Abstract
Altered mRNA translation is one of the effects exerted by the BCR/ABL oncoprotein in the blast crisis phase of chronic myelogenous leukemia (CML). Here, we report that in BCR/ABL+ cell lines and in patient-derived CML blast crisis mononuclear and CD34+ cells, p210BCR/ABL increases expression and activity of the transcriptional-inducer and translational-regulator heterogeneous nuclear ribonucleoprotein K (hnRNP K or HNRPK) in a dose- and kinase-dependent manner through the activation of the MAPKERK1/2 pathway. Furthermore, HNRPK down-regulation and interference with HNRPK translation-but not transcription-regulatory activity impairs cytokine-independent proliferation, clonogenic potential, and in vivo leukemogenic activity of BCR/ABL-expressing myeloid 32Dcl3 and/or primary CD34+ CML-BC patient cells. Mechanistically, we demonstrate that decreased internal ribosome entry site (IRES)-dependent Myc mRNA translation accounts for the phenotypic changes induced by inhibition of the BCR/ABL-ERK-dependent HNRPK translation-regulatory function. Accordingly, MYC protein but not mRNA levels are increased in the CD34+ fraction of patients with CML in accelerated and blastic phase but not in chronic phase CML patients and in the CD34+ fraction of marrow cells from healthy donors. Thus, BCR/ABL-dependent enhancement of HNRPK translation-regulation is important for BCR/ABL leukemogenesis and, perhaps, it might contribute to blast crisis transformation. (Blood. 2006;107:2507-2516)
Introduction
The p210BCR/ABL oncoprotein induces chronic myelogenous leukemia (CML) and contributes to transition from the indolent chronic phase (CML-CP) to the aggressive blast crisis (CML-BC)1,2 through its deregulated kinase activity, which enhances proliferation and survival, and arrests differentiation of hematopoietic progenitors by aberrantly modulating signaling pathways controlling transcription, protein function, and mRNA translation.2-4 The latter results from the effects of BCR/ABL on the translation machinery5,6 and the expression of RNA binding proteins with translation-regulatory function.7 Specifically, high levels of BCR/ABL activity, as those observed in CML-BC,8-10 suppress differentiation and increase resistance to apoptosis by enhancing the translation-regulatory activity of hnRNP E2 and La/SSB, respectively.11,12
By assessing the effect of the Abl kinase inhibitor imatinib mesylate on the transcriptome of BCR/ABL-expressing myeloid progenitors, we observed a marked down-regulation of Hnrpk mRNA levels. Like hnRNP E2, HNRPK is a nucleocytoplasmic shuttling poly(rC)-binding protein that, upon binding to DNA and RNA in a sequence-specific manner, regulates gene transcription and mRNA metabolism.13,14 In fact, besides enhancing the transcription of genes (eg, MYC) containing a cis regulatory CT-rich promoter element,15,16 HNRPK also functions as a substrate and docking platform for kinases,17-21 some of which control its translation-regulatory function. For example, ERK-dependent HNRPK phosphorylation negatively regulates translation of mRNAs (eg, 15-LOX) containing the CT-rich differentiation control element (DICE) in the 3′UTR.21,22 By contrast, HNRPK positively regulates translation of mRNAs (ie, MYC) containing an IRES with an HNRPK-binding site.23 In this report, we show that in BCR/ABL+ cell lines and in patient-derived CML-BC mononuclear and CD34+ cells, BCR/ABL increases HNRPK expression/function in a dose- and kinase-dependent manner through MAPKERK1/2 activation. Accordingly, HNRPK down-regulation and interference with its translation activity impairs proliferation, clonogenic potential, and leukemogenesis of BCR/ABL-transformed cells and/or CD34+ CML-BC patient cells through inhibition of the IRES-dependent translation of MYC mRNA.
Materials and methods
Cell cultures and primary cells
The IL-3-dependent 32Dcl3 myeloid precursor, and 32D-BCR/ABL, K562, and TonB210.1 (G. Daley; Harvard University, Cambridge, MA) cells were maintained in culture in IMDM, 10% FBS, 2 mM l-glutamine (Gibco, Carlsbad, CA). Where indicated, WEHI-conditioned medium (10%) was used as source of mIL-3. 32D-BCR/ABL-derivative cell lines were generated by retroviral infection followed by antibiotic-mediated selection or fluorescence-activated cell sorter (FACS)-mediated sorting of the GFP+ (green fluorescent protein) cells.11,24 For cell starvation, cells were washed 4 times in PBS and incubated (12 hours) in IMDM/10% FBS. Samples of mononuclear hematopoietic cells from bone marrow of unidentifiable CML patients were kept overnight in IMDM, 50% FBS, 2 mM glutamine, and rhIL-3 (20 ng/mL), rhIL-6 (20 ng/mL), rhFlt-3 ligand (100 ng/mL), and rhKL (100 ng/mL) (Stem Cell Technologies, Vancouver, BC, Canada) and used for Western blot analysis or for CD34+ enrichment (CD34 MultiSort kit; Miltenyi Biotec, Auburn, CA). CD34+ normal bone marrow (NBM) cells from healthy donors were from AllCells (Berkeley, CA). Studies performed with human specimens from The Ohio State University Leukemia Tissue Bank (Columbus, OH), the Division of Hematology, Maisonneuve-Rosemont Hospital (Montreal, QC, Canada), and C. Gambacorti-Passerini (Division of Experimental Oncology, NCI, Milan, Italy) were approved by The Ohio State University Institutional Review Board. Survival of BCR/ABL-expressing cells was assessed by trypan-blue exclusion test. Methylcellulose-colony formation assays were carried out by plating 103 BCR/ABL-expressing cell lines or 104 CML-BCCD34+ cells in 0.9% MethoCult H4230 (Stem Cell Technologies). Colonies from cell lines and primary cells were scored 7 and 15 days later, respectively.
Plasmids
MigR1-HNRPK-HA, MigR1-(S284/353A)HNRPK-HA, and MigR1-(S284/353D)HNRPK-HA. The HA-tagged wild-type and S284/353A-HNRPK and S284/353D-HNRPK cDNAs21 (Z. Ronai, Mount Sinai School of Medicine, New York, NY) were subcloned blunted into the HpaI-digested MigR1 retroviral vector.
MigR1-HNRPK-C299-HA. The HA-tagged HNRPK-C299 fragment (amino acids 1-299) was polymerase chain reaction (PCR) generated using CMV-HA-C29915 (P. Raychaudhuri, University of Illinois, Chicago, IL) and subcloned into the HpaI-digested MigR1.
MigR1-HNRPK-AS. HNRPK cDNA from pSP72-K (G. Dreyfuss, UPENN, Philadelphia, PA) was blunted and subcloned in antisense orientation into MigR1.
pSuper.retro-Hnrpk shRNA. The shRNA Hnrpk was obtained by subcloning the mouse Hnrpk nucleotide sequence (5′-tggtgaatttggtaaacgc-3′) into the pSUPER.retro.neo+GFP vector (OligoEngine, Seattle, WA).
MigR1-Flag-ERK1, MigR1-Flag-ERK1K71R, MigR1-Flag-ERK2, and MigR1-Flag-ERK2K52R. The wild-type and dominant-negative ERK1 and ERK2 were PCR generated using the pCEP4-ERK1-WT, pCEP4-K71R-ERK1, pCMV5M-ERK2-WT, and pCMV5M-K52R-ERK2 plasmids (M. Cobb, University of Texas South Western Medical Center, Dallas, TX) and 5′-primers containing a XhoI site, an ATG, the Flag-tag and the first 17 nucleotides of ERK1 or ERK2 cDNAs, and 3′-primers containing an EcoRI site linked to the last 16 nucleotides of ERK1 or ERK2 cDNAs. The fragments were XhoI-EcoRI digested and subcloned into MigR1.
MSCV-puro-MYC. MSCV-based vector contains the MYC cDNA (G. Leone, Ohio State University, Columbus, OH).
pRF and the pRMF(IRES-MYC). pRF and the pRMF(IRES-MYC) were previously described.23
Northern blot analysis and RT-PCR
For Northern blot analysis, acid-phenol/guanidinium-extracted total RNA (5 μg) was hybridized to 32P-labeled mouse Hnrpk and MYC cDNAs. For reverse-transcriptase (RT)-PCR of Hnrpk pre-mRNA, 1 μg nuclear RNA obtained from sucrose gradient-purified nuclei by acid-phenol/guanidinium-mediated extraction was DNAseI treated and reverse transcribed using 200 U Moloney murine leukemia virus (M-MLV) reverse transcriptase (Gibco), 200 μM dNTPs, and 0.25 U/mL random hexamers (Pharmacia, Piscataway, NJ). cDNA was used as template for PCR. Hnrpk pre-mRNA primers were as follows: 5′-atggtgaatttggtgagaagatact-3′ and 5′-ggcgtttacctagaaattaaagttta-3′. As control, cDNA samples were adjusted to yield relatively equal amplification of GRB2. For RT-PCR amplification of MYC transcripts, cDNA was synthesized from total RNA of the CD34+ fraction of marrow from healthy donors and CML patients. hMYC mRNA was amplified using oligomers corresponding to nucleotides 1004 to 1023 and 1286 to 1306 of MYC cDNA. GRB2 mRNA levels were used to normalize levels of MYC mRNA among samples.
Western blot and metabolic labeling
Cells were lysed (107 cells/100 μL) in isotonic buffer (150 mM NaCl, 20 mM Hepes [pH 7.0], 1% NP-40, 10% glycerol) supplemented with protease and phosphatase inhibitors. For direct lysis, cells (2-3 × 105) were lysed in 20 μL Laemmli buffer. Antibodies used were as follows: monoclonal anti-HNRPK (G. Dreyfuss, UPENN, Philadelphia, PA), polyclonal anti-HNRPK (H-300l; Santa Cruz Biotechnology, Santa Cruz, CA), anti-MYC (Ab-2) and antiphosphotyrosine (Calbiochem, San Diego, CA), anti-GRB2 (Transduction Laboratories, Lexington, KY), anti-HA (Covance, Princeton, NJ), anti-Flag (Sigma, St Louis, MO), and anti-pERKthr202/tyr204 and anti-ERK (Cell Signaling, Danvers, MA). For metabolic labeling, cells were PBS rinsed, cultured (2 × 106 cells/mL) for 60 minutes in methionine/cysteine-free RPMI-1640/10% dialyzed FBS (Gibco) and 2 ng/mL rmIL-3 (R&D Systems, Minneapolis, MN), and resuspended (5 × 106 cells/mL) in medium containing 200 μCi/mL (7.4 MBq) [35S]-methionine/cysteine (NEN Life Science Products, Boston, MA). After 90 minutes, cells were lysed in isotonic buffer and lysates were incubated (4°C, 2 hours) with protein G agarose-coupled anti-MYC antibody. Immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto nitrocellulose, and visualized by autoradiography.
Immunofluorescence microscopy
Cytospins were fixed in 3.7% formaldehyde, PBS rinsed, permeabilized (10 minutes) in PBS/0.05% Triton X-100 (Sigma), rinsed, and blocked (10 minutes) in PBS/4% goat serum. Incubation with the anti-HA and anti-HNRPK antibodies (1:250 dilution) and with the fluorophore-labeled goat anti-mouse Texas Red and IgG Alexa 488 A-11001 (1:200 dilution; Molecular Probes, Carlsbad, CA), respectively, were carried out at room temperature (30 minutes). Slides were PBS rinsed, antifade treated (Molecular Probes), and analyzed by confocal microscopy (Zeiss LSM510, 40×/_ NA magnification, and Image software, both from Carl Zeiss, Thornwood, NY).
Luciferase assay
32D-BCR/ABL cells were transduced with the pRF or pRMF (IRES-MYC) plasmids by nucleofection using the manufacturer's protocol (Amaxa, Gaithersburg, MD). After 48 hours, firefly and renilla luciferase activities were assessed by the Dual Luciferase reporter system (Promega, Madison, WI) and detected with the Fluoroskan Ascent-FL luminometer (Thermo, Waltman, MA).
Myc mRNA association with HNRPK
Immunoprecipitation of the mRNAs associated with endogenous and exogenous HNRPK in BCR/ABL-expressing cells was performed as described25 using the monoclonal anti-HNRPK and anti-HA antibodies, respectively. Anti-Flag immunoprecipitation served as negative control. The presence of Myc transcripts was assessed by RT-PCR on equal amounts of RNA, extracted from RNP-enriched lysates (input) and from the anti-HNRPK and anti-HA immunoprecipitates. The mMyc primers used were as follows: 5′-ctggcctcctaccaggctg-3′ and 5′-ctggtggtgggcggtgtct-3′. PCR products were gel fractionated and visualized by ethidium bromide staining.
REMSA and UV cross-linking
Cytoplasmic extracts were prepared and used in REMSA (RNA electrophoretic mobility shift assay) as described.11 Briefly, 107 cells were lysed in 100 μL 10-mM HEPES-KOH (pH 7.5), 14 mM KCl, 3 mM MgCl2, 5% glycerol, 0.2% NP-40, 1 mM DTT, and 1 mM PMSF, and the cytoplasmic fraction was purified at 8500g (2 minutes; 4°C). Proteins (20 μg) were incubated (30 minutes; 25°C) with IRES-MYC oligoribonucleotide (6.25 × 103 cpm) corresponding to nucleotides 406 to 431 (5′-gacgcgacucucccgacgcggggagg-3′) of hMYC mRNA. Reactions were loaded on 5% PAGE or UV cross-linked (3500 μJ × 100; 30 minutes) and SDS-PAGE fractionated after incubation (20 minutes) with 10 mg/mL heparin.
Leukemogenesis in SCID mice
BCR/ABL+ cells were injected (5 × 106 cells/mouse) subcutaneously into 5-week-old ICR-SCID mice (5 mice/group; Taconic, Germantown, NY), which were killed 15 days after injection for tumor excision. To determine the effect of S284/353A-HNRPK on BCR/ABL leukemogenesis, 6-week-old ICR-SCID mice were intravenously injected with 5 × 105 32D-BCR/ABL cells expressing the wild-type or S284/353A-HNRPK (10 mice/group). After 4 weeks, 3 mice from each group were killed and organs were analyzed for the presence of leukemia. The remaining mice were used for survival studies, which were terminated 8 weeks after injection.
Results
Effect of BCR/ABL kinase activity on HNRPK expression
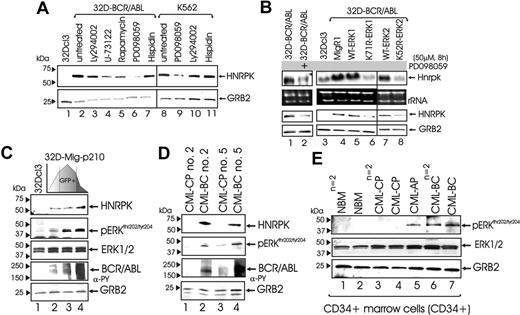
Using Affymetrix U74Av.2 arrays (Affymetrix, Santa Clara, CA), we found that Hnrpk mRNA levels were down-regulated (∼ 9-fold inhibition) in imatinib-treated (1 μM; 24 hours) compared with untreated BCR/ABL-expressing myeloid cells (not shown). Accordingly, Northern blot analysis revealed that Hnrpk mRNA levels were higher in IL-3-cultured 32D-BCR/ABL than parental 32Dcl3 cells, and that Hnrpk expression dropped off at levels similar to that of parental cells upon imatinib treatment (Figure 1Ai). To determine whether up-regulation of Hnrpk mRNA levels in 32D-BCR/ABL cells results from increased transcription and/or enhanced mRNA stability, we carried out RT-PCR on DNAse-I-treated nuclear RNA using primers corresponding to the exon 2-intron 2 and intron 2-exon 3 junctions, and Northern blot on actinomycin D-treated (5 μg/mL; 12 hours) parental and BCR/ABL-expressing 32Dcl3 cells. As shown, Hnrpk pre-mRNA transcripts are higher in 32D-BCR/ABL than IL-3-cultured 32Dcl3 cells (Figure 1Aii). Furthermore, Hnrpk mRNA levels are only marginally affected by inhibition of RNA synthesis in 32D-BCR/ABL cells, whereas they progressively decrease in actinomycin D-treated 32Dcl3 cells (Figure 1Aiii). As expected, HNRPK protein levels are upregulated by BCR/ABL expression in 32Dcl3 cells (Figure 1B, lanes 1-2), and in the doxycycline-treated (2 μg/mL; 3 days) TonB210.1 lymphoid precursors in which BCR/ABL is tetracycline inducible (Figure 1B, lanes 4-5). Accordingly, imatinib (48 hours, 1 μM) down-regulated HNRPK protein levels in IL-3-cultured 32D-BCR/ABL and Ph1 K562 cells (Figure 1B, lanes 3,7). However, HNRPK expression was similar in untreated and imatinibtreated 32D-BCR/ABL cells IL-3-cultured for 24 hours with the protein synthesis inhibitor cycloheximide (CHX, 20 μg/mL) (Figure 1C), suggesting that BCR/ABL does not affect HNRPK protein stability.
Increased HNRPK expression and its dependence on BCR/ABL kinase activity were also evident when we assessed HNRPK levels in mononuclear cells (MNCs) (Figure 1Di) and in the CD34+ bone marrow fraction (Figure 1Dii) from CML patients and healthy donors (NBM). As shown, HNRPK expression was up-regulated more in CML-BC (nCD34+ = 3, nMNC = 7) and CML-AP (nCD34+ = 1) than in CML-CP (nCD34+ = 2, nMNC = 8) and NBM (nCD34+ = 2) samples. Moreover, imatinib treatment of CML-BCCD34+ cells resulted in a marked decrease in HNRPK expression (Figure 1Diii). Note that the percentage of Ph1 cells in the CML-CP samples was 95% or more, and that BCR/ABL activity was higher in CML-BCCD34+ than CML-CPCD34+ cells (Figure 1Dii, inset).
Effect of BCR/ABL on HNRPK mRNA and protein expression. (Ai) Northern blot shows expression of Hnrpk mRNA in parental, untreated, and imatinib-treated BCR/ABL-expressing myeloid progenitor 32Dcl3 cells. (Aii) RT-PCR on nuclear RNA shows levels of Hnrpk pre-mRNA in parental and BCR/ABL-expressing myeloid progenitor 32Dcl3 cells (+RT). The amplified PCR product is comprehensive of part of exon 2, intron 2, and part of exon 3 of the mouse Hnrpk gene.-RT indicates that PCR was performed on non-reverse-transcribed nuclear RNA. GRB2 mRNA levels were measured as control. (Aiii) Effect of actinomycin D on Hnrpk mRNA levels in parental and BCR/ABL-expressing 32Dcl3 cells. Cells were incubated for the indicated time with actinomycin D used at a concentration (5 μg/mL) that reportedly inhibits RNA polII-dependent transcription. (Bi) Western blots show HNRPK protein levels in parental 32Dcl3 cells (lane 1) and in untreated (lane 2) and imatinib-treated (lane 3) 32D-BCR/ABL cells, in BCR/ABL-induced (doxycycline 1 mg/mL, 24 hours) TonB210.1 lymphoid progenitors (lanes 4-5), and in untreated and imatinib-treated Ph1 K562 cells (lanes 6-7). (C) HNRPK protein stability was determined by anti-HNRPK Western blots in untreated and imatinib-treated 32D-BCR/ABL cells exposed to the protein synthesis inhibitor cycloheximide (CHX). GRB2 levels were monitored as control for equal loading. (Di) Plot shows HNRPK protein levels expressed as log of arbitrary densitometric units after normalization with GRB2 levels in mononuclear cells from CML patients in the chronic (CML-CP) and blast crisis (CML-BC) phases. (P = .001; Wicoxon rank sum test). (Dii) Graphs shows GRB2-normalized HNRPK protein levels expressed as densitometric units in the CD34+ fraction of bone marrow from 2 healthy donors (NBM), and 2 CML-CP, 1 CML-AP, and 2 CML-BC patients. The Western blot in the inset shows total tyrosine phosphorylation, and the arrow indicates levels of active BCR/ABL. (Diii) HNRPK protein expression in untreated and imatinib-treated CD34+ CML-BC patient marrow cells. GRB2 was used as control for equal loading.
Effect of BCR/ABL on HNRPK mRNA and protein expression. (Ai) Northern blot shows expression of Hnrpk mRNA in parental, untreated, and imatinib-treated BCR/ABL-expressing myeloid progenitor 32Dcl3 cells. (Aii) RT-PCR on nuclear RNA shows levels of Hnrpk pre-mRNA in parental and BCR/ABL-expressing myeloid progenitor 32Dcl3 cells (+RT). The amplified PCR product is comprehensive of part of exon 2, intron 2, and part of exon 3 of the mouse Hnrpk gene.-RT indicates that PCR was performed on non-reverse-transcribed nuclear RNA. GRB2 mRNA levels were measured as control. (Aiii) Effect of actinomycin D on Hnrpk mRNA levels in parental and BCR/ABL-expressing 32Dcl3 cells. Cells were incubated for the indicated time with actinomycin D used at a concentration (5 μg/mL) that reportedly inhibits RNA polII-dependent transcription. (Bi) Western blots show HNRPK protein levels in parental 32Dcl3 cells (lane 1) and in untreated (lane 2) and imatinib-treated (lane 3) 32D-BCR/ABL cells, in BCR/ABL-induced (doxycycline 1 mg/mL, 24 hours) TonB210.1 lymphoid progenitors (lanes 4-5), and in untreated and imatinib-treated Ph1 K562 cells (lanes 6-7). (C) HNRPK protein stability was determined by anti-HNRPK Western blots in untreated and imatinib-treated 32D-BCR/ABL cells exposed to the protein synthesis inhibitor cycloheximide (CHX). GRB2 levels were monitored as control for equal loading. (Di) Plot shows HNRPK protein levels expressed as log of arbitrary densitometric units after normalization with GRB2 levels in mononuclear cells from CML patients in the chronic (CML-CP) and blast crisis (CML-BC) phases. (P = .001; Wicoxon rank sum test). (Dii) Graphs shows GRB2-normalized HNRPK protein levels expressed as densitometric units in the CD34+ fraction of bone marrow from 2 healthy donors (NBM), and 2 CML-CP, 1 CML-AP, and 2 CML-BC patients. The Western blot in the inset shows total tyrosine phosphorylation, and the arrow indicates levels of active BCR/ABL. (Diii) HNRPK protein expression in untreated and imatinib-treated CD34+ CML-BC patient marrow cells. GRB2 was used as control for equal loading.
HNRPK expression depends on the BCR/ABL-regulated MAPK pathway
To investigate the mechanism(s) whereby BCR/ABL enhances HNRPK expression, 32D-BCR/ABL and K562 cells were exposed to inhibitors of BCR/ABL-activated pathways.2,4 HNRPK protein levels were significantly reduced by 8-hour treatment with the MAPK inhibitor PD098059 (50 μM) but not with inhibitors of PI-3K-dependent (25 μM LY204002), PLCγ-dependent (1 μM U73122), mTOR/S6K-dependent (15 nM rapamycin), and PKCβ-dependent (5 μM hispidin) signals (Figure 2A), suggesting that enhanced HNRPK gene expression may depend on BCR/ABL-induced activation of MAPK (ie, ERK1/2). Indeed, Hnrpk mRNA levels were inhibited by PD098059 treatment and expression of ERK1K71R and ERK2K52R dominant-negative mutants26 (Figure 2B). Conversely, Hnrpk mRNA and protein levels remained unchanged upon expression of wild-type ERK1 and ERK2 (Figure 2B).
To determine whether increased HNRPK expression requires high levels of BCR/ABL activity, 32Dcl3 cells were infected with MigRI-GFP-BCR/ABL11 and sorted for low, medium, and high GFP expression. As expected, GFP levels correlated with BCR/ABL activity (Figure 2C). Of interest, graded BCR/ABL expression resulted in a dose-dependent induction of MAPKERK1/2 activity and HNRPK expression (Figure 2C). Note that total MAPKERK1/2 levels were not affected by BCR/ABL (Figure 2C, third panel). Similarly, HNRPK levels correlated with BCR/ABL and MAPK activity in mononuclear cells from sequential CP and BC samples of 2 CML patients (Figure 2D). Similarly, MAPKERK1/2 activity was higher in CML-BCCD34+ (n = 2) and CML-APCD34+ (n = 1) than in CML-CPCD34+ (n = 2) and NBMCD34+ (n = 2) cells (Figure 2E). Conversely, ERK1/2 expression slightly increased in CML-CPCD34+ and CML-BCCD34+ compared with NBMCD34+, but no significant differences were observed between CML-CPCD34+ and CML-BCCD34+ patient samples (Figure 2E).
In vitro and in vivo HNRPK requirement for BCR/ABL oncogenic activity
To investigate the requirement of HNRPK for BCR/ABL oncogenic potential, we knocked-down HNRPK in BCR/ABL-expressing cells and assessed their growth factor-independent proliferation, clonogenic potential, and ability to undergo granulocytic differentiation and induce tumors in SCID mice. Reduced HNRPK expression was achieved by infecting 32D-BCR/ABL cells with a MigR1-GFP containing either the Hnrpk cDNA in antisense orientation or an shRNA expression cassette containing a 19-bp sequence corresponding to nucleotides from 46 to 65 of the Hnrpk cDNA. Western blots performed on GFP-sorted antisense-expressing (32D-B/A-K-AS) and shRNA-expressing (32D-B/A-K-shRNA) 32D-BCR/ABL cells showed an approximately 60% decrease in HNRPK protein levels (Figure 3Ai, lanes 3,5). Accordingly, levels of MYC, a BCR/ABL- and HNRPK-regulated oncogene,16,23,27 were markedly down-regulated upon shRNA-mediated inhibition of HNRPK (Figure 3Aii).
Down-regulation of HNRPK significantly inhibited growth factor-independent proliferation (Figure 3B) and colony formation (50%-65% inhibition) (Figure 3C). Seemingly, HNRPK knockdown resulted in decreased tumorigenic potential, as 32D-B/A-KAS cells formed tumors with an increased latency time and a 50% decreased weight (not shown). By contrast, HNRPK down-regulation only partially restored G-CSF-driven maturation of 32D-BCR/ABL cells, as cells at the intermediate stages of maturation but not neutrophil cells were detectable in G-CSF-cultured (9 days) 32D-B/A-K-shRNA cells (Figure 3D).
HNRPK transcription-regulatory activity is dispensable for BCR/ABL leukemogenesis
To investigate the importance of HNRPK transcriptional activity for BCR/ABL leukemogenesis, we transduced 32D-BCR/ABL cells with the MigR1-HNRPK-C299-HA retrovirus (Figure 4A, lane 3) containing a C-terminal truncated HNRPK that is retained in the nucleus (Figure 4B) where it binds DNA but is deficient in promoter transactivation.15 Cytokine-independent or -dependent proliferation in liquid culture (not shown) and colony formation (Figure 4C) was similar in vector-transduced and C299-HNRPK-transduced 32D-BCR/ABL cells. As expected, ectopic expression of HA-tagged wild-type HNRPK only slightly increased 32D-BCR/ABL clonogenic potential (Figure 4C). Similarly, C299-HNRPK expression did not impair the tumorigenic potential of 32D-BCR/ABL cells (not shown).
Role of MAPK in the regulation of HNRPK expression in BCR/ABL+ cells. (A) Effect of different chemical inhibitors of known BCR/ABL-activated pathways on HNRPK protein levels in 32D-BCR/ABL and K562 cells. (B) Effect of the MEK1/MAPK inhibitor PD098059 (lane 2) on Hnrpk mRNA (first row) and protein (third row) expression. Hnrpk mRNA (first row) and protein (third row) levels in parental 32Dcl3 cells (lane 3), vector-transduced 32D-BCR/ABL (lane 4), and 32D-BCR/ABL cells ectopically expressing wild-type MAPK ERK1 (lane 5), the K71R dominant-negative MAPK ERK1 mutant (lane 6), wild-type ERK2 (lane 7), and dominant-negative K52R ERK2 mutant (lane 8). rRNA and GRB2 are shown as controls for equal loading. (C) Effect of BCR/ABL tyrosine kinase on MAPK/ERK activity. pERK1/2 indicates phosphorylated (active) ERK1 and ERK2 mitogen-activated protein kinases. Western blots show levels of HNRPK, phospho-ERK1/2, total ERK1/2, BCR/ABL, and GRB2 in 32Dcl3 myeloid precursors transduced with a GFP/BCR/ABL bicistronic retrovirus and sorted for low, medium, and high GFP levels. (D) Levels of HNRPK, phospho-ERK1/2, BCR/ABL, and GRB2 in mononuclear marrow cells from paired chronic phase (CP) and blast crisis (BC) CML patient samples. (E) Levels of pERK1/2, ERK1/2, and GRB2 in NBMCD34+, CML-CPCD34+, CML-APCD34+, and CML-BCCD34+samples.
Role of MAPK in the regulation of HNRPK expression in BCR/ABL+ cells. (A) Effect of different chemical inhibitors of known BCR/ABL-activated pathways on HNRPK protein levels in 32D-BCR/ABL and K562 cells. (B) Effect of the MEK1/MAPK inhibitor PD098059 (lane 2) on Hnrpk mRNA (first row) and protein (third row) expression. Hnrpk mRNA (first row) and protein (third row) levels in parental 32Dcl3 cells (lane 3), vector-transduced 32D-BCR/ABL (lane 4), and 32D-BCR/ABL cells ectopically expressing wild-type MAPK ERK1 (lane 5), the K71R dominant-negative MAPK ERK1 mutant (lane 6), wild-type ERK2 (lane 7), and dominant-negative K52R ERK2 mutant (lane 8). rRNA and GRB2 are shown as controls for equal loading. (C) Effect of BCR/ABL tyrosine kinase on MAPK/ERK activity. pERK1/2 indicates phosphorylated (active) ERK1 and ERK2 mitogen-activated protein kinases. Western blots show levels of HNRPK, phospho-ERK1/2, total ERK1/2, BCR/ABL, and GRB2 in 32Dcl3 myeloid precursors transduced with a GFP/BCR/ABL bicistronic retrovirus and sorted for low, medium, and high GFP levels. (D) Levels of HNRPK, phospho-ERK1/2, BCR/ABL, and GRB2 in mononuclear marrow cells from paired chronic phase (CP) and blast crisis (BC) CML patient samples. (E) Levels of pERK1/2, ERK1/2, and GRB2 in NBMCD34+, CML-CPCD34+, CML-APCD34+, and CML-BCCD34+samples.
Because the C299-HNRPK mutant is still competent for DNA binding,15 we investigated whether its expression alters Myc mRNA expression, which is transcriptionally induced by the HNRPK binding to a CT-element within MYC promoter.16 Northern and Western blot analyses revealed that regulation of Myc expression in BCR/ABL-transformed cells does not involve HNRPK transcriptional activity, as C299-HNRPK expression did not affect Myc mRNA and protein levels (Figure 4A, second and fourth panels).
Requirement of HNRPK translation-regulatory activity for BCR/ABL leukemogenesis
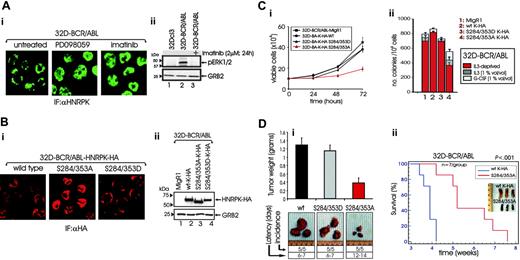
MAPKERK1/2 are activated by BCR/ABL in a tyrosine kinase-dependent manner28,29 (Figure 5Aii), and MAPKERK1/2-dependent HNRPK phosphorylation drives HNRPK cytoplasmic accumulation in HeLa cells.21 Thus, inhibition of MAPK signaling might influence HNRPK subcellular localization also in BCR/ABL-transformed cells. Indeed, confocal microscopy on anti-HNRPK-stained cells showed a clear nuclear accumulation of HNRPK in PD098059- or imatinib-treated 32D-BCR/ABL cells (Figure 5A), suggesting that BCR/ABL kinase activity not only enhances HNRPK expression but also induces HNRPK cytoplasmic accumulation in a MAPK-dependent manner.
In vitro and in vivo requirement of HNRPK for BCR/ABL oncogenic potential. (Ai) HNRPK levels in parental 32Dcl3, 32D-BCR/ABL, and 32D-BCR/ABL cells transduced with an antisense (lane 3) or an shRNA (lane 5) HNRPK retroviral construct. (Aii) Effect of HNRPK down-regulation by shRNA (lane 3) on the levels of the HNRPK- and BCR/ABL-regulated Myc protein. (B) Effect of HNRPK down-regulation on the growth factor-independent proliferation of 32D-BCR/ABL cells. (C) Effect of Hnrpk antisense and shRNA on the growth factor-independent clonogenic efficiency of 32D-BCR/ABL cells. Bars represent mean ± SE of data from 3 independent experiments. (D) 32D-BCR/ABL and GFP+ 32D-BCR/ABL cells retrovirally transduced with pSuper-retroneo-Hnrpk shRNA were induced to differentiate with G-CSF. Cells were subjected to cytospin and were May-Grünwald/Giemsa stained. 32Dcl3 parental cells were used as a control for granulocyte colony-stimulating factor (G-CSF)-induced granulocytic differentiation. Images were taken with a Zeiss Axioskope 2 Plus and a 40×/0.75 NA objective, with a Canon Powershot A70 (Canon, Lake Success, NY) and Canon Capture software.
In vitro and in vivo requirement of HNRPK for BCR/ABL oncogenic potential. (Ai) HNRPK levels in parental 32Dcl3, 32D-BCR/ABL, and 32D-BCR/ABL cells transduced with an antisense (lane 3) or an shRNA (lane 5) HNRPK retroviral construct. (Aii) Effect of HNRPK down-regulation by shRNA (lane 3) on the levels of the HNRPK- and BCR/ABL-regulated Myc protein. (B) Effect of HNRPK down-regulation on the growth factor-independent proliferation of 32D-BCR/ABL cells. (C) Effect of Hnrpk antisense and shRNA on the growth factor-independent clonogenic efficiency of 32D-BCR/ABL cells. Bars represent mean ± SE of data from 3 independent experiments. (D) 32D-BCR/ABL and GFP+ 32D-BCR/ABL cells retrovirally transduced with pSuper-retroneo-Hnrpk shRNA were induced to differentiate with G-CSF. Cells were subjected to cytospin and were May-Grünwald/Giemsa stained. 32Dcl3 parental cells were used as a control for granulocyte colony-stimulating factor (G-CSF)-induced granulocytic differentiation. Images were taken with a Zeiss Axioskope 2 Plus and a 40×/0.75 NA objective, with a Canon Powershot A70 (Canon, Lake Success, NY) and Canon Capture software.
Reportedly, the HNRPK phosphomimetic S284/353D mutant, which carries the MAPKERK1/2 phosphoacceptor sites (serines 284 and 353) mutated to aspartic acid, has cytoplasmic localization in HeLa cells and a constitutively enhanced translational-regulatory activity.21 By contrast, the nonphosphatable serine to alanine (S284/353A) mutant is nucleus localized and competes with the endogenous HNRPK for mRNA binding and nuclear export.21 Thus, to determine whether HNRPK-dependent regulation of mRNA translation is important for the phenotype of BCR/ABL-transformed myeloid progenitors, 32D-BCR/ABL cells were transduced with HA-tagged wild-type, S284/353A-HNRPK, or S284/353D-HNRPK cDNAs. 32D-BCR/ABL-derivative cell lines expressed the ectopic HNRPK proteins at similar levels (Figure 5Bii). Moreover, confocal microscopy on anti-HA-stained HA-HNRPK-expressing 32D-BCR/ABL cells revealed that the S284/353A-HNRPK and S284/353D-HNRPK mutants were exclusively localized in the nucleus and in the cytoplasm, respectively, while the wild-type HNRPK was prevalently cytoplasmic (Figure 5Bi).
HNRPK transcription-regulatory activity is dispensable for BCR/ABL oncogenic potential. (A) Western and Northern blots show levels of Myc and/or HA-tagged wild-type and mutant C299-HNRPK in 32D-BCR/ABL cells transduced with the empty MigR1 vector (lane 1), wild-type HNRPK (lane 2), and C299-HNRPK dominant-negative mutant (lane 3). (B) Confocal microscopy of anti-HA-stained wild-type and C299-HNRPK-expressing 32D-BCR/ABL cells. (C) Effect of wild-type and C299-HNRPK on growth factor-independent clonogenic efficiency of 32D-BCR/ABL cells.
HNRPK transcription-regulatory activity is dispensable for BCR/ABL oncogenic potential. (A) Western and Northern blots show levels of Myc and/or HA-tagged wild-type and mutant C299-HNRPK in 32D-BCR/ABL cells transduced with the empty MigR1 vector (lane 1), wild-type HNRPK (lane 2), and C299-HNRPK dominant-negative mutant (lane 3). (B) Confocal microscopy of anti-HA-stained wild-type and C299-HNRPK-expressing 32D-BCR/ABL cells. (C) Effect of wild-type and C299-HNRPK on growth factor-independent clonogenic efficiency of 32D-BCR/ABL cells.
In 32D-BCR/ABL cells, cytokine-independent growth (Figure 5Ci) and colony formation (Figure 5Cii) were impaired by S284/353A-HNRPK but not by wild-type or S284/353D-HNRPK expression. Note that the clonogenic activity of S284/353A-HNRPK-expressing 32D-BCR/ABL cells was only partially restored by IL-3 and G-CSF (Figure 5Cii). Consistent with their effects in vitro, wild-type (wt) and S284/353D-HNRPK-HA-expressing 32D-BCR/ABL cells formed tumors (incidence: 5/5) in 6 to 7 days, whereas tumors from S284/353A-HNRPK-expressing 32D-BCR/ABL cells were palpable (incidence: 5/5) only after 12 to 14 days after injection. At 15 days after injection, S284/353A-HNRPK-expressing 32D-BCR/ABL cells developed tumors that weighed 70% to 80% less than the tumors formed by injection of the wild-type or S284/353D-HNRPK-expressing 32D-BCR/ABL cells (`Figure 5Di).
HNRPK translation-regulatory activity is essential for BCR/ABL oncogenic potential. (Ai) Confocal microscopy on anti-HNRPK-stained 32D-BCR/ABL cells untreated and treated with the MAPK inhibitor PD098059 or with imatinib. (Aii) Effect of imatinib on the BCR/ABL-induced phosphorylation of MAPK ERK1/2. (Bi) Confocal microscopy on anti-HA-stained 32D-BCR/ABL cells ectopically expressing HA-tagged wild-type, dominant-active S284/353D, and dominant-negative S284/353A mutant HNRPK proteins. (Bii) Levels of ectopic wild-type and mutant HNRPK proteins. (C) Effect of wild-type, S284/353D, and S284/353A mutant HNRPK on growth factor-independent proliferation (i) and cytokine-dependent and independent clonogenic efficiency (ii) of 32D-BCR/ABL cells. Bars represent the mean ± SE of values from 3 different experiments. (Di) Effect of wild-type and HNRPK mutants on the tumorigenic potential of 32D-BCR/ABL cells. Cells (106) were injected subcutaneously into SCID mice; tumor appearance and growth were monitored daily and mice were killed 15 days after injection. Bars represent the mean ± SE from the weight of 5 tumors for each group. (Dii) Effect of wild-type and S284/353A-HNRPK on the leukemogenic process induced in SCID mice by intravenous injection of BCR/ABL-expressing cells. Kaplan-Meier plot shows survival of mice injected with 32D-BCR/ABL cells expressing wild-type or S284/353A-HNRPK. The log-rank test evaluated the differences among survival distributions (P < .001). Inset shows spleens of 3 mice of each group killed 4 weeks after cell injection.
HNRPK translation-regulatory activity is essential for BCR/ABL oncogenic potential. (Ai) Confocal microscopy on anti-HNRPK-stained 32D-BCR/ABL cells untreated and treated with the MAPK inhibitor PD098059 or with imatinib. (Aii) Effect of imatinib on the BCR/ABL-induced phosphorylation of MAPK ERK1/2. (Bi) Confocal microscopy on anti-HA-stained 32D-BCR/ABL cells ectopically expressing HA-tagged wild-type, dominant-active S284/353D, and dominant-negative S284/353A mutant HNRPK proteins. (Bii) Levels of ectopic wild-type and mutant HNRPK proteins. (C) Effect of wild-type, S284/353D, and S284/353A mutant HNRPK on growth factor-independent proliferation (i) and cytokine-dependent and independent clonogenic efficiency (ii) of 32D-BCR/ABL cells. Bars represent the mean ± SE of values from 3 different experiments. (Di) Effect of wild-type and HNRPK mutants on the tumorigenic potential of 32D-BCR/ABL cells. Cells (106) were injected subcutaneously into SCID mice; tumor appearance and growth were monitored daily and mice were killed 15 days after injection. Bars represent the mean ± SE from the weight of 5 tumors for each group. (Dii) Effect of wild-type and S284/353A-HNRPK on the leukemogenic process induced in SCID mice by intravenous injection of BCR/ABL-expressing cells. Kaplan-Meier plot shows survival of mice injected with 32D-BCR/ABL cells expressing wild-type or S284/353A-HNRPK. The log-rank test evaluated the differences among survival distributions (P < .001). Inset shows spleens of 3 mice of each group killed 4 weeks after cell injection.
To determine if expression of the translation-defective21 HNRPK mutant (S284/353A) can attenuate the acute leukemia-like disease process induced in mice by intravenous injection of BCR/ABL+ cells, SCID mice (10 mice per group) were injected with 32D-BCR/ABL cells (5 × 105 cells/mouse) expressing either wild-type K-HA or S284/353A-HNRPK-HA. Three and half weeks later, various organs obtained from 3 mice per group were evaluated by visual inspection and light microscopy. All 3 mice injected with wt-K-HA-expressing 32D-BCR/ABL cells showed massive splenomegaly (Figure 5Dii, inset). By contrast, morphology of spleens from mice injected with S284/353A-expressing 32D-BCR/ABL cells appeared normal (Figure 5D) and resembled that of control agematched mice (not shown). Consistent with these findings, the survival of mice injected with S284/353A-K-HA-expressing 32D-BCR/ABL cells was significantly longer than that of mice injected with wild type K-HA-expressing 32D-BCR/ABL cells (Figure 5Dii). However, modest to severe splenomegaly and leukemic infiltration of hematopoietic organs was observed in all mice that died 5 or more weeks after cell injection (not shown).
HNRPK-dependent regulation of Myc expression is important for BCR/ABL oncogenic potential
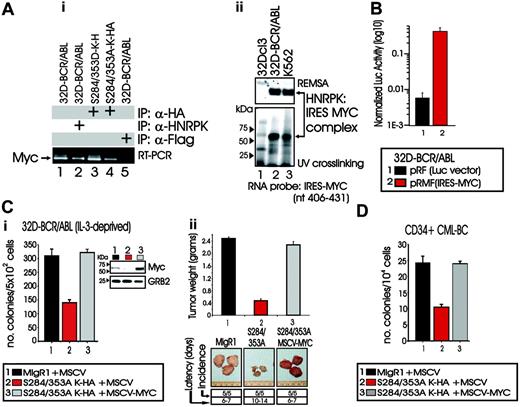
By RT-PCR, we detected Myc mRNA in anti-HNRPK and anti-HA immunoprecipitates from lysates of 32D-BCR/ABL cells expressing either the wild-type HNRPK or the HA-tagged serine 284 and 353 HNRPK mutants (Figure 6Ai, lanes 2-4), but not in anti-Flag immunoprecipitates used as a negative control (Figure 6Ai, lane 5). Accordingly, REMSA with extracts of parental 32Dcl3, 32D-BCR/ABL, and K562 cells and 32P-labeled RNA oligonucleotide corresponding to nucleotides 406 to 431 of the IRES element present in the human MYC mRNA (HSMYC1; V00568), and containing the UCCCGA HNRPK binding site,23 showed that an RNA/protein complex is formed with lysates of BCR/ABL-expressing (Figure 6Aii, top panel, lanes 2-3) but not 32Dcl3 (Figure 6Aii, top panel, lane 1) cells. Moreover, UV cross-linking showed that the protein present in 32D-BCR/ABL and K562 lysates and binding to the MYC IRES RNA-oligo has an apparent molecular weight identical to that of HNRPK (Figure 6Aii, bottom panel, lanes 2-3), suggesting that HNRPK up-regulates MYC expression upon binding to MYC IRES. Indeed, the presence of the MYC IRES element strongly enhanced expression of firefly luciferase in 32D-BCR/ABL transiently transduced with the pRMF (IRES-MYC)23 construct containing the renilla luciferase cDNA and the MYC IRES element fused in frame to the firefly luciferase reporter gene (wt-IRES) (Figure 6B).
Because decreased proliferation and leukemogenic potential of S284/353A-HNRPK-expressing 32D-BCR/ABL cells (Figure 5) may, in part, depend on inhibition of HNRPK-dependent MYC mRNA translation,23 S284/353A-expressing 32D-BCR/ABL cells were transduced with MSCV-puro-MYC and tested for cytokine-independent colony formation and tumorigenesis in SCID mice. Ectopic MYC expression (Figure 6C, inset) completely counteracted the effects of S284/353A-HNRPK (lane 2 in Figure 6Ci and 6Cii), as clonogenic potential and weight of tumors from S284/353A-MYC-expressing BCR/ABL cells were similar to those of vector-transduced 32D-BCR/ABL cells (Figure 6C, lanes 1-3). Similarly, CML-BCCD34+ patient cells transduced with the (S284/353A)HNRPK construct formed approximately 60% fewer colonies than vector-transduced cells (Figure 6D). Furthermore, forced MYC expression counteracted the effect of S284/353A-HNRPK expression and restored CML-BCCD34+ clonogenicity (Figure 6D). Thus, HNRPK-dependent regulation of MYC expression is important in vitro and in vivo for BCR/ABL oncogenic potential.
HNRPK binding to Myc mRNA and effect of ectopic Myc expression on the leukemic phenotype of S284/353A-HNRPK-expressing 32D-BCR/ABL cells. (Ai) RT-PCR shows presence of Myc mRNA in anti-HNRPK and anti-HA immunoprecipitates from lysates of parental (lane 2) and S284/353(A/D) (lanes 3-4) HNRPK-expressing 32D-BCR/ABL cells, respectively. Lane 1 shows levels of Myc in total mRNA, whereas lane 5 represents RT-PCR of mRNA immunoprecipitated with a nonrelated (anti-Flag) antibody; (Aii) REMSA (top) and UV cross-linking (bottom) with cytoplasmic extracts of 32Dcl3, 32D-BCR/ABL, and K562 cells show binding of HNRPK to the HNRPK binding site contained in the MYC IRES element. (B) Graphs shows firefly luciferase activity after normalization with Renilla luciferase activity in 32D-BCR/ABL cells transduced with the empty pRF and with the pRMF(IRES-MYC) plasmid containing the MYC IRES element cloned in front of the firefly luciferase cDNA. (C) Effect of ectopic MYC expression on the growth factor-independent colony formation (i) and tumorigenic potential (ii) of S284/353A-HNRPK-expressing 32D-BCR/ABL cells. Inset in panel Ci shows total Myc protein levels in vector-transduced, S284/353A-HNRPK/MYC-transduced, and S284/353A-HNRPK/MYC-transduced 32D-BCR/ABL cells. (D) Effect of MYC overexpression on the growth factor-independent clonogenic potential of CD34+ bone marrow CML-BC patient cells ectopically expressing the S284/353A-HNRPK mutant. Primary CD34+ CML-BC marrow cells were transduced with the MigR1-S284/353A-HNRPK-HA construct, selected for GFP+, and infected again with the MSCV-puro-MYC retrovirus. In panels B-D, bars represent mean ± SE of values from 3 different experiments.
HNRPK binding to Myc mRNA and effect of ectopic Myc expression on the leukemic phenotype of S284/353A-HNRPK-expressing 32D-BCR/ABL cells. (Ai) RT-PCR shows presence of Myc mRNA in anti-HNRPK and anti-HA immunoprecipitates from lysates of parental (lane 2) and S284/353(A/D) (lanes 3-4) HNRPK-expressing 32D-BCR/ABL cells, respectively. Lane 1 shows levels of Myc in total mRNA, whereas lane 5 represents RT-PCR of mRNA immunoprecipitated with a nonrelated (anti-Flag) antibody; (Aii) REMSA (top) and UV cross-linking (bottom) with cytoplasmic extracts of 32Dcl3, 32D-BCR/ABL, and K562 cells show binding of HNRPK to the HNRPK binding site contained in the MYC IRES element. (B) Graphs shows firefly luciferase activity after normalization with Renilla luciferase activity in 32D-BCR/ABL cells transduced with the empty pRF and with the pRMF(IRES-MYC) plasmid containing the MYC IRES element cloned in front of the firefly luciferase cDNA. (C) Effect of ectopic MYC expression on the growth factor-independent colony formation (i) and tumorigenic potential (ii) of S284/353A-HNRPK-expressing 32D-BCR/ABL cells. Inset in panel Ci shows total Myc protein levels in vector-transduced, S284/353A-HNRPK/MYC-transduced, and S284/353A-HNRPK/MYC-transduced 32D-BCR/ABL cells. (D) Effect of MYC overexpression on the growth factor-independent clonogenic potential of CD34+ bone marrow CML-BC patient cells ectopically expressing the S284/353A-HNRPK mutant. Primary CD34+ CML-BC marrow cells were transduced with the MigR1-S284/353A-HNRPK-HA construct, selected for GFP+, and infected again with the MSCV-puro-MYC retrovirus. In panels B-D, bars represent mean ± SE of values from 3 different experiments.
BCR/ABL-induced Myc expression is translationally regulated by HNRPK in a MAPK-dependent manner
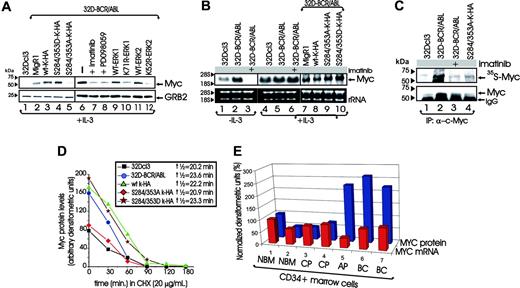
In IL-3-cultured 32Dcl3 myeloid progenitors, p210BCR/ABL augmented Myc protein levels (Figure 7A, lanes 1-2) without significantly affecting Myc mRNA expression (Figure 7B, lanes 4-5). Accordingly, treatment of IL-3-cultured 32D-BCR/ABL cells with imatinib (1 μM, 24 hours) inhibited Myc protein expression without affecting Myc mRNA levels (Figure 7A and B, lanes 7 and 6, respectively), suggesting that BCR/ABL posttranscriptionally up-regulates Myc expression. Of note, Myc mRNA down-regulation was evident only upon IL-3 deprivation (12 hours) in 32Dcl3 and imatinib-treated 32D-BCR/ABL cells (Figure 7B, lanes 1-3) and in K562 cells growth arrested by imatinib (not shown).
Because MYC expression is required for proliferation and colony formation of CML-BC cells,30 and HNRPK knock-down induces Myc down-regulation (Figure 3), we assessed the effects of ectopic expression of wild-type, S284/353D-, and S284/353A-HNRPK on Myc protein and mRNA levels. Myc levels were markedly reduced in S284/353A-HNRPK-expressing 32D-BCR/ABL (Figure 7A, lane 5) and K562 (not shown) cells compared with vector-transduced cells (Figure 7A, lane 2). By contrast, Myc levels were increased in wild-type and S284/353D-HNRPK-expressing 32D-BCR/ABL (Figure 7A, lanes 3-4) and K562 (not shown) cells. Accordingly, inhibition of MAPK activity by PD098059 and ectopic expression of ERK1K71R and ERK2K52R dominant-negative, but not wild-type, MAPKs also down-regulated Myc protein levels (Figure 7A, lanes 8-12). As expected, Myc mRNA levels remained unchanged in IL-3-cultured (Figure 7B, lanes 7-10) and IL-3-deprived (not shown) 32D-BCR/ABL cells expressing wild-type or mutant HNRPK proteins.
To assess whether the BCR/ABL-induced up-regulation of Myc expression reflects an HNRPK-dependent increase in Myc synthesis, we pulse labeled IL-3-cultured 32Dcl3 cells, and untreated and imatinib-treated 32D-BCR/ABL and S284/353A-HNRPK-expressing 32D-BCR/ABL cells with 35[S]-methionine/cysteine and determined the levels of newly translated Myc after immunoprecipitation. Newly synthesized Myc is barely detected in 32Dcl3 cells, readily detected in BCR/ABL-expressing cells, and substantially inhibited in imatinib-treated or S284/353A-HNRPK-expressing 32D-BCR/ABL cells (Figure 7C). As expected, total Myc levels reflected its synthesis rate (Figure 7C, lower panel). In addition, in CHX-treated (20 μg/mL; 0-180 minutes) IL-3-cultured 32Dcl3, 32D-BCR/ABL, and derivative wild-type-expressing, S284/353D-expressing, and S284/353A-HNRPK-expressing cell lines, Myc protein stability is only slightly affected by expression of BCR/ABL or wild-type and mutant HNRPK proteins (Figure 7D).
Furthermore, MYC protein but not mRNA levels were higher in CD34+ CML-BC and CML-AP than CD34+ CML-CP patient-derived marrow cells and CD34+ normal marrow cells (NBM) (Figure 7E). As expected, MYC levels correlated with those of HNRPK and with BCR/ABL kinase activity in CD34+ CML patient cells (Figure 1Dii). Thus, MYC overexpression in CML-BC depends in part on the ability of BCR/ABL to activate MAPKs, which, in turn, enhance IRES-dependent MYC protein synthesis through the HNRPK translation-stimulatory activity.
Discussion
Cytogenetic and molecular changes occur in the vast majority of CML patients during transition to blast crisis2 ; however, the mechanism(s) whereby each specific secondary genetic alteration contributes to disease progression is still largely unclear. Conversely, there is growing evidence attributing an important role to the BCR/ABL oncoprotein in determining the phenotype of CML-BC cells.2 In fact, increased BCR/ABL expression is a feature of CML-BC progenitors,8-10,31 and unrestrained BCR/ABL activity in CML-BC alters the expression of factors important for proliferation, survival, and maturation of myeloid progenitors.2 For example, in CML-BC, high BCR/ABL levels suppress C/EBPα-dependent differentiation, down-regulate G-CSFR, and enhance the expression of the antiapoptotic Bcl-XL and of the p53-ubiquitin ligase MDM2 by posttranslationally enhancing the expression/activity of several RNA binding proteins (ie, hnRNP E2, hnRNP A1, TLS/FUS, and La).11,12,24,32 Furthermore, increased expression and activity of BCR/ABL oncoprotein in CML-BC may result from the functional inactivation of protein phosphatase 2A (PP2A), which depends on the BCR/ABL-hnRNP A1-dependent regulation of the leukemia-associated factor and PP2A inhibitor SET.33 In fact, molecular or pharmacologic restoration of PP2A tumor suppressor activity impairs the leukemogenic potential of CML-BC and imatinib-sensitive and -resistant (T315I included) BCR/ABL+ cells by inducing BCR/ABL dephosphorylation and proteasome-dependent degradation.33
BCR/ABL- and MAPK-dependent phosphorylation of HNRPK regulates Myc mRNA translation in BCR/ABL-transformed myeloid cells. (A) Western blots show Myc levels in parental 32Dcl3 cells (lane 1), untreated 32D-BCR/ABL cells (lane 6); or cells treated with imatinib (lane 7) or with the MEK1 inhibitor PD098059 (lane 8); or cells transduced with the empty retrovirus (lane 2) or with wild-type and mutant HNRPK (lanes 3-5) or with MAPK ERK1 (lanes 9-10) and ERK2 (lanes 11-12) cDNAs. All cell lines were cultured in the presence of IL-3. (B) Northern blots show Myc mRNA levels in the indicated cell lines either maintained in IL-3 or IL-3 deprived for 12 hours in the presence or absence of imatinib. (C) Autoradiography and anti-MYC Western blot show levels of 35S-labeled newly synthesized and total immunoprecipitated Myc protein in parental 32Dcl3, untreated, and imatinib-treated 32D-BCR/ABL and S284/353A expressing 32D-BCR/ABL cells. (D) Levels of Myc protein, expressed as arbitrary densitometric units after normalization with GRB2 levels, in cycloheximide-treated parental 32Dcl3, 32D-BCR/ABL, and its derivative cell lines expressing wild-type or mutant HNRPK proteins. Half-life was calculated using the algorithm t½ = 0.693tn/ln(C0/Cn), where n represents the last time point. (E) Histograms show levels of MYC mRNA (red bars) and protein (blue bars) after normalization with GAPDH mRNA and GRB2 protein levels, respectively. Values in samples 2 to 7 represent arbitrary densitometric units expressed as percentage of those of sample 1. Western blot and RT-PCR were performed with lysate and total RNA, respectively, of CD34+marrow cells from healthy individuals (lanes 1-2) and from CML-CP (lanes 3-4), CML-AP (lane 5), and CML-BC (lanes 6-7) patients.
BCR/ABL- and MAPK-dependent phosphorylation of HNRPK regulates Myc mRNA translation in BCR/ABL-transformed myeloid cells. (A) Western blots show Myc levels in parental 32Dcl3 cells (lane 1), untreated 32D-BCR/ABL cells (lane 6); or cells treated with imatinib (lane 7) or with the MEK1 inhibitor PD098059 (lane 8); or cells transduced with the empty retrovirus (lane 2) or with wild-type and mutant HNRPK (lanes 3-5) or with MAPK ERK1 (lanes 9-10) and ERK2 (lanes 11-12) cDNAs. All cell lines were cultured in the presence of IL-3. (B) Northern blots show Myc mRNA levels in the indicated cell lines either maintained in IL-3 or IL-3 deprived for 12 hours in the presence or absence of imatinib. (C) Autoradiography and anti-MYC Western blot show levels of 35S-labeled newly synthesized and total immunoprecipitated Myc protein in parental 32Dcl3, untreated, and imatinib-treated 32D-BCR/ABL and S284/353A expressing 32D-BCR/ABL cells. (D) Levels of Myc protein, expressed as arbitrary densitometric units after normalization with GRB2 levels, in cycloheximide-treated parental 32Dcl3, 32D-BCR/ABL, and its derivative cell lines expressing wild-type or mutant HNRPK proteins. Half-life was calculated using the algorithm t½ = 0.693tn/ln(C0/Cn), where n represents the last time point. (E) Histograms show levels of MYC mRNA (red bars) and protein (blue bars) after normalization with GAPDH mRNA and GRB2 protein levels, respectively. Values in samples 2 to 7 represent arbitrary densitometric units expressed as percentage of those of sample 1. Western blot and RT-PCR were performed with lysate and total RNA, respectively, of CD34+marrow cells from healthy individuals (lanes 1-2) and from CML-CP (lanes 3-4), CML-AP (lane 5), and CML-BC (lanes 6-7) patients.
Here we reported that in BCR/ABL-expressing myeloid and lymphoid progenitor cells and in CML-BCCD34+ patient cells, BCR/ABL kinase activity induces HNRPK expression by enhancing Hnrpk gene transcription and mRNA stability. Enhanced expression of this hnRNP is not restricted to CML-BC, as HNRPK overexpression has also been associated with tumor progression and resistance to therapeutic drug-induced apoptosis in different types of cancer.34-37
By assessing the mechanism whereby BCR/ABL augments HNRPK levels, we found that Hnrpk mRNA expression depends on the BCR/ABL-regulated activity of MAPKERK1/2. In fact, BCR/ABL-graded expression activates MAPKERK1/2 and increases HNRPK levels in a dose-dependent manner. Consistent with the role of HNRPK as a target of MAPK activity, we showed that activation of MAPKERK1/2 is readily detectable in CML-BCCD34+ but not in CML-CPCD34+ or NBMCD34+ primary cells, and that HNRPK knock-down inhibits growth factor-independent proliferation, colony formation, and tumorigenesis of BCR/ABL-expressing myeloid progenitor 32Dcl3 cells. The link between BCR/ABL, MAPKERK1/2 activation, and increased HNRPK expression is also supported by the evidence showing that BCR/ABL constitutively enhances proliferation of hematopoietic cells by circumventing cytokine-generated signals leading to MAPK activation, and that MAPK inhibition impairs CML progenitor growth.28,29,38,39 Moreover, HNRPK down-regulation resulted in reduced levels of Myc, which is not only essential for BCR/ABL transformation and proliferation of CML progenitors27,30 but is also transcriptionally and translationally induced by HNRPK.16,23 Notably, growth factors (eg, EGF) that elicit mitogenic signals by potentiating MAPKERK1/2 activity40 also enhance HNRPK, which, in turn, increases MYC expression.34
In BCR/ABL-transformed cells, HNRPK transcriptional activating function seems to be dispensable for BCR/ABL oncogenic potential, as the nuclear localized C299-HNRPK mutant, which binds DNA but lacks transactivation activity,15 affected neither Myc gene transcription nor the in vitro and in vivo growth of BCR/ABL-expressing myeloid cells. Accordingly, we found HNRPK prevalently localized in the cytoplasm of 32D-BCR/ABL cells. Although in other cell types, HNRPK, though capable of nucleocytoplasmic shuttling, is localized primarily within the nucleus,41 it also has been reported that sustained MAPKERK1/2 activity drives cytoplasmic accumulation of HNRPK in HeLa cells.21 Indeed, inhibition of MEK1 and short-term treatment of BCR/ABL cells with imatinib turned off BCR/ABL-dependent MAPKERK1/2 activity and induced HNRPK nuclear accumulation. Thus, HNRPK is not only transcriptionally but also posttranslationally regulated by BCR/ABL through the activity of MAPKERK1/2. Since MAPK activation is important for BCR/ABL leukemogenic potential,28,42,43 it was reasonable to speculate that in BCR/ABL-expressing cells, like in HeLa cells,21 HNRPK translation-regulatory activity depends on phosphorylation of HNRPK on serines 284 and 353 by the BCR/ABL-activated MAPKERK1/2, and that MAPKERK1/2-dependent HNRPK translation regulation may be important for BCR/ABL leukemogenesis. Indeed, in BCR/ABL-transformed myeloid progenitor 32Dcl3 cells, expression of the nuclear-localized nonphosphatable S284/353A-HNRPK mutant recapitulated the effects of HNRPK down-regulation. In fact, marked inhibition of cytokine-independent proliferation and colony formation (∼ 60% decrease) and suppression of in vivo BCR/ABL leukemogenic potential were the characteristics of a 32D-BCR/ABL cell line and primary CD34+ CML-BC cells transduced with the S284/353A-HNRPK mutant. Since this mutant still binds mRNAs containing a DICE element in their 3′UTR21 or an IRES in the 5′UTR (ie, MYC;Figure 6), it is plausible that its dominantnegative activity may derive from its ability to sequester mRNAs in the nucleus and, therefore, decrease the HNRPK-dependent nuclear export of the mRNA cargo available for translation. Indeed, we provided evidence showing that either wild-type or the nucleuslocalized S284/353A mutant HNRPK binds Myc mRNA and that levels of an IRES-containing reporter gene were enhanced in myeloid progenitor cells by BCR/ABL expression. Note that, in reporter assays, the presence of 2 repeats of the 15-LOX-DICE element21 at the 3′-end of dsRED also altered the reporter gene expression in 32D-BCR/ABL cells (not shown). Consistent with our results, it has been shown that S284/353A-HNRPK expression in HeLa cells overrides HNRPK translation silencing of a DICE-bearing luciferase reporter construct.21 Although we cannot exclude the possibility that the detrimental effects of S284/353A-HNRPK expression on the in vitro and in vivo leukemic potential of BCR/ABL cells might, in part, depend on interference with the DICE-dependent HNRPK translation regulatory activity, our data suggest that inhibition of ERK-dependent phosphorylation of HNRPK on serines 284/353 impairs HNRPK-dependent translation regulation of IRES-bearing mRNAs (ie, MYC) in BCR/ABL-transformed cells. In fact, we showed that restoration of MYC expression is sufficient to rescue factor-independent colony formation and tumorigenic potential of 32D-BCR/ABL and primary CD34+ CML-BC cells from the inhibitory effects of S284/353A-HNRPK expression.
Steady-state and newly synthesized Myc protein levels were suppressed by S284/353A-HNRPK expression and inhibition of BCR/ABL or MAPKERK1/2 kinase activity. Mechanistically, it appears that BCR/ABL positively regulates MYC translation but not transcription by inducing ERK1/2-dependent phosphorylation of HNRPK. Further, expression of S284/353A-HNRPK mutant did not affect Myc protein stability. Consistent with the existence of a BCR/ABL-MAPK-HNRPK network positively regulating MYC mRNA translation in the advanced phase of CML, MYC protein but not mRNA expression was higher in the CD34+ fraction of CML-BC and CML-AP marrow cells than in the CD34+ fraction of normal and CML-CP patient marrow cells. These findings, which are consistent with the reported MYC overexpression found in CML-BC and with the requirement of MYC for BCR/ABL leukemogenesis,27,30,44-47 are only apparently in contrast with the reported ability of BCR/ABL to enhance MYC expression at transcriptional level,48,49 as most of the reported experiments showing BCR/ABL-dependent transcriptional induction of MYC mRNA have been performed either in growth factor-independent cell lines maintained in the absence of cytokines or by comparing total marrow cells from CML-CP and CML-BC patients.50,51 In these experiments, treatment of growth factor-independent cell lines (eg, K562) with imatinib results in proliferation arrest and apoptosis, 2 conditions in which MYC levels rapidly decline. Similarly, the differences in MYC mRNA levels detected in total CML-CP and CMP-BC marrow cells may reflect differences in the percentage of Ph1 and postmitotic cells within the CML samples. Indeed, increased MYC mRNA was not found in microarrays that used CD34+ or AC133+ fraction of CML-CP and CML-BC marrow cells.52 Thus, one of the molecular mechanisms whereby BCR/ABL enhances MYC expression involves the MAPK-dependent regulation of HNRPK translation regulatory activity. However, increased MYC transcription can still be found in those CML-BC patients with amplification of the MYC gene,53-55 and we also cannot exclude the possibility that transcriptional, translational, and posttranslational mechanisms such as those involving the activity of Jak2 kinase49 may all participate in the regulation of MYC expression in primary CML-BC cells.
Prepublished online as Blood First Edition Paper, November 17, 2005; DOI 10.1182/blood-2005-09-3732.
Supported by the National Cancer Institute CA095512 (D.P.), and CA16058 and GRT8230100 (OSU-CCC); the United States Army, Chronic Myelogenous Leukemia (CML) Research Program DAMD17-03-1-0184 (D.P.); The Elsa Pardee Foundation for Cancer Research (D.P.); The Lauri Strauss Leukemia Research Foundation (D.P.); and Reserche en Sante du Quebec (D.C.R.).
M.N. performed research and wrote the paper; P.N. performed research; R.S. performed research; B.W.B. performed research; J.-S.C. performed research; A.G. performed research; A.E.W. contributed vital material; D.C.R. contributed vital material; M.A.C. contributed vital material and analyzed data; G.M. analyzed data; and D.P. designed research, analyzed data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs C. D. Bloomfield and S. P. Whitman for scientific discussion, Ms A. Carter for editorial assistance, and Novartis Oncology for providing imatinib mesylate.
The authors do not have financial interests related to this work that need to be disclosed.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal