Abstract
Classical von Willebrand disease (VWD) type 2A, the most common qualitative defect of VWD, is caused by loss of high-molecular-weight multimers (HMWMs) of von Willebrand factor (VWF). Underlying mutations cluster in the A2 domain of VWF around its cleavage site for ADAMTS13. We investigated the impact of mutations commonly found in patients with VWD type 2A on ADAMTS13-dependent proteolysis of VWF. We used recombinant human ADAMTS13 (rhuADAMTS13) to digest recombinant full-length VWF and a VWF fragment spanning the VWF A1 through A3 domains, harboring 13 different VWD type 2A mutations (C1272S, G1505E, G1505R, S1506L, M1528V, R1569del, R1597W, V1607D, G1609R, I1628T, G1629E, G1631D, and E1638K). With the exception of G1505E and I1628T, all mutations in the VWF A2 domain increased specific proteolysis of VWF independent of the expression level. Proteolytic susceptibility of mutant VWF in vitro closely correlated with the in vivo phenotype in patients. The results imply that increased VWF susceptibility for ADAMTS13 is a constitutive property of classical VWD type 2A, thus explaining the pronounced proteolytic fragments and loss of HMWM seen in multimer analysis in patients.
Introduction
Von Willebrand factor (VWF) is a complex multimeric glycoprotein synthesized by megakaryocytes and endothelial cells. It consists of multiple copies of the 2050-residue mature VWF protein. The VWF subunits are linked through disulfide bridges that are localized both at their C-terminal and N-terminal regions. Polymerization of VWF generates protein complexes of different sizes, referred to as VWF multimers, displaying molecular weights between 500 kDa and 20 MDa (for a review, see Ruggeri and Ware1 ). After secretion into plasma, VWF is cleaved by the specific VWF cleaving protease ADAMTS13, a liver-synthesized metalloprotease that belongs to the disintegrin and metalloproteinase with thrombospondin motif family of proteases.2-5 ADAMTS13 cleaves VWF at a specific cleavage site in the A2 domain between residues Y1605 and M1606.3,6 As a consequence of the complex processing, plasma VWF displays a mixture of intact multimers and proteolytic fragments of different sizes, which is reflected in typical multimer patterns of plasma VWF seen after SDS gel electrophoresis.7
The molecular size of VWF is critical for its physiologic function as a mediator of platelet adhesion and aggregation in primary hemostasis. It is the high-molecular-weight multimers (HMWMs) that are biologically most active and that are essential to maintain hemostasis under conditions of high shear stress in the microvasculature.8 Therefore, defects in synthesis or processing of VWF can cause severe coagulation disorders. The persistence of ultralarge multimers of VWF due to hereditary or acquired deficiency of ADAMTS13 gives rise to thrombotic thombocytopenic purpura (TTP),9,10 a life-threatening microangiopathic disease with extensive formation of clots consisting of VWF and platelets11 (for a review, see Moake12 ).
The loss of HMWMs of VWF results in hemorrhagic diathesis, as seen in classical von Willebrand disease (VWD) type 2A, the most common qualitative defect of VWD.13 There is accumulated evidence that the loss of HMWMs in classical VWD type 2A is related to ADAMTS13-dependent proteolysis of VWF. VWF multimer analysis, as a diagnostic tool for VWD, demonstrated that cleavage of VWF is increased in VWD type 2A (IIA) and 2B (IIB), resulting in prominent subbands in the triplet structure of individual VWF oligomers.7,14 Dent et al demonstrated that these subbands represent proteolytic fragments originating from specific cleavage of VWF at the Y1605-M1606 bond.6 The majority of mutations in patients with classical VWD type 2A cluster in the A2 domain of VWF close to the cleavage site,15 and expression studies suggest that some mutations make VWF more susceptible to ADAMTS13-dependent proteolysis (group 2), while others impair secretion of HMWMs (group 1).16 However, the authors of the latter study did not directly examine the role of VWF proteolysis.16 Although the nature of the VWF-cleaving protease as a metalloproteinase had been recognized earlier,9,10 its molecular characterization as ADAMTS13 has been reported just recently.2-5 Therefore, earlier attempts to investigate VWF cleavage in VWD type 2A have been restricted to the use of less defined plasma fractions17 and have not examined VWD type 2A systematically.18
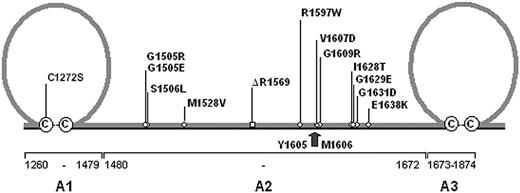
Distribution of VWD type 2A mutations. A schematic representation of the A1 through A3 domain of von Willebrand factor (VWF) is shown. The A1 and A3 domains are characterized by a loop structure of 187 amino acids formed by disulfide bonds between C1272 and C1458 (A1 domain) and C1686 and C1872 (A3 domain), respectively. With the exception of C1272S all VWD type 2A mutations are located within the A2 domain of VWF. The arrow marks the proteolytic cleavage site for ADAMTS13 between residues Y1605 and M1606. The cysteines involved in loop-structure formation are shown in circles.
Distribution of VWD type 2A mutations. A schematic representation of the A1 through A3 domain of von Willebrand factor (VWF) is shown. The A1 and A3 domains are characterized by a loop structure of 187 amino acids formed by disulfide bonds between C1272 and C1458 (A1 domain) and C1686 and C1872 (A3 domain), respectively. With the exception of C1272S all VWD type 2A mutations are located within the A2 domain of VWF. The arrow marks the proteolytic cleavage site for ADAMTS13 between residues Y1605 and M1606. The cysteines involved in loop-structure formation are shown in circles.
Stressing the clinical relevance of the interaction between these molecules, recent reports suggest that increased ADAMTS13-dependent proteolysis of VWF may also contribute to the loss of VWF in a subset of patients with VWD type 1.19,20 Interestingly, the increased susceptibility has been correlated with a polymorphism that is located in the VWF A2 domain (eg, Y/C1584).19 These findings indicate that increased susceptibility of VWF for ADAMTS13-dependent proteolysis could represent an important pathomechanism in different types of VWD. The VWF A2 domain accommodating the proteolytic cleavage site may have an impact on the regulated interaction between VWF and ADAMTS13. We used recombinant human ADAMTS13 (rhuADAMTS13) as a tool to pinpoint how mutations in the VWF A2 domain influence ADAMTS13-dependent proteolysis of VWF.
Materials and methods
Mutations
We investigated 13 different VWD type 2A mutations comprising 7 previously described mutations (G1505E, G1505R, S1506L, R1597W, V1607D, G1609R, and I1628T) and 6 novel mutations (C1272S, M1528V, R1569del, G1629E, G1631D, and E1638K) (Figure 1). The novel mutations and 4 previously described mutations derived from a mutation screening that we had performed in 45 nonrelated patients with VWD type 2A. All patients were heterozygous for the particular mutations and did not show any other mutations in the screening by gene sequencing. We did not find any of the mutations in 100 normal alleles of the VWF gene. With the exception of the patient with the deletion R1569del, who lacked pronounced subbands in multimer analysis, all patients presented with classical VWD type 2A (subtype IIA).7,21 Three additional mutations (G1505E, G1505R, and V1607D) have been included in our study becausee they represent mutations commonly described in the database of VWF mutations.22 C1272S is the only mutation located in the A1 domain, while all others are located in the A2 domain of VWF. All patients and/or their parents were informed about the nature of the study and gave their consent.
Full-length VWF
In general, VWF expression studies were performed as described before.23 In brief, the expression vector pRc.CMV containing full-length mutant and wild-type (WT) VWF cDNA (designated pRc.CMV-VWF), respectively, was used to transform Top10 supercompetent cells (Invitrogen, Karlsruhe, Germany). After purification of the plasmids, 10 μg mutant or WT pRc.CMV-VWF was used to transiently transfect 293 EBNA cells (2 × 106) by liposomal transfer. The cells were grown for 72 hours (24 hours in Dulbecco modified Eagle medium [DMEM; Invitrogen] with 10% [vol/vol] fetal bovine serum [FBS] and 48 hours in serum-free Iscove modified Dulbecco medium [IMDM; Sigma, St Louis, MO] plus 2% [vol/vol] Ultroser G [Invitrogen]). VWF product was secreted in the medium and concentrated in Centricon tubes to one-tenth of the original volume prior to digestion by rhuADAMTS13 or multimer analysis that was performed by SDS agarose gel electrophoresis24 with luminescent visualization.25 The luminescent blot was stored on electronic media by means of photo imaging (FluorChem 8000; Alpha Innotech, San Leandro, CA). To analyze intracellular VWF the transfected cells were lysed by 3 rounds of freezing (–80°C) and thawing in lysis buffer (0.1 M Tris/HCl [pH 8.0] and 0.6% [vol/vol] Triton X-100). The same methods were used for cotransfection of supercompetent 293 EBNA cells with WT and mutant pRc.CMV-VWF (5 μg of each).
VWF A1-A2-A3 domain fragments
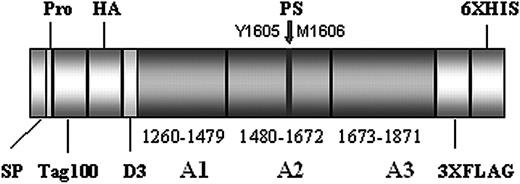
To obtain VWF fragments that correspond to the A1 through A3 domain (VWFA1-A2-A3), we designed a novel expression vector containing the coding sequence from amino acid V1229 through L1871 of VWF plus an N-terminal Tag100 and HA-sequence and a C-terminal (3 ×) FLAG and (6 ×) HIS sequence (Figure 2). Consecutively, the cDNA of the signal peptide and the first 3 amino acids of the propeptide of VWF and an oligonucleotide with the Tag100 and HA-sequence were introduced into the pIRES neo2 vector (BD-Clontech, Heidelberg, Germany) using the NheI and BamHI restriction site followed by cloning of another oligonucleotide with the (3 ×) FLAG and (6 ×) HIS sequence using an EcoRI restriction site. The resulting vector was designated pIRES neo2 T2. Subsequently, VWF cDNA spanning nucleotides 3685 through 5613 (corresponding to amino acids V1229 through L1871) was amplified by polymerase chain reaction (PCR) using pRc.CMV-VWF as a template. The primers were designed to contain restriction sites (shown underlined) for BamHI (sense-primer) and EcoRI (antisense-primer), respectively (sense primer 5′-TATGGATCCGTTGTCAACCTCACCTGTGAAGCCTGCCAGG-3′; antisense primer 5′-TATGAATTCCAGTTTGTGGAGGAAGGAATTGCCCAAGGT). These restriction sites were used for cloning of the VWFA1-A2-A3 sequence into the pIRES neo2 T2 vector. The resulting plasmid was designated pIRES neo2 T2 A1-A2-A3WT and was used for mutagenesis by means of a commercial kit (QuickchangeKit; Stratagene, Amsterdam, the Netherlands) according to the manufacturer's instructions. All cloned sequences were verified by automated sequencing (ABI Prism 310; ABI, Foster City, CA). Transfection of 293 EBNA cells and further tissue culture and protein sampling were performed as described. Due to low expression levels of mutant VWF, conditioned media of C1272S-, R1569del-, and V1607D-transfected cells had to be further concentrated by a second centrifugation in Centricon tubes. All transfection experiments were done in triplicate.
rhuADAMTS13
ADAMTS13 cDNA was obtained from normal liver tissue at the margin of a benign human liver tumor (haemangioendothelioma) by reverse transcriptase (RT)–PCR. Full-length ADAMTS13 cDNA was cloned into the expression vector pIRES neo2 for stable transfection of HEK 293 cells by liposomal transfer under selective pressure by G418, 48 hours after transfection. Wild-type rhuADAMTS13 secreted in the medium was used for subsequent proteolysis assays. The activity of rhuADAMTS13 in the medium was about 400% compared with a normal human plasma pool.
Outline of the pIRES neo 2 A1-A2-A3 WT construct. The protein contains the A1 through A3 domain of von Willebrand factor (VWF) plus a short segment of the D3 domain (residues V1229 through L1871). The VWF fragment is enveloped by a N-terminal Tag100 and HA sequence and a C-terminal (3 ×) FLAG and (6 ×) HIS sequence, respectively. The proteolytic cleavage site (PS) for ADAMTS13 is located in the A2 domain between residues Y1605 and M1606 (marked by an arrow). SP indicates signal peptide; pro, first 3 residues of the VWF propeptide.
Outline of the pIRES neo 2 A1-A2-A3 WT construct. The protein contains the A1 through A3 domain of von Willebrand factor (VWF) plus a short segment of the D3 domain (residues V1229 through L1871). The VWF fragment is enveloped by a N-terminal Tag100 and HA sequence and a C-terminal (3 ×) FLAG and (6 ×) HIS sequence, respectively. The proteolytic cleavage site (PS) for ADAMTS13 is located in the A2 domain between residues Y1605 and M1606 (marked by an arrow). SP indicates signal peptide; pro, first 3 residues of the VWF propeptide.
ADAMTS13 assays
The ADAMTS13-dependent proteolysis of VWF was tested essentially as described by Furlan et al.9 Our assay was free of plasma because we used both recombinant VWF and recombinant human ADAMTS13. rhuADAMTS13 was adjusted to 0.05 U/mL with Tris/HCl buffer (5 mM, pH 8.0). All experiments were carried out by adding 100 μL of this solution to 200 μL conditioned media containing rhuVWF (80 U/dL). After incubation with 10 mM barium chloride final concentration the aliquots were dialyzed against buffer solution (1.5 M urea, 5 mM Tris-HCL [pH 8.0]) and incubated at 37°C before the reaction was stopped by adding EDTA to a final concentration of 10 mM. Full-length rhuVWF was digested for 5 hours and further analyzed by multimer analysis as described. To assess susceptibility to ADAMTS13-dependent proteolysis, we visually compared the multimer patterns of mutant and WT rhuVWF digested under the same conditions using photoimaging (FluorChem 8000). The VWFA1-A2-A3 domain fragments were digested overnight followed by Western blotting on a 4% to 12% SDS-PAGE gradient gel (NuPage; Invitrogen) and immunostaining using anti-HA and anti-FLAG–M2 antibodies (Sigma-Aldrich, Steinheim, Germany). The results obtained from different mutants by photo imaging were visually compared with the result from VWFA1-A2-A3-WT. To test cleavage under nondenaturing conditions, additional assays were performed in the absence of urea (5 mM Tris-HCL [pH 8.0]).
A quantitative analysis of proteolysis focused on 3 representative mutations: the group 1 mutation S1506L, and the group 2 mutations G1629E and E1638K. The latter differ in their cleavage pattern in the VWFA1-A2-A3 assay. G1629E is proteolyzed even in the absence of urea while E1638K is proteolyzed only in the presence of urea. Digestion of full-length mutant VWF and VWF-WT was performed as described using serial dilutions of rhuADAMTS13 (1:10, 1:20, 1:40, 1:80, and 1:160 dilutions of rhuADAMTS13 that had been adjusted to an activity of 0.5 U/mL in 5 mM Tris/HCl buffer [pH 8.0] containing 1.5 M urea). A quantitative readout for the proteolytic degradation was achieved by measuring the decrease of ristocetin induced binding of digested VWF to immobilized recombinant glycoprotein Ib (GpIb) according to a published method26 with the following modifications: we added a FLAG tag to the carboxyterminal of the GpIb fragment and used an anti-FLAG antibody instead of an anti-GpIb antibody to capture the GpIb fragment to the microtiter plate. For this assay we used a 1:5 dilution of digested VWF in phosphate-buffered saline (PBS) buffer, added ristocetin to a final concentration of 1 mg/mL and incubated 100 μL reaction mix on GpIb fragment–coated microtiter plates for 1 hour at room temperature. After washing, the bound VWF was detected by a horseradish peroxidase (HRP)–labeled polyclonal rabbit anti-VWF antibody as described.26 The protease sensitivity of the VWF mutants was assessed by comparing their binding curves with that of VWF-WT.
For quantitative evaluation of VWFA1-A2-A3 proteolytic susceptibility, which was carried out in duplicate, we assessed the concentrations of unproteolyzed VWFA1-A2-A3 and the FLAG-tagged proteolytic fragment by photoimaging. The ratios between the areas under the curve of the undigested fragment and the digestion product, respectively, were taken as measures for proteolysis.
Results
Expression studies
Full-length rhuVWF-WT and rhuVWF harboring the mutations G1505E, M1528V, R1597W, G1609R, I1628T, G1629E, G1631D, or E1638K, respectively, were similar in expression level and in multimer distribution (Figure 3; Table 1). In contrast, introduction of the mutations C1272S, G1505R, S1506L, R1569del, and V1607D resulted in decreased expression of high- and intermediate-molecular-weight multimers of VWF, consistent with a group 1 mechanism. Coexpression of the WT allele of VWF to mimic the heterozygous condition seen in patients attenuated the impaired expression (Table 1). When expressed in the VWFA1-A2-A3 constructs, 3 of the latter mutations (C1272S, R1569del, and V1607D) strongly impaired expression of VWF. To date, only in the case of C1272S were we able to obtain sufficient quantities of VWF for further experiments by using a second round of medium concentration.
Data from expression studies
. | . | . | . | . | . | . | Proteolysis full-length rhuVWF‡ . | . | Proteolysis rhuVWFA1-A2-A3 . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Group* . | Cell-lysate VWF/Ag, % WT† . | SD, % . | Medium VWF/Ag, % WT† . | SD, % . | Ratio medium: cell-lysate . | + Urea . | - Urea . | + Urea . | - Urea . | ||
| WT | 100.0 | 1.0 | 100.0 | 1.2 | 1.00 | Cleavage | Cleavage | Yes | No | |||
| C1272S§ | 1 | 98.1 | 3.5 | 40.9 | 4.1 | 0.42 | - | ++ | No | No | ||
| C1272S + WT§ | 1 | 82.7 | 1.1 | 71.0 | 1.5 | 0.86 | - | + | NA | NA | ||
| G1505E | 2 | 53.4 | 3.2 | 72.6 | 1.5 | 1.36 | = | = | Yes | Yes | ||
| G1505E + WT | 2 | 87.5 | 2.8 | 89.7 | 2.9 | 1.02 | - | - | NA | NA | ||
| G1505R | 1 | 58.8 | 3.6 | 8.2 | 2.1 | 0.14 | ++ | ++ | Yes | Yes | ||
| G1505R + WT | 1 | 74.8 | 2.5 | 50.6 | 1.7 | 0.67 | - | + | NA | NA | ||
| S1506L | 1 | 20.9 | 1.7 | 1.3 | 0.5 | 0.06 | ++ | ++ | Yes | No | ||
| S1506L + WT | 1 | 68.1 | 0.2 | 40.3 | 1.6 | 0.59 | = | + | NA | NA | ||
| M1528V§ | 2 | 75.9 | 3.8 | 86.3 | 1.3 | 1.14 | + | ++ | Yes | No | ||
| M1528V + WT§ | 2 | 86.7 | 3.0 | 77.2 | 1.8 | 0.89 | - | + | NA | NA | ||
| Del R1569§ | 1 | 27.1 | 1.3 | 0.9 | 0.02 | 0.03 | ++ | ++ | NA | NA | ||
| Del R1569 + WT§ | 1 | 67.5 | 1.1 | 46.8 | 1.6 | 0.69 | - | - | NA | NA | ||
| R1597W | 2 | 94.0 | 4.2 | 81.7 | 5.0 | 0.87 | ++ | ++ | Yes | No | ||
| R1597W + WT | 2 | 68.6 | 3.4 | 80.3 | 1.2 | 1.17 | + | + | NA | NA | ||
| V1607D | 1 | 47.0 | 2.4 | 0.1 | 0.0 | 0.00 | ++ | ++ | NA | NA | ||
| V1607D + WT | 1 | 66.8 | 1.8 | 35.7 | 2.0 | 0.53 | - | + | NA | NA | ||
| G1609R | 2 | 30.0 | 0.9 | 84.4 | 2.2 | 2.81 | ++ | ++ | Yes | Yes | ||
| G1609R + WT | 2 | 70.7 | 1.7 | 118.0 | 1.2 | 1.67 | + | + | NA | NA | ||
| I1628T | 2 | 103.3 | 4.5 | 87.5 | 2.1 | 0.85 | (-) | - | Yes | Yes | ||
| I1628T + WT | 2 | 95.0 | 2.7 | 89.0 | 1.8 | 0.94 | = | - | NA | NA | ||
| G1629E§ | 2 | 43.2 | 2.7 | 78.5 | 3.0 | 1.82 | ++ | +++ | Yes | Yes | ||
| G1629E + WT§ | 2 | 56.7 | 3.0 | 90.9 | 0.3 | 1.60 | + | ++ | NA | NA | ||
| G1631D§ | 2 | 60.4 | 3.4 | 63.7 | 1.2 | 1.06 | ++ | ++ | Yes | Yes | ||
| G1631D + WT§ | 2 | 72.1 | 1.6 | 66.7 | 2.1 | 0.93 | = | + | NA | NA | ||
| E1638K | 2 | 77.6 | 0.9 | 79.1 | 2.1 | 1.02 | ++ | ++ | Yes | No | ||
| E1638K + WT | 2 | 87.5 | 2.5 | 84.6 | 3.0 | 0.97 | + | + | NA | NA | ||
. | . | . | . | . | . | . | Proteolysis full-length rhuVWF‡ . | . | Proteolysis rhuVWFA1-A2-A3 . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation . | Group* . | Cell-lysate VWF/Ag, % WT† . | SD, % . | Medium VWF/Ag, % WT† . | SD, % . | Ratio medium: cell-lysate . | + Urea . | - Urea . | + Urea . | - Urea . | ||
| WT | 100.0 | 1.0 | 100.0 | 1.2 | 1.00 | Cleavage | Cleavage | Yes | No | |||
| C1272S§ | 1 | 98.1 | 3.5 | 40.9 | 4.1 | 0.42 | - | ++ | No | No | ||
| C1272S + WT§ | 1 | 82.7 | 1.1 | 71.0 | 1.5 | 0.86 | - | + | NA | NA | ||
| G1505E | 2 | 53.4 | 3.2 | 72.6 | 1.5 | 1.36 | = | = | Yes | Yes | ||
| G1505E + WT | 2 | 87.5 | 2.8 | 89.7 | 2.9 | 1.02 | - | - | NA | NA | ||
| G1505R | 1 | 58.8 | 3.6 | 8.2 | 2.1 | 0.14 | ++ | ++ | Yes | Yes | ||
| G1505R + WT | 1 | 74.8 | 2.5 | 50.6 | 1.7 | 0.67 | - | + | NA | NA | ||
| S1506L | 1 | 20.9 | 1.7 | 1.3 | 0.5 | 0.06 | ++ | ++ | Yes | No | ||
| S1506L + WT | 1 | 68.1 | 0.2 | 40.3 | 1.6 | 0.59 | = | + | NA | NA | ||
| M1528V§ | 2 | 75.9 | 3.8 | 86.3 | 1.3 | 1.14 | + | ++ | Yes | No | ||
| M1528V + WT§ | 2 | 86.7 | 3.0 | 77.2 | 1.8 | 0.89 | - | + | NA | NA | ||
| Del R1569§ | 1 | 27.1 | 1.3 | 0.9 | 0.02 | 0.03 | ++ | ++ | NA | NA | ||
| Del R1569 + WT§ | 1 | 67.5 | 1.1 | 46.8 | 1.6 | 0.69 | - | - | NA | NA | ||
| R1597W | 2 | 94.0 | 4.2 | 81.7 | 5.0 | 0.87 | ++ | ++ | Yes | No | ||
| R1597W + WT | 2 | 68.6 | 3.4 | 80.3 | 1.2 | 1.17 | + | + | NA | NA | ||
| V1607D | 1 | 47.0 | 2.4 | 0.1 | 0.0 | 0.00 | ++ | ++ | NA | NA | ||
| V1607D + WT | 1 | 66.8 | 1.8 | 35.7 | 2.0 | 0.53 | - | + | NA | NA | ||
| G1609R | 2 | 30.0 | 0.9 | 84.4 | 2.2 | 2.81 | ++ | ++ | Yes | Yes | ||
| G1609R + WT | 2 | 70.7 | 1.7 | 118.0 | 1.2 | 1.67 | + | + | NA | NA | ||
| I1628T | 2 | 103.3 | 4.5 | 87.5 | 2.1 | 0.85 | (-) | - | Yes | Yes | ||
| I1628T + WT | 2 | 95.0 | 2.7 | 89.0 | 1.8 | 0.94 | = | - | NA | NA | ||
| G1629E§ | 2 | 43.2 | 2.7 | 78.5 | 3.0 | 1.82 | ++ | +++ | Yes | Yes | ||
| G1629E + WT§ | 2 | 56.7 | 3.0 | 90.9 | 0.3 | 1.60 | + | ++ | NA | NA | ||
| G1631D§ | 2 | 60.4 | 3.4 | 63.7 | 1.2 | 1.06 | ++ | ++ | Yes | Yes | ||
| G1631D + WT§ | 2 | 72.1 | 1.6 | 66.7 | 2.1 | 0.93 | = | + | NA | NA | ||
| E1638K | 2 | 77.6 | 0.9 | 79.1 | 2.1 | 1.02 | ++ | ++ | Yes | No | ||
| E1638K + WT | 2 | 87.5 | 2.5 | 84.6 | 3.0 | 0.97 | + | + | NA | NA | ||
SD indicates standard deviation; NA, not applicable; +, increased proteolysis compared with rhuVWF-WT; ++, strongly increased proteolysis compared with rhuVWF-WT; +++, very strongly increased proteolysis compared with rhuVWF-WT; -, reduced proteolysis compared with rhuVWF-WT; (-), moderately reduced proteolysis compared with rhuVWF-WT; =, proteolysis equal to rhuVWF-WT.
Group classification according to Lyons et al16 (1 indicates decreased expression; 2, increased cleavage).
Expression experiments were done in triplicate; data represent mean values.
Cleavage of recombinant human VWF (rhuVWF) mutants was assessed in comparison with rhuVWF-WT digested under the same conditions.
Novel mutations.
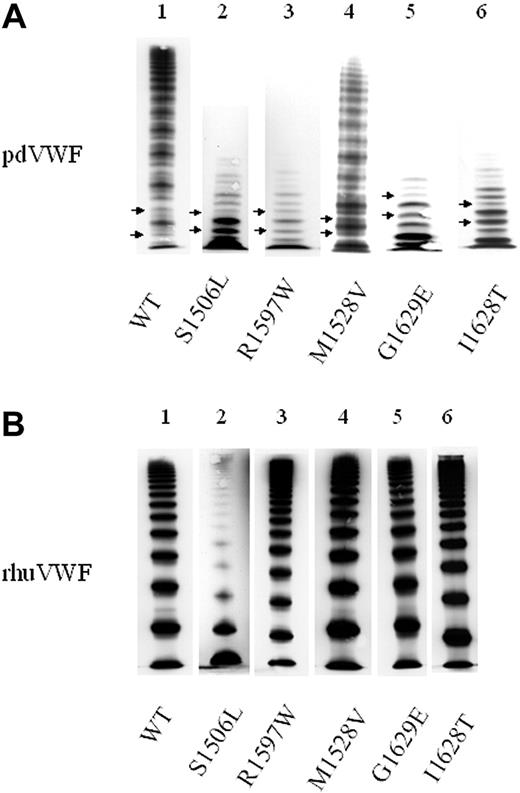
Typical VWF multimer patterns in classical VWD type 2A. VWF multimers were separated by SDS gel electrophoresis and visualized by immunostaining. (A) Plasma-derived VWF (pdVWF). Lane 1 shows wild-type VWF (VWF-WT); lane 2, VWF of a patient with a group 1 mutation (S1506L); and lanes 3-6, VWF of patients with group 2 mutations (R1597W, M1528V, G1629E, and I1628T). Compared with VWF-WT, patient samples lack HMWMs and show pronounced subbands of VWF (marked by arrows). (B) “Homozygous” rhuVWF. Lane 1 shows rhuVWF-WT; lane 2, group 1 mutant rhuVWF1506L lacking high- and intermediate-molecular-weight multimers of VWF; and lanes 3-6, group 2 mutants rhuVWF1597W, rhuVWF1528V, rhuVWF1629E, and rhuVWF1628T with multimer patterns indistinguishable from rhuVWF-WT. Note that in the recombinant VWF proteins there is no triplet pattern and no subbands, respectively, due to the absence of ADAMTS13 in the expression system. Patient samples were collected over several years and analyzed on different gels. Images were adjusted for size and brightness (Microsoft PowerPoint 2000; Redmond, WA).
Typical VWF multimer patterns in classical VWD type 2A. VWF multimers were separated by SDS gel electrophoresis and visualized by immunostaining. (A) Plasma-derived VWF (pdVWF). Lane 1 shows wild-type VWF (VWF-WT); lane 2, VWF of a patient with a group 1 mutation (S1506L); and lanes 3-6, VWF of patients with group 2 mutations (R1597W, M1528V, G1629E, and I1628T). Compared with VWF-WT, patient samples lack HMWMs and show pronounced subbands of VWF (marked by arrows). (B) “Homozygous” rhuVWF. Lane 1 shows rhuVWF-WT; lane 2, group 1 mutant rhuVWF1506L lacking high- and intermediate-molecular-weight multimers of VWF; and lanes 3-6, group 2 mutants rhuVWF1597W, rhuVWF1528V, rhuVWF1629E, and rhuVWF1628T with multimer patterns indistinguishable from rhuVWF-WT. Note that in the recombinant VWF proteins there is no triplet pattern and no subbands, respectively, due to the absence of ADAMTS13 in the expression system. Patient samples were collected over several years and analyzed on different gels. Images were adjusted for size and brightness (Microsoft PowerPoint 2000; Redmond, WA).
Cleavage of full-length VWF by rhuADAMTS13
Full-length rhuVWF is a complex molecule when used in ADAMTS13 assays. However, it permits a direct comparison with proteolytic patterns of VWF multimers obtained from plasma. Due to the lack of proteolytic activity and shear stress in the culture medium, rhuVWF is fully multimerized and lacks the proteolytically generated triplet structure seen in plasma. Figure 4 illustrates the specific proteolytic pattern and loss of HMWMs of full-length rhuVWF after incubation with rhuADAMTS13.
In all experiments, proteolytic degradation of mutant rhuVWF under denaturing and nondenaturing conditions was assessed in comparison to rhuVWF-WT (Table 1; Figure 5). In the “homozygous” expression experiment, 10 mutations (G1505R, S1506L, M1528V, R1569del, R1597W, V1607D, G1609R, G1629E, G1631D, and E1638K) caused increased proteolysis compared with rhuVWF-WT in both the presence and absence of urea. Coexpression of the wild-type allele attenuated the proteolysis-permissive phenotype (Table 1; Figure 5). The mutations G1629E and R1597W resulted in a very pronounced loss of HMWMs (Figure 5). Three mutations (C1272S, G1505E, and I1628T) failed to promote ADAMTS13-dependent proteolysis of VWF in general, although C1272S increased VWF cleavage under nondenaturing conditions.
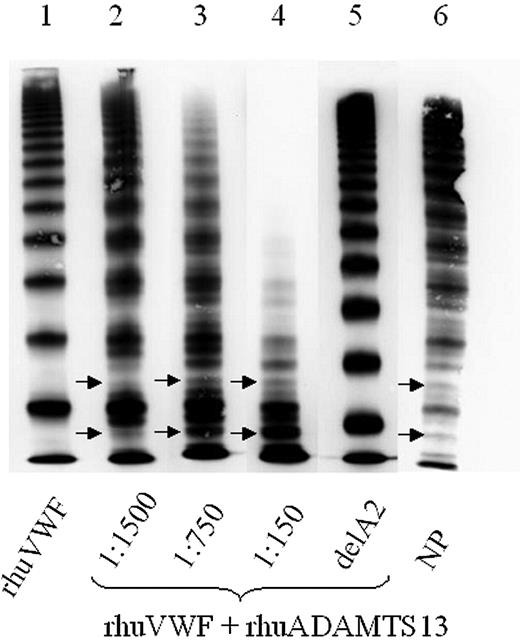
ADAMTS13 assay using wild-type full-length von Willebrand factor (VWF). Wild-type rhuVWF-WT was digested by increasing concentrations of rhuADAMTS13. Although the undigested control (lane 1) lacks a triplet structure of the individual oligomers, these are specifically generated by ADAMTS13 digestion in a dose-dependent manner, paralleled by loss of HMWMs (lanes 2-4). Arrows mark the characteristic outer subbands of VWF. Deletion of the A2 domain of VWF, which contains the specific ADAMTS13 cleavage site, abrogates VWF proteolysis by ADAMTS13 (lane 5). NP indicates normal plasma pool (lane 6).
ADAMTS13 assay using wild-type full-length von Willebrand factor (VWF). Wild-type rhuVWF-WT was digested by increasing concentrations of rhuADAMTS13. Although the undigested control (lane 1) lacks a triplet structure of the individual oligomers, these are specifically generated by ADAMTS13 digestion in a dose-dependent manner, paralleled by loss of HMWMs (lanes 2-4). Arrows mark the characteristic outer subbands of VWF. Deletion of the A2 domain of VWF, which contains the specific ADAMTS13 cleavage site, abrogates VWF proteolysis by ADAMTS13 (lane 5). NP indicates normal plasma pool (lane 6).
The quantitative analysis of proteolytic susceptibility of 3 representative mutants by GpIb binding enabled us to determine a 4- and 5.6-fold increase in ADAMTS13 suceptibility in the group 2 mutations G1629E and G1638K, respectively. However, for the group 1 mutation S1506L, the GpIb-binding assay did not allow quantification of proteolytic susceptibility. The lack of VWF HMWMs due to impaired secretion of the aberrant translation product markedly reduced GpIb binding of rhuVWF1506L even before digestion with ADAMTS13. The level of binding was below the dynamic range of this assay, thereby disqualifying the test as a valid method for this particular mutant.
Cleavage of VWFA1-A2-A3 by rhuADAMTS13
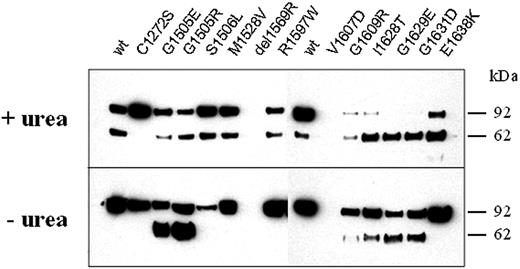
The recombinant VWFA1-A2-A3 fragment used in this study (rhuVWFA1-A2-A3) provides a simple readout for the specific action of ADAMTS13 on VWF. Two fragments are generated by specific proteolysis and can be independently visualized by Western blotting and tag-specific antibodies (Figure 6). Using denaturing conditions, wild-type rhuVWFA1-A2-A3 (rhuVWFA1-A2-A3-WT) and all rhuVWFA1-A2-A3 constructs with mutations in the A2 domain were cleaved by rhuADAMTS13 (Figure 6). Under nondenaturing conditions, 6 mutant rhuVWFA1-A2-A3 constructs (G1505E, G1505R, G1609R, I1628T, G1629E, and G1631D) were cleaved by rhuADAMTS13, but not the rhuVWFA1-A2-A3-WT construct (Figure 6). This suggests that these mutations make VWF more susceptible to ADAMTS13-dependent proteolysis. Two mutations (G1629E and G1631D) promoted complete cleavage of the VWFA1-A2-A3 fragment. Five mutations did not allow cleavage under nondenaturing conditions (C1272S, S1506L, M1528V, R1597W, and E1638K; Figure 6). The mutation C1272S resulted in resistance of the recombinant fragment to ADAMTS13-dependent proteolysis, even in the presence of urea. Due to a very low expression level, 2 mutants (R1569del and V1607D) were not used for this test.
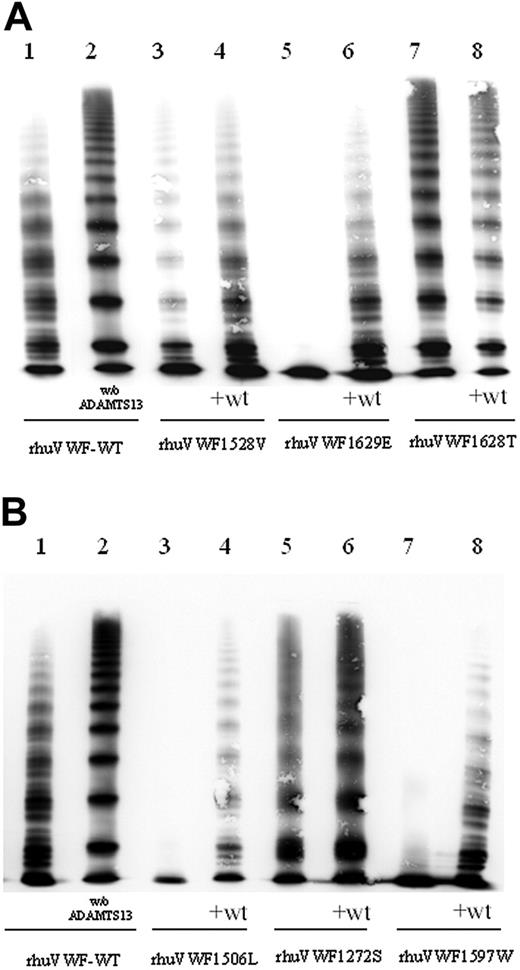
ADAMTS13 assay using mutant full-length VWF. rhuVWF-WT and mutant VWF were digested by ADAMTS13 and subject to multimer analysis. The specific susceptibility to ADAMTS13-dependent proteolysis can be assessed by comparing the loss of HMWMs of VWF and the appearance of the proteolytic subbands between wild-type and mutant VWF. (A) Compared with rhuVWF-WT (lane 1) there is a moderately increased (rhuVWF1528V, lanes 3 and 4), highly increased (rhuVWF1629E, lanes 5 and 6), or reduced (rhuVWF1628T, lanes 7 and 8) degradation of mutant VWF. Increased cleavage is less prominent in the coexpression experiments (+wt, lanes 4, 6, and 8). (B) G1506L represents a group 1 mutation and has an increased susceptibility for ADAMTS13 (lanes 3 and 4); C1272 causes almost no change in proteolytic degradation (lanes 5 and 6); and R1597W is a classical group 2 mutation and exhibits increased susceptibility for ADAMTS13-dependent proteolysis (lanes 7 and 8). w/o ADAMTS13 indicates the undigested control of rhuVWF-WT (lane 2 of panels A and B).
ADAMTS13 assay using mutant full-length VWF. rhuVWF-WT and mutant VWF were digested by ADAMTS13 and subject to multimer analysis. The specific susceptibility to ADAMTS13-dependent proteolysis can be assessed by comparing the loss of HMWMs of VWF and the appearance of the proteolytic subbands between wild-type and mutant VWF. (A) Compared with rhuVWF-WT (lane 1) there is a moderately increased (rhuVWF1528V, lanes 3 and 4), highly increased (rhuVWF1629E, lanes 5 and 6), or reduced (rhuVWF1628T, lanes 7 and 8) degradation of mutant VWF. Increased cleavage is less prominent in the coexpression experiments (+wt, lanes 4, 6, and 8). (B) G1506L represents a group 1 mutation and has an increased susceptibility for ADAMTS13 (lanes 3 and 4); C1272 causes almost no change in proteolytic degradation (lanes 5 and 6); and R1597W is a classical group 2 mutation and exhibits increased susceptibility for ADAMTS13-dependent proteolysis (lanes 7 and 8). w/o ADAMTS13 indicates the undigested control of rhuVWF-WT (lane 2 of panels A and B).
Digestion of the A1-A2-A3 domain fragments of VWF (rhuVWFA1-A2-A3) by rhuADAMTS13. WT and mutant VWF fragments were incubated with rhuADAMTS13 either under denaturing (+urea; top gel) or nondenaturing (– urea; bottom gel) conditions and visualized by SDS-PAGE and immunostaining (anti-HA antibody; numbers on the right side refer to a molecular weight marker). Although rhuVWFA1-A2-A3-WT is cleaved under denaturing conditions only, rhuVWFA1-A2-A3 harboring either the mutation G1505E, G1505R, G1609R, I1628T, G1629E, or G1631D is cleaved also under nondenaturing conditions as indicated by the appearance of smaller fragments.
Digestion of the A1-A2-A3 domain fragments of VWF (rhuVWFA1-A2-A3) by rhuADAMTS13. WT and mutant VWF fragments were incubated with rhuADAMTS13 either under denaturing (+urea; top gel) or nondenaturing (– urea; bottom gel) conditions and visualized by SDS-PAGE and immunostaining (anti-HA antibody; numbers on the right side refer to a molecular weight marker). Although rhuVWFA1-A2-A3-WT is cleaved under denaturing conditions only, rhuVWFA1-A2-A3 harboring either the mutation G1505E, G1505R, G1609R, I1628T, G1629E, or G1631D is cleaved also under nondenaturing conditions as indicated by the appearance of smaller fragments.
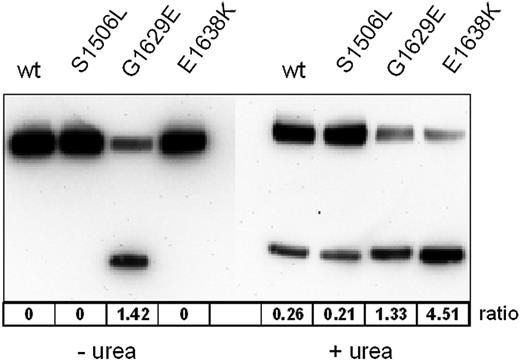
Quantification of the protease sensitivity of VWF mutants was assessed by measuring the ratios between the areas under the curve of the undigested fragment and the digestion product, respectively. The most sensitive mutant in this assay was E1638K, which showed a ratio of 4.51 between the digested and the undigested fragment. Although this ratio was lower for G1629E (1.33), it was cleaved to a similar extent (ratio, 1.42) even in the absence of urea (Figure 7).
Discussion
Using recombinant VWF and ADAMTS13 for the cleavage assay allowed us to focus directly on the interactions between these 2 molecules. This approach guarantees that the assay is not biased by proteolyzed plasma VWF. In other publications, ADAMTS13 activity is usually measured by means of HMWM loss alone.9,10 However, specificity can only be deduced from the appearance of the typical triplet structure (Figures 3, 4). The use of unproteolyzed rhuVWF allowed us to monitor not only the loss of large multimers, but also the appearance of the characteristic proteolytic subbands (Figure 5).
With this assay we could distinguish between mutants with different levels of ADAMTS13 susceptibility. While the majority of VWD type 2A mutations enhanced VWF cleavage to a similar degree, M1528V resulted only in a minor increase, whereas G1629E and R1597W promoted proteolysis very strongly. The in vitro results correlated with the phenotype in the affected patients in vivo. Compared to the average proteolysis signs seen in VWD type 2A,7 the patient with the mutation G1629E showed very prominent VWF subbands, suggestive of a markedly increased proteolysis of VWF in vivo (Figure 3). Two patients with the mutation M1528V displayed only slightly pronounced subbands of VWF, suggestive of less prominent VWF cleavage in vivo (Figure 3). The differences in susceptibility, as displayed by G1629E and M1528V, suggest that cleavage of VWF by ADAMTS13 is probably not controlled by an on/off mechanism but can be modulated by the conformation of the A2 domain.
Quantitative analysis of ADAMTS13-mediated proteolysis of representative mutant and WT VWFA1-A2-A3 fragments. Ratios of the FLAG-tagged proteolytic fragment to the uncleaved substrate calculated from the areas under the curve were taken as proteolytic susceptibility.
Quantitative analysis of ADAMTS13-mediated proteolysis of representative mutant and WT VWFA1-A2-A3 fragments. Ratios of the FLAG-tagged proteolytic fragment to the uncleaved substrate calculated from the areas under the curve were taken as proteolytic susceptibility.
Pronounced VWF subbands were not seen in plasma of the patient with the deletion of R1569. Interestingly, homozygous expression of R1569del in full-length rhuVWF leads to increased ADAMTS13 susceptibility and pronounced subbands. In contrast, coexpression of the wild-type allele with R1569del, which reflects the heterozygote situation in patients, abrogates increased proteolysis completely. Since rhuVWFR1569del is expressed at a very low level, this suggests a strong underrepresentation of the mutant monomer in secreted VWF. Therefore, VWF cleavage appears not to be increased and no pronounced subbands can be detected in plasma. R1569del might represent the only mutation investigated here that takes effect by an exclusive group 1 mechanism in the affected patients. Our results underline both the importance of increased VWF proteolysis for a classical VWD type 2A phenotype and the correlation between the occurrence of VWF subbands and the susceptibility of VWF to ADAMTS13-dependent proteolysis.
Recently, it has been reported that a polymorphism in the A2 domain (eg,Y/C1584) is causing increased VWF susceptibility for ADAMTS13-dependent proteolysis.19 Interestingly, in some cases this is correlated with VWD type 1, a partial quantitative deficiency of VWF that displays a normal multimer distribution. It is controversial why the affected patients present a VWD type 1 rather than a VWD type 2A phenotype.27 It has to be emphasized that even the “mildest” VWD type 2A group 2 mutation investigated in this study (eg, M1528V) correlates with loss of VWF HMWMs and pronounced VWF subbands. The Y/C1584 polymorphism could result in only slightly increased susceptibility to ADAMTS13-dependent proteolysis. In part, this might be due to underrepresentation of the aberrant molecule, which is the likely reason why patients with R1569del also lack pronounced subbands. Bowen estimated that VWF Y/C1584 is 18- to 30-fold less susceptible to ADAMTS13-dependent proteolysis than VWF in classical VWD type 2A patients.27 Therefore, the effects of proteolysis (loss of HMWMs, pronounced subbands) are less prominent or are being compensated for by constitutive synthesis of new VWF and elimination of fragments from the plasma. Either way, multimer analysis might not be sensitive enough to resolve aberrations in this range.
Surprisingly, 2 mutations (G1505E and I1628T, respectively), when introduced into full-length VWF, did not increase ADAMTS13-dependent proteolysis of VWF in vitro. Although these findings are in contrast to the patient data, we have no doubt that G1505E and I1628T are causing VWD type 2A, since we did not find them in 100 normal alleles of the VWF gene and mutation screening in the affected patients did not reveal any other mutations of the VWF gene. We hypothesize that the discrepancy between in vitro and in vivo data are due to our test conditions. In vivo, shear stress is crucial for the cleavage of VWF by ADAMTS13.28 In vitro conditions, however, might result in a different exposure of the VWF cleavage site to ADAMTS13. This notion is further underlined by our observation that urea is not sufficient to promote increased proteolysis of other full-length mutants when coexpressed with WT (G1505R, S1506L, M1528V, V1607D, and G1631D). Due to the lack of a crystallographic model of the VWF under these distinct conditions, the molecular basis of this phenomenon remains unclear. At least under nondenaturing conditions it is unlikely a matter of sensitivity, since the “mildest” mutation in our study M1528V did show increased proteolysis in the full-length VWF assay.
To further elucidate to role of the mutations in the A2 domain, we aimed to reduce test complexity and generated tag-labeled VWF monomers containing the A1 through A3 domains. This enabled analysis of the impact of VWD type 2A mutations on cleavage of a single Y1605-M1606 bond. ADAMTS13-dependent proteolysis of rhuVWFA1-A2-A3 results in the appearance of 2 characteristic proteolytic fragments (Figure 6). Under denaturing conditions, rhuVWFA1-A2-A3 is a good substrate of rhuADAMTS13, whether it contains the WT sequence or mutations in the VWF A2 domain (Figure 6). Discriminative power arises from this assay when it is performed under nondenaturing conditions, which restricts cleavage to VWF mutants with significantly increased susceptibility to ADAMTS13. Since 6 mutants are cleaved under nondenaturing conditions, but 5 are not, this assay seems less sensitive than the assay using full-length VWF. At least in part, this might be due to the configuration of the A3 domain that lacks an intact loop between C1686 and C1872 since the fragment ends at L1871. According to a recent publication, assigning an inhibitory effect of the A1 domain on ADAMTS13 mediated proteolysis,29 using a VWFA2-A3 fragment as substrate could provide higher sensitivity.
Visually detected differences of mutants in susceptibility to proteolysis were quantified for 3 representative mutations: the group 1 mutation S1506L, the group 2 mutation G1629E that is proteolyzed even in the absence of urea, and the group 2 mutation E1638K that is proteolyzed only in the presence of urea. Whereas significant differences could be shown between the group 2 mutations and VWF-WT and between the 2 group 2 mutations themselves, proteolysis of the S1506L VWFA1-A2-A3 fragment was not different from VWF-WT. These deviations of the applied in vitro tests from the observed S1506L associated in vivo phenotype with enhanced proteolysis remain unclear.
In conclusion, we investigated a representative spectrum of commonly described and novel mutations causing classical VWD type 2A.22 At least in 1 of the applied assays, each of the mutants increased VWF cleavage by ADAMTS13, including G1505R, S1506L, and V1607D, previously classified as group 1 mutations.16 Although we could confirm a decrease in biosynthesis/secretion of VWF in these cases, secreted VWF was nevertheless more susceptible to proteolysis. Therefore, increased cleavage of mutant VWF by ADAMTS13 seems to be a more general property of VWD type 2A (IIA). These data explain the pronounced satellite bands seen in multimer analysis in patients of both groups.14
Our results broaden our knowledge of VWD type 2A pathogenesis. Based on a proposed 3-dimensional structural model of the VWF A2 domain, a recent study suggests that group 1 mutations cause severe structural changes of VWF, thereby promoting intracellular retention. In contrast, changes caused by group 2 mutations are minor and do not impair processing of VWF.30 Our results suggest that most of the VWD type 2A mutations result in increased cleavage of VWF in plasma, independent of the expression level. This effect could result from 2 different mechanisms. First, the mutations could cause a more unspecific conformational change within the A2 domain, resulting in increased exposure of the VWF cleavage site to ADAMTS13. Second, they could enhance specific binding of ADAMTS13 to VWF, thereby causing a gain of cleavage function.
The methods described here can be useful to further investigate the specific interaction between VWF and ADAMTS13. Targeting ADAMTS13-dependent proteolysis of VWF might be a potential therapeutic approach in VWD.17,31
Prepublished online as Blood First Edition Paper, December 1, 2005; DOI 10.1182/blood-2005-04-1758.
Supported in part by research funding from the Deutsche Forschungsgemeinschaft to R.S. (DFG grants Schn 325/4-1, Schn 325/4-2).
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Deirdre O'Sullivan and Zaverio Ruggeri for careful reading of the manuscript and helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal