernatant for 8 hours and cells were cultured for 2 days with M-CSF (30 ng/mL) or M-CSF (30 ng/mL) and TRANCE (100 ng/mL). Fluorescein-conjugated zymosan A (Saccharomyces cerevisiae) Bio Particle (Molecular Probes, Eugene, OR) was added to cultured cells in 96-well culture plates (20 μg/0.2 mL/well). After 1 hour of incubation, cells were washed with PBS to remove the particles that were not incorporated by the cells. Cells were fixed and observed with ultraviolet (UV) illumination under the microscope (Leica, Wetzlar, Germany).
Cell culture and FACS analysis
DCs were prepared from BMMs with modification as previously described.22 The harvested cells were resuspended in RPMI 1640 medium supplemented with 5% FBS, and BMMs were seeded in 24-well plates with granulocyte-macrophage (GM)–CSF (10 ng/mL). Cells were cultured for 4 more days with replacement of fresh medium containing the same concentration of GM-CSF on day 2. On day 4, 1 mL fresh medium containing 1 μg/mL of LPS (Sigma-Aldrich) was added to cultures to stimulate the maturation of DCs. The next day, the cells were harvested and analyzed with anti-CD86, anti-CD11b, anti–I-Ab, and anti-CD11c antibodies. For macrophages, BMMs were transduced with pMX-puro (control) or pMX-puro-Id2. Transduced BMMs were selected with puromycin for 2 days. Selected cells were cultured for 2 days in the presence of M-CSF (30 ng/mL) or M-CSF (30 ng/mL)/TRANCE (100 ng/mL). Cultured cells were stained with anti-F4/80 and anti–c-fms.
Semiquantitative RT-PCR
RT-PCR analysis was performed as described.23 Primers used were the following: 5′-NFATc1, 5′-CTC GAA AGA CAG CAC TGG AGC AT-3′; 3′-NFATc1, 5′-CGG CTG CCT TCC GTC TCA TAG-3′; 5′-OSCAR, 5′-CTG CTG GTA ACG GAT CAG CTC CCC AGA-3′; 3′-OSCAR, 5′-CCA AGG AGC CAG AAC CTT CGA AAC T-3′; 5′-TRAP, 5′-CTG GAG TGC ACG ATG CCA GCG ACA-3′; 3′-TRAP, 5′-TCC GTG CTC GGC GAT GGA CCA GA -3′;5′-FcRγ,5′-ATC TCA GCC GTG ATC TTG TTC TTG-3′; 3′-FcRγ, 5′-TCT CAT GCT TCA GAG TCT CAT ATG-3′; 5′-DAP12, 5′-TTC CTT CCT GTC CTC CTG ACT GTG -3′; 3′-DAP12, 5′-TGC CTC TGT GTG TTG AGG TCA CTG-3′; 5′-TREM2, 5′-GGC CTC TAC CAG TGT CAG AGT CTC-3′; 3′-TREM2, 5′-GGT GGG TGG GAA GGA GGT CTC TTG-3′; 5′-c-fms, 5′-AGT GTG GGT AAC AGC TCT CAG TAC-3′; 3′-c-fms, 5′-TCC TAG AGT CTT ACC AAA CTG CAG-3′; 5′-CD14, 5′-TTC AGA ATC TAC CGA CCA TGG AGC-3′; 3′-CD14, 5′-TAT CAG TGA ACT GCC CCA GAT CTG -3′; 5′-F4/80, 5′-TAT CTT TTC CTC GCC TGC TTC TTC-3′; 3′-F4/80, 5′-GTT TAT CGT GAT GAT CAT GCA GAC-3′; 5′-HPRT, 5′-GTA ATG ATC AGT CAA CGG GGG AC-3′; and 3′-HPRT, 5′-CCA GCAAGC TTG CAA CCT TAA CCA-3′.
Northern hybridization and Western blot analysis
Northern blot analysis was performed as previously described.17 For immunoblotting analysis, 293T cells were cotransfected with pEBG, pEBG-Id1, pEBG-Id2, or pEBG-Id3 and Flag-Mitf. For osteoclasts, BMMs were transduced with pMX or pMX-Id2 for the indicated times in the presence of M-CSF and TRANCE. Cells were then washed with ice-cold PBS and lysed in extraction buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and protease inhibitors). Cell lysates were subjected to SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting. Signals were detected and analyzed by LAS3000 luminescent image analyzer (Fuji, Tokyo, Japan).
In vitro pull-down assay
35S-labeled Id1, Id2, or Id3 protein was synthesized using the TNT-coupled rabbit reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions. 293T cells was transfected with Flag-tagged Mitf DNA and harvested. To isolate Flag-tagged Mitf protein, the lysate from the transfectants was incubated with anti-Flag bead and washed extensively with lysis buffer. 35S-labeled Id1, Id2, or Id3 protein (10 μL) was incubated with Flag-Mitf protein bound to M2 beads, and binding reaction was carried out overnight at 4°C. Beads were washed for 3 times with lysis buffer and boiled for 5 minutes in sample loading buffer. Samples were subjected to SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Millipore, Bedford, MA). X-ray film was then exposed by the membrane overnight.
Transfection and luciferase assay
For transfection of reporter plasmids, 293T cells were plated on 6-well plates at a density of 2 × 105 cells/well 1 day before transfection. A total of 2.47 μg of plasmid DNA, including 0.1 μg of luciferase reporter, 0.25 μgof Mitf or PU.1 expression vector, 0.2 to 2 μg of Id1, Id2, or Id3 expression vector, and 20 ng of pcDNA3–β-galactosidase was transfected. The amount of transfected DNA was held constant to 2.47 μg by addition of empty-vector DNA where necessary. After 48 hours of transfection, the cells were washed twice with PBS and then lysed in reporter lysis buffer (Promega). Luciferase activity was measured with a luciferase assay system (Promega) according to the manufacturer's instructions. Luciferase activity was measured in triplicate, averaged, and then normalized to β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) as a substrate.
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as previously described.17 E-box wild- and mutant-type sequences used were as follows: E-box 2 wild-type, 5′-CAG GAC TCT CAC ATG GCT TTC TG-3′ (sense) and 5′-CA GAAAGC CAT GTG AGA GTC CTG-3′ (antisense); and E-box 2 mutant type, 5′-CAG GAC TCT GAG ATG GCT TTC TG-3′ (sense) and 5′-CA GAA AGC CAT CTC AGA GTC CTG-3′ (antisense).
Results
TRANCE down-regulates the expression of Id genes
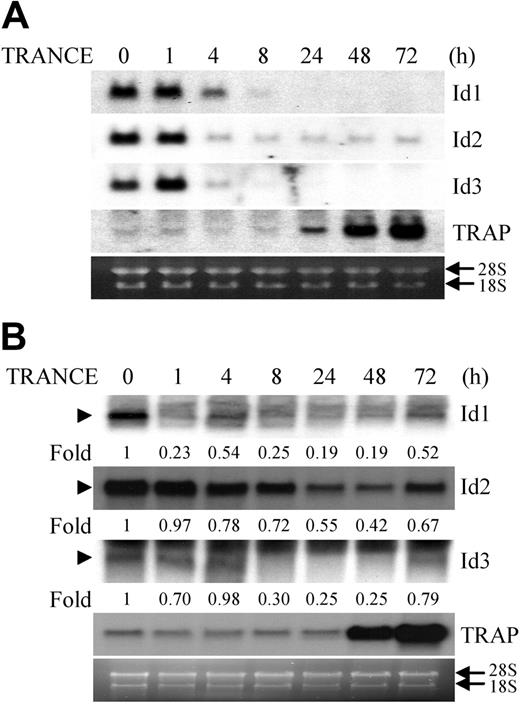
TRANCE induces activation and induction of transcription factors including NF-κB, c-fos, Mitf, and NFATc1, and thereby supports osteoclast differentiation from precursors.4-6 To identify important genes for osteoclast differentiation, we performed suppression-subtractive hybridization between osteoclasts and macrophages derived from common precursors (BMMs), and found that Id2 was down-regulated in osteoclasts compared with macrophages. To confirm the differential expression of the Id2 gene in both cells, we examined the expression pattern of Id genes during TRANCE-mediated osteoclastogenesis. Among the 4 known members of the Id family, Id1, Id2, and Id3 are expressed in the RAW264.7 cell line, which is derived from the monocyte/macrophage lineage and is capable of differentiating into osteoclasts and resorbing bone. The expression of these genes was significantly reduced 1 hour after TRANCE treatment and was maintained at that low level throughout the osteoclastogenic process, whereas the expression of TRAP, a marker for mature osteoclasts, increased as the cells differentiated (Figure 1A). However, Id4 was not detected in any stage of osteoclastogenesis mediated by TRANCE (data not shown). In addition, when BMMs were stimulated with TRANCE to induce osteoclast differentiation, the expression of Id genes was reduced by TRANCE treatment and slightly increased at the end of osteoclastogenesis (Figure 1B).
Mitf, a bHLH transcription factor, is known to regulate osteoclastogenesis mediated by TRANCE.7-9 It has been shown that TRANCE signaling in osteoclasts results in Mitf phosphorylation mediated by a MKK6/p38 signaling cascade and also induces expression of the Mitf gene.11,12 HLH transcription factors such as Id genes form heterodimers with members of the bHLH family of transcription factors and regulate cell growth and differentiation.24,25 They act as dominant-negative inhibitors of their binding partners. Previously, we showed that TRANCE treatment increased Mitf expression in RAW264.7 cells and in BMMs12 These results suggest that TRANCE signaling regulates the expression of bHLH and HLH transcription factors during osteoclast differentiation, and that the balance of expression of these genes may be important for the process of osteoclastogenesis.
Overexpression of Ids inhibits osteoclast formation from BMMs, but does not affect phagocytosis or DC differentiation
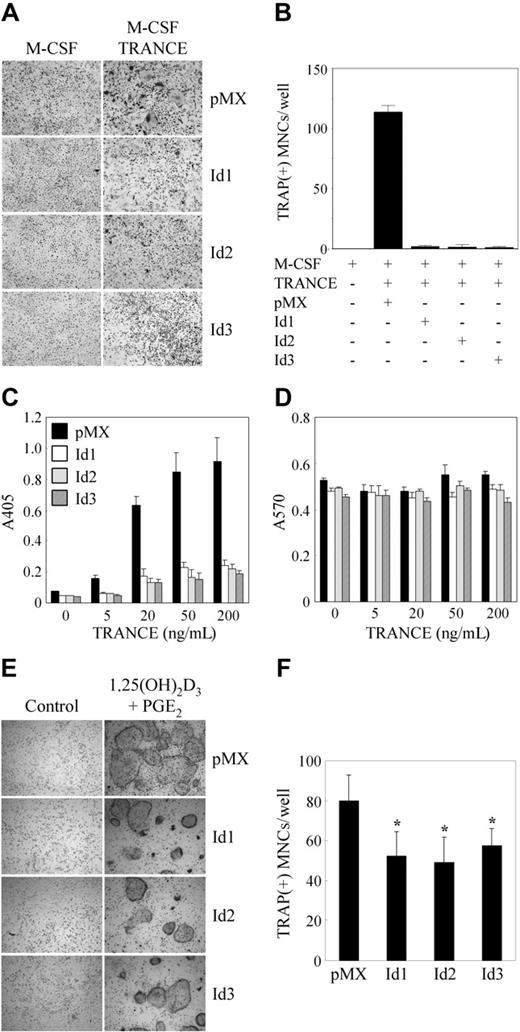
To investigate the role of Id genes in osteoclastogenesis mediated by TRANCE, we overexpressed Id genes in BMMs using a retroviral vector. Transduced BMMs were cultured with M-CSF alone or M-CSF and TRANCE, and were stained for TRAP (Figure 2A). Although a small number of TRAP+ mononuclear cells were detected, overexpression of Id1, Id2, or Id3 gene largely inhibited the formation of TRAP+ MNCs mediated by M-CSF and TRANCE, whereas control vector-infected BMMs could mature into osteoclasts (Figure 2A-B). Moreover, whereas TRANCE treatment of control vector–infected BMMs increased TRAP activity in a dose-dependent manner, overexpression of Id genes in BMMs attenuated this TRANCE-mediated TRAP activity (Figure 2C). We confirmed that overexpression of Id genes did not affect the proliferation or survival of BMMs by means of an MTT assay (Figure 2D). When transduced BMMs were cocultured with primary osteoblasts in the presence of 1,25(OH)2D3 and PGE2, the number of TRAP+ MNCs was significantly reduced by overexpression of Id genes compared with that of control vector, although the efficiency of inhibition by overexpression of Id genes in coculture was not strong as that in BMM culture (Figure 2E-F). These data suggest that Id genes may have a role for osteoclast differentiation mediated by TRANCE.
TRANCE down-regulates the expression of Id genes during osteoclastogenesis. RAW264.7 cells (A) and BMMs (B) were cultured for the indicated time in the presence of TRANCE (A) and M-CSF, and TRANCE (B), respectively. Total RNA was collected from each time point and analyzed by Northern blot using probes for Id1, Id2, Id3, and TRAP. The relative amounts of Ids were shown under the figures (B). Arrows indicate ribosomal RNA; arrowheads, the bands representing Id1, Id2, or Id3.
TRANCE down-regulates the expression of Id genes during osteoclastogenesis. RAW264.7 cells (A) and BMMs (B) were cultured for the indicated time in the presence of TRANCE (A) and M-CSF, and TRANCE (B), respectively. Total RNA was collected from each time point and analyzed by Northern blot using probes for Id1, Id2, Id3, and TRAP. The relative amounts of Ids were shown under the figures (B). Arrows indicate ribosomal RNA; arrowheads, the bands representing Id1, Id2, or Id3.
Overexpression of Id genes in BMMs inhibits osteoclastogenesis. (A-B) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cultured for 3 days with M-CSF alone or M-CSF and TRANCE. (A) Cultured cells were fixed and stained for TRAP (original magnification, × 100). (B) Numbers of TRAP-positive multinucleated cells were counted. (C-D) BMMs were transduced with control (pMX-IRES-EGFP), Id1, Id2, or Id3 retrovirus and cultured for 3 days with M-CSF and various concentrations of TRANCE as indicated. (C) TRAP activity was determined by measuring OD values at 405 nm. (D) MTT assay results were determined by measuring OD values at A570. (E-F) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cocultured for 6 days with osteoblasts in the presence of 1,25(OH)2D3 and PGE2. (E) Cultured cells were fixed and stained for TRAP (original magnification, × 100). (F) Numbers of TRAP-positive multinucleated cells were counted (*P < .05). Results are representative of at least 3 independent sets of similar experiments (A-F). Data represent means ± SDs of triplicate experiments. Cells were observed under a Leica DMIRB microscope equipped with an N Plan 10×/0.25 objective lens (Leica, Wetzlar, Germany). Images were obtained through a Leica IM50 using Leica IM 4.0 software (Leica, Cambridge, United Kingdom).
Overexpression of Id genes in BMMs inhibits osteoclastogenesis. (A-B) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cultured for 3 days with M-CSF alone or M-CSF and TRANCE. (A) Cultured cells were fixed and stained for TRAP (original magnification, × 100). (B) Numbers of TRAP-positive multinucleated cells were counted. (C-D) BMMs were transduced with control (pMX-IRES-EGFP), Id1, Id2, or Id3 retrovirus and cultured for 3 days with M-CSF and various concentrations of TRANCE as indicated. (C) TRAP activity was determined by measuring OD values at 405 nm. (D) MTT assay results were determined by measuring OD values at A570. (E-F) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cocultured for 6 days with osteoblasts in the presence of 1,25(OH)2D3 and PGE2. (E) Cultured cells were fixed and stained for TRAP (original magnification, × 100). (F) Numbers of TRAP-positive multinucleated cells were counted (*P < .05). Results are representative of at least 3 independent sets of similar experiments (A-F). Data represent means ± SDs of triplicate experiments. Cells were observed under a Leica DMIRB microscope equipped with an N Plan 10×/0.25 objective lens (Leica, Wetzlar, Germany). Images were obtained through a Leica IM50 using Leica IM 4.0 software (Leica, Cambridge, United Kingdom).
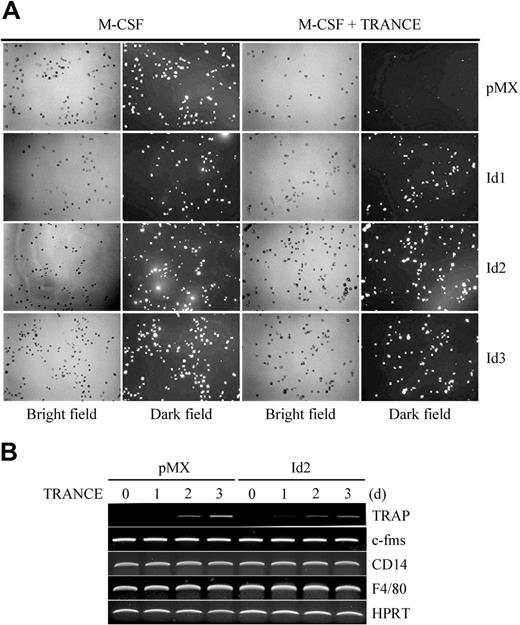
It is believed that macrophages, osteoclasts, and DCs originate from the same precursors. Recently, we showed that BMMs have phagocytic activity, characteristic of macrophages, but that their phagocytic activity was abolished by stimulation with M-CSF and TRANCE in vitro.20 Since the overexpression of Id genes inhibited osteoclastogenesis, we reasoned that Id gene–infected BMMs might retain phagocytic activity. When we cultured retrovirally infected BMMs for 2 days with M-CSF and TRANCE, BMMs overexpressing Id genes did retain their phagocytic activity, whereas control vector–infected BMMs lost their phagocytic activity as a result of stimulation with M-CSF and TRANCE. When BMMs were cultured with M-CSF alone, neither overexpression of Id genes nor pMX control vector inhibited phagocytic activity (Figure 3A). In addition, although the gene expression of TRAP, a marker for osteoclasts, was attenuated by overexpression of Id2, no significant changes in expression of c-fms, CD14, and F4/80, markers for macrophages, were detected (Figure 3B).
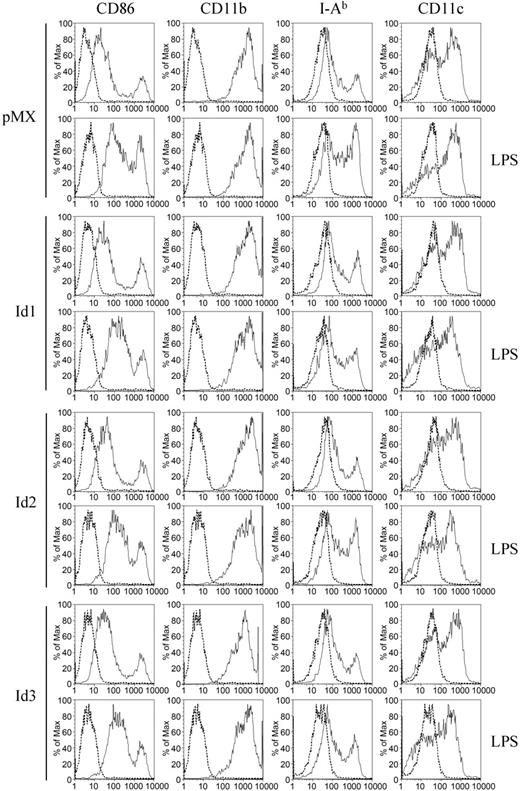
Previously, we demonstrated that BMMs could also become DCs through stimulation with GM-CSF in vitro.20 Therefore, we tested whether overexpression of Id genes affected the differentiation of BMMs into DCs. After retrovirally infected BMMs were cultured with GM-CSF for 4 days, immature DCs were treated with LPS overnight. When cells were harvested and stained for DC markers, including CD11b, CD11c, CD86, and I-Ab. DCs from control vector–infected BMMs expressed those marker genes. However, overexpression of the Id genes affected neither DC differentiation nor LPS-mediated DC maturation (Figure 4). These data suggest that down-regulation of Id genes may play a role in determining destiny of common precursors toward osteoclasts rather than macrophages or DCs.
Overexpression of Id genes in BMMs does not affect phagocytosis. (A) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cultured for 2 days with M-CSF alone or M-CSF and TRANCE. Cultured cells were incubated with fluorescein-conjugated zymosan particles for 1 hour and washed with PBS. Cells were fixed and observed with UV illumination under a microscope. Fluorescein-conjugated zymosan particles incorporated by the cells appear as bright dots in the dark field (original magnification, ×100). (B) BMMs were transduced with control (pMX-IRES-EGFP) or Id2 retrovirus and cultured for the indicated days with M-CSF and TRANCE. RT-PCR was performed for the expression of TRAP, c-fms, CD14, F4/80, and HPRT. Images were acquired as described for Figure 2.
Overexpression of Id genes in BMMs does not affect phagocytosis. (A) BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus and cultured for 2 days with M-CSF alone or M-CSF and TRANCE. Cultured cells were incubated with fluorescein-conjugated zymosan particles for 1 hour and washed with PBS. Cells were fixed and observed with UV illumination under a microscope. Fluorescein-conjugated zymosan particles incorporated by the cells appear as bright dots in the dark field (original magnification, ×100). (B) BMMs were transduced with control (pMX-IRES-EGFP) or Id2 retrovirus and cultured for the indicated days with M-CSF and TRANCE. RT-PCR was performed for the expression of TRAP, c-fms, CD14, F4/80, and HPRT. Images were acquired as described for Figure 2.
Id proteins regulate the gene expression of OSCAR, through interaction with Mitf, as well as that of NFATc1
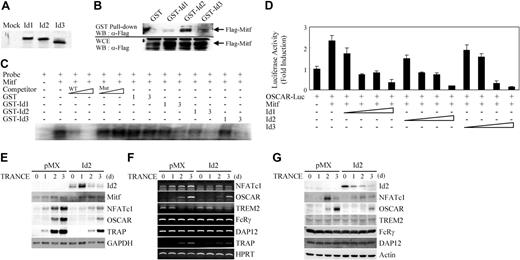
Mitf, a bHLH transcription factor, plays an important role in osteoclast differentiation. Since HLH transcription factors can bind to bHLH proteins and inhibit transactivation of target genes, we tested whether Id proteins could bind Mitf. The direct association between Mitf and Id proteins was demonstrated using an in vitro pull-down assay. A Flag-tagged Mitf construct was transfected into 293T cells and Flag-Mitf protein was immobilized on anti-Flag M2 agarose beads. Beads were incubated with 35S-labeled Id1, Id2, or Id3 protein and subjected to SDS-PAGE and autoradiography. Id proteins bound to Flag-Mitf but not to the control vector (Figure 5A). To demonstrate the interaction between Mitf and Id proteins in mammalian cells, we performed an immunoprecipitation assay. 293T cells were cotransfected with GST, GST-Id1, GST-Id2, or GST-Id3 together with Flag-Mitf, and cell lysates were immunoprecipitated with glutathione beads. Beads were subjected to SDS-PAGE and Western blotting by anti-Flag antibody. Flag-Mitf bound to GST-Id proteins but not to GST alone (Figure 5B). These data demonstrate that Id proteins interact with Mitf in mammalian cells.
Overexpression of Id genes in BMMs does not affect the expression level of characteristic dendritic-cell markers. BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus. Cells were cultured for 4 more days with GM-CSF to generate DCs. LPS (1 μg/mL) was added to cultures to induce DC maturation. The cells were harvested the next day and stained for FACS analysis with anti-CD86, anti-CD11b, anti–I-Ab, anti-CD11c antibodies (solid line), or control IgG (dotted line).
Overexpression of Id genes in BMMs does not affect the expression level of characteristic dendritic-cell markers. BMMs were transduced with pMX-IRES-EGFP (control), Id1, Id2, or Id3 retrovirus. Cells were cultured for 4 more days with GM-CSF to generate DCs. LPS (1 μg/mL) was added to cultures to induce DC maturation. The cells were harvested the next day and stained for FACS analysis with anti-CD86, anti-CD11b, anti–I-Ab, anti-CD11c antibodies (solid line), or control IgG (dotted line).
To determine whether Id proteins can modulate the binding of Mitf to E-box of OSCAR promoter, EMSA was performed in the presence of GST-Id fusion proteins. When the purified GST-Id fusion proteins were added to the reaction mixture, we observed a significant, dose-dependent decrease of Mitf binding to labeled probe (E-box 2 of OSCAR promoter) caused by increasing doses of GST-Id proteins, but not by GST alone (Figure 5C). This result suggests that Id proteins decrease Mitf binding to E-box of OSCAR promoter by association with Mitf.
We have previously shown that Mitf induces expression of OSCAR, which is a modulator of osteoclastogenesis.12 Since Id proteins could bind Mitf in vitro, we tested whether overexpression of Ids could inhibit transactivation of the OSCAR promoter by Mitf. When 293T cells were cotransfected with a luciferase reporter plasmid containing OSCAR 1.7-kb promoter together with Mitf vector, Mitf induced an approximately 2.5-fold increase in OSCAR promoter activity compared with control (Figure 5D). Induction of OSCAR promoter activity by Mitf was decreased in a dose-dependent manner by Id expression. Although Ids could also bind to PU.1, a positive regulator of OSCAR gene expression, it is interesting that overexpression of Ids did not suppress the transactivation of the OSCAR gene by PU.1 (data not shown). These results suggest that Id proteins interact with Mitf and inhibit Mitf transactivation of the OSCAR gene.
Since Ids directly bind to Mitf and inhibit its transactivation of the OSCAR gene by blocking the binding of Mitf to the promoter region of OSCAR, we next investigated whether overexpression of Id2 in BMMs could regulate OSCAR gene expression. TRANCE stimulation in BMMs increased the expression of NFATc1 and OSCAR in mRNA and protein levels. (Figure 5E,G). Compared with the control, exogenous overexpression of Id2 gene attenuated the expression of NFATc1 as well as of OSCAR during TRANCE-mediated osteoclastogenesis. However, the expression of Mitf was not affected by Id2 overexpression (Figure 5E). These results indicate that Ids can regulate the gene expression of OSCAR as well as of NFATc1.
Ids down-regulate the gene expression of OSCAR through interaction with Mitf, as well as that of NFATc1. (A) 35S-labeled mock, Id1, Id2, or Id3 protein was incubated with Flag-Mitf immobilized on M2-sepharose beads, respectively. After washing, retained 35S-labeled mock, Id1, Id2, or Id3 protein was examined by SDS-PAGE and autoradiography. (B) 293T cells were cotransfected with GST, GST-Id1, GST-Id2, or GST-Id3 plasmid together with Flag-Mitf plasmid. After 36 hours of transfection, cell lysates were immunoprecipitated with Flag-Mitf immobilized on M2-sepharose beads. The beads were washed, resolved by SDS-PAGE, and detected by Western blotting using anti-Flag antibody (top panel). Whole-cell extracts (WCE) were also subjected directly to Western blot (WB) analysis with same antibody (bottom panel). (C) Full-length murine Mitf cDNA was prepared from TNT rabbit reticulocyte lysate as described in “Methods and materials.” Oligonucleotides spanning E-box 2 in the murine OSCAR promoter was used as probe for EMSA. Specific binding was determined by cold competition using unlabeled wild- or mutant-type probe at 10- and 100-fold molar excess concentrations (lanes 3-6). Mitf lysate and probe were incubated with indicated amounts (1-3 μg) of GST, GST-Id1, GST-Id2, or GST-Id3 (lanes 7-14). Results are representative of at least 2 independent sets of similar experiments. (D) 293T cells were cotransfected with 0.1 μg OSCAR 1.7-kb promoter luciferase reporter and 0.25 μg Mitf plasmid together with indicated amounts (0.2-2 μg) of plasmid expressing Id1, Id2, or Id3. Each well was also cotransfected with 20 ng β-galactosidase expression vector to control for transfection efficiency. Luciferase activities were normalized to β-galactosidase activity as expressed by the cotransfected plasmid. The level of 1.7-kb OSCAR reporter construct in the presence of empty expression vector was set to 1. Data represent the mean and the SE of triplicate samples. Results are representative of at least 3 independent sets of similar experiments. (E-G) BMMs were transduced with control (pMX-IRES-EGFP) or Id2 retroviruses and cultured for the indicated days with M-CSF and TRANCE. Northern blot analysis (E), RT-PCR (F), and Western blot analysis (G) were performed to assess the expression of the indicated genes.
Ids down-regulate the gene expression of OSCAR through interaction with Mitf, as well as that of NFATc1. (A) 35S-labeled mock, Id1, Id2, or Id3 protein was incubated with Flag-Mitf immobilized on M2-sepharose beads, respectively. After washing, retained 35S-labeled mock, Id1, Id2, or Id3 protein was examined by SDS-PAGE and autoradiography. (B) 293T cells were cotransfected with GST, GST-Id1, GST-Id2, or GST-Id3 plasmid together with Flag-Mitf plasmid. After 36 hours of transfection, cell lysates were immunoprecipitated with Flag-Mitf immobilized on M2-sepharose beads. The beads were washed, resolved by SDS-PAGE, and detected by Western blotting using anti-Flag antibody (top panel). Whole-cell extracts (WCE) were also subjected directly to Western blot (WB) analysis with same antibody (bottom panel). (C) Full-length murine Mitf cDNA was prepared from TNT rabbit reticulocyte lysate as described in “Methods and materials.” Oligonucleotides spanning E-box 2 in the murine OSCAR promoter was used as probe for EMSA. Specific binding was determined by cold competition using unlabeled wild- or mutant-type probe at 10- and 100-fold molar excess concentrations (lanes 3-6). Mitf lysate and probe were incubated with indicated amounts (1-3 μg) of GST, GST-Id1, GST-Id2, or GST-Id3 (lanes 7-14). Results are representative of at least 2 independent sets of similar experiments. (D) 293T cells were cotransfected with 0.1 μg OSCAR 1.7-kb promoter luciferase reporter and 0.25 μg Mitf plasmid together with indicated amounts (0.2-2 μg) of plasmid expressing Id1, Id2, or Id3. Each well was also cotransfected with 20 ng β-galactosidase expression vector to control for transfection efficiency. Luciferase activities were normalized to β-galactosidase activity as expressed by the cotransfected plasmid. The level of 1.7-kb OSCAR reporter construct in the presence of empty expression vector was set to 1. Data represent the mean and the SE of triplicate samples. Results are representative of at least 3 independent sets of similar experiments. (E-G) BMMs were transduced with control (pMX-IRES-EGFP) or Id2 retroviruses and cultured for the indicated days with M-CSF and TRANCE. Northern blot analysis (E), RT-PCR (F), and Western blot analysis (G) were performed to assess the expression of the indicated genes.
It has been reported that Ig-like receptors, including triggering receptor expressed in myeloid cells (TREM) 2 and OSCAR, can bind to DAP12 or FcRγ and that the costimulatory signals mediated by immunoreceptor tyrosine-based activation motif (ITAM)–harboring adaptors, such as DAP12 and FcRγ, cooperate with TRANCE for osteoclastogenesis, and that their activation enhances the induction of NFATc1.26 Since overexpression of Id2 attenuated the gene expression of NFATc1 and OSCAR, we performed RT-PCR to investigate the effect of expression level of Ig-like receptors and ITAM-harboring adaptors by Id2 overexpression. Although the gene expressions of NFATc1, OSCAR, and TRAP were inhibited by overexpression of Id2, no significant changes in expression of TREM2, FcRγ, and DAP12 were detected (Figure 5F). Similar results were obtained from Western blot analysis (Figure 5G). These results suggested that Id proteins regulate the gene expression of OSCAR, through interaction with Mitf, as well as that of NFATc1.
Deficiency of Id2 enhances TRANCE-induced osteoclast formation
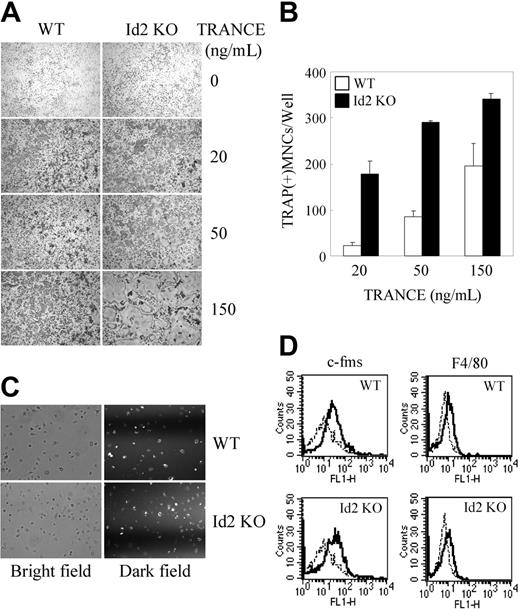
Since Ids act as negative regulators for osteoclastogenesis, we investigated their physiologic role in osteoclastogenesis. BMMs from WT or Id2 knockout (KO) mice were cultured with M-CSF and TRANCE. As expected, TRANCE induced osteoclast formation in a dose-dependent manner in WT mice (Figure 6A-B). Intriguingly, the number of TRAP+ MNCs was higher in Id2 KO mice compared with WT mice. However, the deficiency of Id2 did not significantly affect the phagocytic activity or the expression level of F4/80 and c-fms, markers for macrophages (Figure 6C-D). These results suggest that Ids may have a role in osteoclastogenesis as negative regulators.
Discussion
In this study, we present evidence that Ids are responsible for the negative regulation of TRANCE-induced osteoclast differentiation, and that TRANCE promotes osteoclastogenesis, in part, by downregulating these repressors. Various transcription factors, including Mitf, PU.1, and NFATc1, are important for osteoclast differentiation. It has been shown that PU.1 is required for the correct development of both myeloid and lymphoid lineages, including macrophages, osteoclasts, DCs, neutrophils, and mast cells.27-30 Osteoclast differentiation fails at an early stage in PU.1-deficient mice. Mutation of Mitf results in multiple phenotypes including osteopetrosis, lack of pigment, deafness, and small eyes.31 Although the expression of Mitf and PU.1 is not restricted to osteoclast precursor cells, only these monocytes/macrophage precursors can differentiate into mature osteoclasts via treatment with M-CSF and TRANCE. This implies that the other molecules might be involved in osteoclastogenesis.
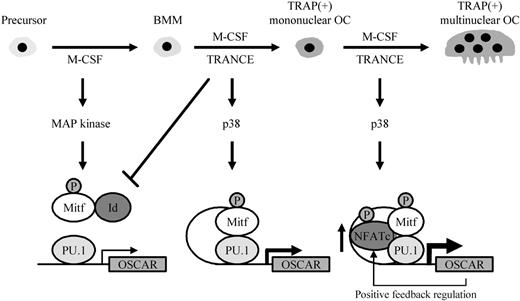
Here, we propose a possible schematic model of OSCAR gene regulation in osteoclastogenesis based on our results and on previous studies (Figure 7). In the early stage of osteoclastogenesis, M-CSF activates phosphorylation of Mitf and also induces expression of RANK in response to TRANCE.32,33 Id proteins, as well as Mitf and PU.1, are abundantly expressed in the committed precursors. Because Id proteins can directly bind to Mitf and inhibit Mitf binding to the E-box of the OSCAR promoter, OSCAR gene expression may be reduced to a level where it can no longer drive osteoclast formation. When TRANCE binds to its cognate receptor RANK, it induces activation of Mitf by phosphorylation of serine residues via the MKK6/p38 mitogen-activated protein (MAP) kinase signaling cascade11 and by down-regulating expression of Id genes within a few hours. Subsequently, down-regulation of Id genes allows Mitf to bind to its cognate E-box site, cooperating with PU.1 to induce the expression of target genes including OSCAR, TRAP, and cathepsin K.12,34,35 Later, TRANCE also activates NFATc1 through MKK6/p38, allowing NFATc1 to induce its target genes in a synergistic manner with other transcription factors, including Mitf and PU.1.17,35 At a late stage, a positive feedback circuit involving TRANCE, NFATc1, and OSCAR induces the efficient differentiation of osteoclasts.17,26,36
HLH transcription factors are required for a multitude of important developmental processes, including neurogenesis, hematopoiesis, myogenesis, and pancreatic development.14 Ids are known to negatively regulate the differentiation of various cell types. The expression of Id genes are down-regulated during differentiation, and the ectopic overexpression of Id genes inhibits the differentiation of various cell types including B cells, myeloid cells, hepatocytes, and muscle cells.37-40 It has been shown that Id proteins can associate specifically with MyoD, E12, and E47, thereby attenuating their ability to bind DNA, and that overexpression of Id inhibits the transactivation of the muscle creatine kinase enhancer by MyoD, a bHLH transcription factor.15 In this paper, we observed that Id proteins directly interacted with Mitf protein and that the binding of Mitf to the E-box of the OSCAR promoter was inhibited by GST-Id proteins. In addition, Id genes decreased Mitf transactivation of the OSCAR gene in a dose-dependent manner. Furthermore, overexpression of Id2 gene in BMMs attenuated the induction of OSCAR gene, as well as osteoclastogenesis mediated by TRANCE. These results suggest that Id proteins can associate directly with Mitf and inhibit its role in osteoclastogenesis. The expression of NFATc1 was also attenuated by Id2 overexpression. Although AP-1 induces the expression of NFATc1,41 Ids did not affect transactivation of c-fos on NFATc1 gene in transient transfection experiments (data not shown). Although the underlying mechanism of the inhibitory effect of Id2 on NFATc1 expression is not clear, a possible mechanism is that down-regulation of OSCAR by Id2 may cause the impairment of a positive feedback regulation between OSCAR and NFATc1.17,36 However, the possibility remains that Id proteins act as negative regulators of osteoclast differentiation via interaction with other molecules.
Deficiency of Id2 enhances TRANCE-mediated osteoclast formation. (A-B) BMMs derived from WT or Id2 KO mice were incubated for 3 days with M-CSF and an increased concentration of TRANCE. (A) Cultured cells were fixed and stained for TRAP (original magnification, ×100). (B) Numbers of TRAP+ MNCs were counted. (C) BMMs from WT or Id2 KO mice were analyzed for phagocytosis as described in Figure 3 (original magnification, ×100). (D) BMMs from WT or Id2 KO mice were stained for FACS analysis with anti-F4/80 and anti–c-fms (solid line) or control IgG (dotted line). Data represent means ± SDs from triplicate experiments. Images were acquired as described for Figure 2.
Deficiency of Id2 enhances TRANCE-mediated osteoclast formation. (A-B) BMMs derived from WT or Id2 KO mice were incubated for 3 days with M-CSF and an increased concentration of TRANCE. (A) Cultured cells were fixed and stained for TRAP (original magnification, ×100). (B) Numbers of TRAP+ MNCs were counted. (C) BMMs from WT or Id2 KO mice were analyzed for phagocytosis as described in Figure 3 (original magnification, ×100). (D) BMMs from WT or Id2 KO mice were stained for FACS analysis with anti-F4/80 and anti–c-fms (solid line) or control IgG (dotted line). Data represent means ± SDs from triplicate experiments. Images were acquired as described for Figure 2.
A schematic model of OSCAR gene regulation via transcription factors during osteoclastogenesis. M-CSF and TRANCE induce the formation of TRAP+ multinuclear osteoclasts from BMM. Upon stimulation, activated MAP kinases regulate transcription factors such as Mitf and NFATc1, subsequently leading to the up-regulation of the OSCAR gene during osteoclastogenesis. OSCAR and NFATc1 show positive feedback regulation.
A schematic model of OSCAR gene regulation via transcription factors during osteoclastogenesis. M-CSF and TRANCE induce the formation of TRAP+ multinuclear osteoclasts from BMM. Upon stimulation, activated MAP kinases regulate transcription factors such as Mitf and NFATc1, subsequently leading to the up-regulation of the OSCAR gene during osteoclastogenesis. OSCAR and NFATc1 show positive feedback regulation.
Macrophages, osteoclasts, and DCs are thought to originate from common precursors. Since Id genes were expressed in macrophage/monocyte lineage cells, we examined whether overexpression of Id genes affected the differentiation of BMMs into each of these cell types. Interestingly, ectopic overexpression of Id genes in BMMs inhibited osteoclastogenesis, but not phagocytic activity or differentiation into DCs. Although Ids are known to regulate PU.1-mediated signals in macrophages,42 overexpression of Id2 in BMMs by retroviral transfection did not significantly affect the expression of markers for macrophages such as c-fms, CD14, and F4/80. A possible explanation is that overexpression using a retroviral vector may not strong enough to induce detectable changes in expression of target genes. Recently, it has been shown that a HLH transcription factor Id2 is up-regulated during DC development in vitro and crucial for the development of distinct DC subsets in vivo.43 These observations imply that the different expression level of Id genes in common precursors can affect cell fate upon stimulation with differentiation factors such as TRANCE, M-CSF, and GM-CSF.
Taken together, our results demonstrate that TRANCE-mediated down-regulation of Ids relieves their inhibitory effect on Mitf and subsequently on OSCAR gene expression, thereby underscoring an important role for Ids in osteoclast differentiation, and that Ids may have a physiologic role in TRANCE-mediated osteoclastogenesis. To our knowledge, Ids are the first repressors in osteoclasts that are down-modulated by TRANCE. Hence, our work reveals both an additional layer of negative regulation, as well as the mechanism whereby TRANCE signaling overcomes it, allowing osteoclastogenesis to proceed. Further study of the detailed mechanism of transcription factor gene regulation will allow for a clearer understanding of the true nature of Id genes, their functions in osteoclast formation, and their utility as molecular targets for therapeutics addressing bone diseases including osteoporosis.
Prepublished online as Blood First Edition Paper, December 1, 2005; DOI 10.1182/blood-2005-07-2798.
Supported in part by National Institutes of Health grants AR49078 and AR48521 (to Y.C.) and the Korea Research Foundation grant R08-2004-000-10303-0 (to N.K.). J.L. was financially supported in part by the Korea Research Foundation grant (KRF-2004-C00141) and by Chonnam National University.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank T. Kitamura for PLAT-E cells, M. Colonna for anti-DAP12 antibody, and C. Perchonock and C. King for critical reading and for providing helpful comments on this manuscript. We also thank Y. Kong (Pohang University of Science and Technology, Korea) and D. Jeong (Seoul National University, Korea) for technical support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal