Abstract
Hematopoietic stem cells (HSCs) display extensive heterogeneity in their behavior even when isolated as phenotypically homogeneous populations. It is not clear whether this heterogeneity reflects inherently diverse subsets of HSCs or a homogeneous population of HSCs diversified by their response to different external stimuli. To address this, we analyzed 97 individual HSCs in long-term transplantation assays. HSC clones were obtained from unseparated bone marrow (BM) through limiting dilution approaches. Following transplantation into individual hosts, donor-type cells in blood were measured bimonthly and the resulting repopulation kinetics were grouped according to overall shape. Only 16 types of repopulation kinetics were found among the HSC clones even though combinatorially 54 groups were possible. All HSC clones, regardless of their origin, could be assigned to this subset of groups, and the probability of finding new patterns is negligible. Thus, the full repertoire of repopulating HSCs was covered. These data indicate that the HSC compartment consists of a limited number of distinct HSC subsets, each with predictable behavior. Enrichment of HSCs (Lin–Rho–SP) changes the representation of HSC types by selecting for distinct subsets of HSCs. These data from the steady-state HSC repertoire could provide a basis for the diagnosis of perturbed patterns of HSCs potentially caused by disease or aging.
Introduction
Hematopoietic stem cells (HSCs), like all stem cells, are defined by their undifferentiated state and their self-renewal capacity. Although HSCs have been extensively studied, the mechanisms that control these HSC functions are not well understood. The slow pace of progress in this area is due to a number of experimental limitations. These include the low numbers of HSCs that can be accessed, the lack of methods for their long-term propagation in vitro, and the functional and phenotypic heterogeneity of HSCs. In particular, the molecular basis of the intriguing functional heterogeneity of HSCs has been difficult to investigate.
In vivo, in vitro, and in silico studies have revealed extensive heterogeneity in the phenotypes and behaviors of HSCs.1-9 HSCs can differ in self-renewal capacity, clone size (number of differentiated progeny per HSC), differentiation capacity (contribution to the hematopoietic lineages), migration patterns, and primitiveness.4,6,9-16 These HSC qualities can be assessed in repopulation assays, by following over time the appearance of mature progeny derived from an engrafted HSC (repopulation kinetics). For example, primitive HSCs are characterized by a slow onset of repopulation leading to sustained production of mature cells.9,17 Thus, repopulation patterns are highly informative for HSC function.
Nevertheless, it is unclear just how heterogeneous the HSC compartment is and the basis of this heterogeneity has remained speculative. According to one model, heterogeneity could be explained as the behavior of a population of homogeneous HSCs where individual HSCs respond to extrinsic stimuli but the response is reversible.18-23 Such flexibility should lead to a continuum of HSC function in the HSC compartment. Similarly, a continuum of HSC functions should arise if stochastic mechanisms control HSC behavior.20-23 Inherent in the idea of an HSC continuum is the idea that every HSC behavior possible should actually be observable. Support for reversible HSC behaviors derives from studies where HSCs exposed to cytokines transiently lose the ability to engraft when the cells proceed through the active phases of the cell cycle.24 Similarly, reversible expression of the cell surface antigen CD34 on HSCs has been documented.25
Yet, there is increasing evidence that HSCs can be more predictable and less flexible than previously appreciated. Many HSC behaviors are fixed intrinsically through genetic or epigenetic mechanisms.4,15,26-31 Genetic differences in HSC behavior were documented when different inbred strains of mice were shown to have different frequencies of HSCs in their BM.32,33 Similarly, genetic manipulation of HSCs, for example through overexpression of homeobox genes, can profoundly affect their behavior.34,35 Epigenetic effects cause heterogeneity in genetically identical cells. A striking example of epigenetically fixed heterogeneity among HSCs is found in myeloid-biased HSCs. These HSCs make typical levels of myeloid cells but generate too few lymphocytes. This is caused by epigenetic changes on the level of the myeloid-biased HSC, which predetermines its lymphoid progeny to have a blunted response to IL-7.4,13 Similar to the ability to differentiate, the overall repopulation patterns can be fixed on the level of each HSC. For example, daughter HSCs (from a clonally derived HSC) behaved remarkably similar to each other in the kinetics of repopulation, their ability to contribute to the different hematopoietic lineages, and their life spans.4,13 Overall, these data support the idea that the HSC compartment is composed of functionally distinct subpopulations of HSCs. This model makes the prediction that HSC behaviors should be predictable, that is, the behaviors of an HSC should cluster in discrete (not continuous) patterns.
To begin to distinguish between these models, we analyzed the in vivo repopulation behavior of 97 individual clonally derived HSCs. The results show that all repopulation kinetics can be grouped into a limited number of families, suggesting that these families represent discrete subpopulations of HSCs with predictable behaviors. The major and novel conclusion is that the HSC compartment consists of a limited number of clonotypes each with predictable behavior. We conclude that the heterogeneity in the HSC compartment is predetermined and not due to the random manifestations of a single type of HSC. The complete description of the HSC repertoire is likely to be valuable as a benchmark for defining the clonal consequences of treatment and disease.
Materials and methods
Generation of clonally repopulated mice
Adult mouse bone marrow (BM) HSCs were isolated by 3 different methods: (1) In vivo limiting dilution (LD) transplants of BM cells: 2000 to 5000 BM cells containing limiting (0.2-0.5) numbers of HSCs36,37 were injected directly into recipients. Mice were considered to be clonally repopulated only if 40% or less of the injected hosts in a group showed repopulation. (2) Transplants of HSCs following in vitro LD: BM cells were seeded onto preformed layers of the stromal line S1738 for 4 weeks in limiting dilution conditions exactly as described.4,39 Conditions were chosen so that less than 20% of the wells contained a colony of small granulocytic cells after 4 weeks of culture. This leads to a conditional probability of 90% or more that each positive well (identified as containing a colony of small granulocytic cells) is initiated by a single cell. Individual positive wells were harvested by vigorous pipetting and injected intravenously into individual mice.4,39 (3) Purified HSCs: Single-lineage Rhodamine-123–Hoechst 33342– side population cells (Lin–Rho–SP) cells isolated from adult mouse BM were injected directly into individual recipients. The approach, evaluation, and results from the single HSC studies were described in detail by Uchida et al.7 For all LD experiments, B6 mice, congenic for either CD45 or GFP, served as donors. Hosts were sublethally irradiated W41/W41 (500 rads [5 Gy]) or lethally irradiated (2 doses of 550 rads [5.5 Gy], 2 hours apart) B6 mice.4,7,39 Whenever B6 hosts were used, a genetically distinguishable source of radioprotecting cells was coinjected as described.39 There were no differences in HSC function between B6 and W41/W41 hosts, and data were pooled from both systems. All mice were bled in regular intervals and the fraction of donor-type cells in the lymphoid and myeloid lineages was measured by flow cytometry as described.4,7,13,39 Briefly, purified white blood cells from each mouse at each time point were split into 3 tubes and cells in each tube were either stained for the CD45.1 donor-type marker or GFP expression was used as donor-type marker. Thus, each time point had 3 measurements of donor-type cells, and the resulting error of measurement is used in the symbolization of the kinetics. At each time point, cells were also stained with Thy-1 to detect T cells, B220 for B cells, or Mac-1 and Gr-1 for myeloid cells. Lin–Rho–SP repopulated mice were analyzed as described.7 Mice were considered to be repopulated if their blood contained 3% or more donor-type cells at least at any one time point and if donor-type T and B lymphocytes and macrophages and granulocytes were found at the same time.39
Classification of clone types and statistics
Based on the level of donor-type cells measured over time in blood, individual HSC clones were classified according to the shape of the repopulation curve as described previously.4,40 Briefly, donor-type cell data were plotted as a function of time. Individual slopes of each of the repopulation curves were then symbolized, where an increase in repopulation is denoted by “+,” a decrease by “–,” and a flat segment by “π.” A segment was called flat if the difference between the adjacent values of the 2 points was equal to or smaller than the standard error of the measurements of corresponding donor-type cells.4,40 The relative Hamming distance41 for all kinetics was then pair-wise calculated, and kinetics with a Hamming distance of 0 were sorted into groups. Therefore, each group contains only kinetics with identical shapes. As described previously in detail,4,40 the Hamming distance measures the number of mismatches in the comparison of 2 aligned strings of equal length formed over a fixed alphabet of symbols. More formally, assigning the value m(x, y) = 0 to a match (x = y) and the value m(x, y) = 1 to a mismatch (x ≠ y), the Hamming distance of 2 strings A = a1 a2...aN, B = b1 b2...bN is given as the sum over m(ai, bi) for 1 ≤ i ≤ N. Hence, with n = 3 and the alphabet (+, –), the sequence “++ +” has Hamming distance 0 to itself, but Hamming distance 1 to the sequence “+–+.”
To approximate the output of differentiated cells per HSC clone during the first 7 months after transplantation, we used the area under the curve (AUC). The AUC values of all clones examined followed a roughly biphasic distribution. Accordingly, AUCs were classified as low (≤ 200; range, 14-196) or high (> 200; range, 227-513).
We used an add-constant estimator to determine the probability of new kinetics occurring after a number of kinetics has already been observed.42 The probability of a seen kinetic x is Pseen (x) = (c(x) + λ)/(N + λX), where λ is the estimator constant, c(x) is the number of times that the kinetic x is observed, N is the total number of events already seen, and X denotes the size of the corpus of all events. Hence, the probability that the next event is an as-yet-unseen kinetic is Punseen = 1 – Pseen. We chose λ= 1/X.
Results
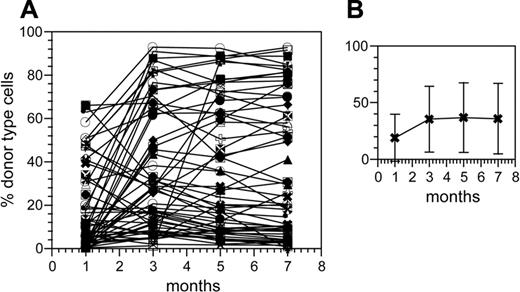
Previously, we described a highly efficient method for isolating repopulating HSCs on the clonal level. For this, BM cells were allowed to repopulate stromal cell–supported cultures in limiting dilution. After 4 to 5 weeks, individual cultures were injected into individual ablated congenic hosts.4,39 Donor-type cells in blood were measured bimonthly by flow cytometry, and the mice were followed for at least 7 months following transplantation. As previously described, a high frequency of the mice showed donor-type cells in blood, indicating that these mice had received an HSC. Here, we used this approach to generate a large panel of clonally repopulated hosts. Fifty-seven clones were followed for at least 7 months. The kinetics of repopulation of all of these 57 HSC clones are depicted in Figure 1A. The data emphasize the extensive fluctuations and heterogeneity expected when individual HSC clones are monitored over time. There appears to be no underlying order or patterns in this set of kinetics.
The average of these clonal kinetics shows a curve that reaches a plateau early and then stays flat (Figure 1B). This is reminiscent of the kinetics of repopulation obtained when mice receive transplants of high numbers of HSCs.43 This supports the interpretation that population-based analyses of HSCs mask the behavior of individual HSC clones. At the same time, the data (Figure 1B) indicate that the behavior of populations of HSCs reflects a reasonable superposition of the behavior of the individual clones.
Individual repopulating kinetics of clonally derived HSCs. Shown are the percent donor-type cells in blood at the indicated time points after the clones received injections. (A) Fifty-seven clonal HSCs isolated after in vitro LD. All clones (repopulated mice) obtained by this approach in our lab that yielded at least 7 months of data are shown (a few clones have been described previously4,13 ). (B) The mean repopulation curve (± SD) of all clonal curves. The arithmetic mean of the data in Figure 1A is shown for each time point.
Individual repopulating kinetics of clonally derived HSCs. Shown are the percent donor-type cells in blood at the indicated time points after the clones received injections. (A) Fifty-seven clonal HSCs isolated after in vitro LD. All clones (repopulated mice) obtained by this approach in our lab that yielded at least 7 months of data are shown (a few clones have been described previously4,13 ). (B) The mean repopulation curve (± SD) of all clonal curves. The arithmetic mean of the data in Figure 1A is shown for each time point.
Families of repopulation kinetics
A visual inspection of the clonal repopulation kinetics (Figure 1A) revealed no obvious patterns. Previously, we described a powerful method to quantify similarities based on the shape of kinetics.4,40 For this approach, individual segments of each repopulation curve (Figure 1) were symbolized. A positive slope is assigned a “+,” a flat segment a “π,” and “–” indicates a negative slope. The resulting symbolized sequences are then pair-wise compared and their Hamming distance was calculated. The Hamming distance is a quantitative classifier of similarities among short time-series—in this case HSC repopulation kinetics. Kinetics that have the same sequence of symbols have a Hamming distance of 0 and are considered to be identical.
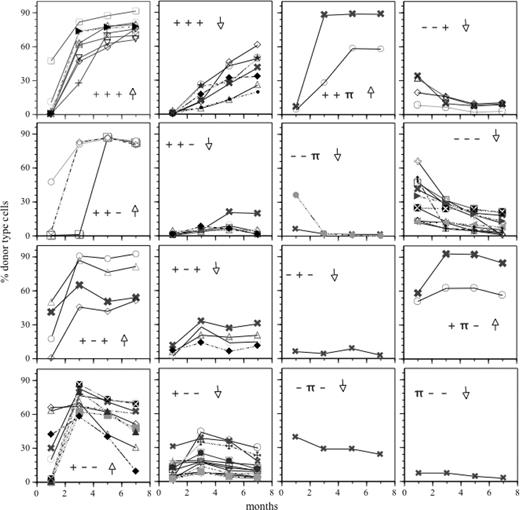
This approach was applied to the repopulation kinetics shown in Figure 1, and HSC kinetics that were identical by these criteria were sorted into groups. Figure 2 depicts the groups of repopulation kinetics identified by this approach. The symbolized shapes of the kinetics are indicated for each group. It is clear that the approach identifies discrete families of kinetics (Figure 2).
The repertoire of clonal HSC repopulation kinetics. Each graph depicts a different family of repopulation kinetics, identified after comparison of symbolized kinetics by Hamming distance. The symbolized slopes for each group of kinetics are indicated in each panel, where the slope of each segment is expressed as positive (+), negative (–), or flat (π). Groups were further subdivided according to whether their AUC value was high (↑) or low (↓). Solid lines indicate data from the same 57 mice shown in Figure 1A. The dashed lines portray similarly analyzed data from 27 HSC clones followed in mice that received injections of freshly isolated BM cells (in vivo LD).
The repertoire of clonal HSC repopulation kinetics. Each graph depicts a different family of repopulation kinetics, identified after comparison of symbolized kinetics by Hamming distance. The symbolized slopes for each group of kinetics are indicated in each panel, where the slope of each segment is expressed as positive (+), negative (–), or flat (π). Groups were further subdivided according to whether their AUC value was high (↑) or low (↓). Solid lines indicate data from the same 57 mice shown in Figure 1A. The dashed lines portray similarly analyzed data from 27 HSC clones followed in mice that received injections of freshly isolated BM cells (in vivo LD).
Because each repopulation kinetic consists of 3 segments or slopes, combinatorially, this approach allows for 27 possible types of kinetics. Yet, only 12 kinetics (ie, about 40%) were identified among the HSC clones tested (Figure 3A). As an additional parameter, we classified clones as large or small based on their AUC value. The AUC was used as a rough measure of the number of differentiated progeny produced from each HSC clone over the 7 months of follow-up. The slopes together with the AUC generated a quadruple of meta data. In this case, the total number of possible families is 54. Strikingly, the kinetics observed did not show all possible behaviors—rather only 16 discrete families of kinetics (29%) were seen (Figure 2).
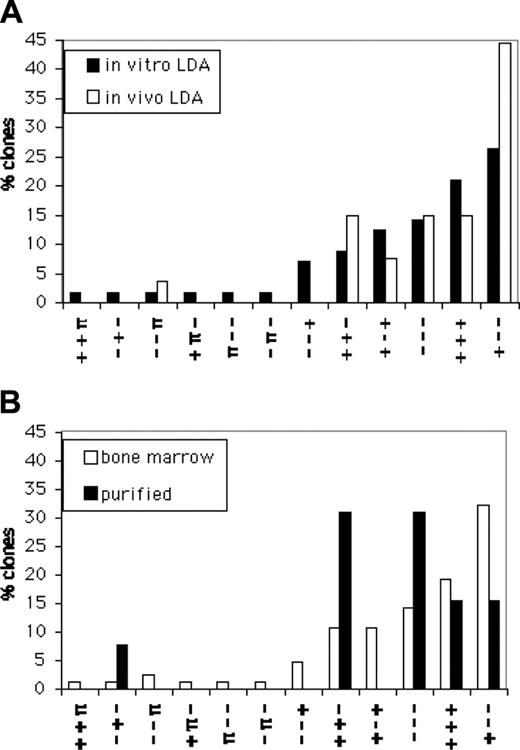
If this classification of the kinetics is a meaningful description of HSC behavior, then the results should be independent of the way the HSCs are isolated. To test this, we generated HSC clones by injecting limiting numbers of freshly explanted BM cells directly into ablated hosts.36 We analyzed 27 HSC clones, obtained from 4 independent in vivo LD experiments. All of these clones (Figure 2, dashed lines) readily fell into the existing families. The distributions of the kinetics of HSC clones from the in vivo and the in vitro LD approaches (Figure 3A) showed a high degree of correlation (r2 = 0.80; P < .001). Thus, the 2 approaches yielded similar results, indicating that both methods identified the same types of HSC. The HSC clones from the in vivo and in vitro LD were combined to create a set of 84 unmanipulated HSC clones (Figure 2).
To estimate the probability that examination of additional clones would reveal new groups, we used the add-constant estimator. This determines the probability of new events occurring after a number of events has already been observed.42 For both classifications (with and without the AUC parameter), the probability of finding new kinetics is approximately 1% (P < .001). In other words, on average one new group would be expected if an additional 100 sets of 84 clones each were analyzed. The negligible probability of finding new groups indicates that the analysis covered the complete repertoire of repopulation behaviors of HSCs in adult B6 mice.
Distribution of HSC groups from different sources. The percent of clones of each type for each origin of HSC studied is shown. The kinetic families as defined in Figure 2 are shown on the horizontal axis. Here, the AUC subclassification was not used because of the low number of clones derived from in vivo LD (n = 27) and from purified HSCs (n = 13). Vertically, each bar represents the percent of kinetics that fall within the indicated family. (A) Comparison of HSC groups derived after in vitro and in vivo LD. (B) Comparison of groups derived from unpurified HSCs (data for in vivo and in vitro LD combined) and purified Lin–Rho–SP HSCs.
Distribution of HSC groups from different sources. The percent of clones of each type for each origin of HSC studied is shown. The kinetic families as defined in Figure 2 are shown on the horizontal axis. Here, the AUC subclassification was not used because of the low number of clones derived from in vivo LD (n = 27) and from purified HSCs (n = 13). Vertically, each bar represents the percent of kinetics that fall within the indicated family. (A) Comparison of HSC groups derived after in vitro and in vivo LD. (B) Comparison of groups derived from unpurified HSCs (data for in vivo and in vitro LD combined) and purified Lin–Rho–SP HSCs.
Purified HSCs
Since we wished to include all types of HSCs found in BM, we had so far avoided a phenotypic definition of HSCs. Next, we examined repopulation data from mice that received transplants of single freshly isolated, but purified, Lin–Rho–SP cells. Single cells were injected into individual mice and mice were followed at a different center.7 All of the 13 clones available for analysis fell into groups that had already been seen (Figure 2), and no new clonal patterns were identified (Figure 3B). Thus, data from 2 different labs and 3 different methods of purification generated HSCs that fell into the same categories. However, the population of Lin–Rho–SP cells contained fewer families of kinetics, and the distribution of the remaining families was significantly skewed when compared with those found in unseparated BM (r2 = 0.31, P = .05). For example, Lin–Rho–SP cells were enriched for HSCs with – – – and + + – kinetics (Figure 3B). Since the Lin–Rho–SP cells were directly injected into the hosts, we also compared their kinetics with the subset of 27 HSC clones isolated following in vivo LD. Again, there is a noticeable lack of correlation (r2 = 0.08, P = .58). Overall, the data suggest that the Lin–Rho–SP purification protocol selectively enriches some subsets of all the HSCs found in BM.
Predicting self-renewal capacity
Our analysis focused on the kinetics of repopulation for the first 7 to 8 months after transplantation. Does the classification of HSC clones during this time have predictive value for their long-term behavior? To test this, we followed the self-renewal capacity of the HSC clones in serial transplantation. The data are summarized in Table 1. The analysis of the + + + groups is perhaps the most complete: All 10 HSC clones examined from these groups were capable of self-renewal and generated daughter HSCs that repopulated secondary hosts, and 70% of these clones repopulated tertiary hosts. A marked reduction (33% of the clones) in the ability to repopulate tertiary hosts was seen for HSCs with kinetics that flattened or declined after 5 months. HSCs with kinetics that flattened even earlier—after 3 months—repopulated secondary hosts, but none self-renewed sufficiently to repopulate tertiary hosts. Clones in the – – – group failed to repopulate even secondary hosts (Table 1). Overall, HSC clones with an initial positive slope are more likely to contribute long term than HSC clones with an initial negative slope. While the number of clones followed is too low to arrive at definitive conclusions at this point, the data support the concept that the initial kinetics predict the long-term behavior of HSCs.
Long-term repopulation behaviors of HSC clones in different groups
. | . | Repopulation* . | . | |
|---|---|---|---|---|
| Group . | No. tested . | Secondary . | Tertiary . | |
| +++↑ | 6 | 6 | 4 | |
| +++↓ | 4 | 4 | 3 | |
| ++π↑ | 2 | 2 | 1 | |
| ++-↑ | 3 | 3 | 1 | |
| ++-↓ | 1 | 1 | 0 | |
| +-+↑ | 4 | 3 | 2 | |
| +-+↓ | 1 | 0 | ND | |
| +--↑ | 2 | 2 | 0 | |
| +--↓ | 2 | 2 | 0 | |
| --+ | 1 | 0 | ND | |
| --- | 1 | 0 | ND | |
. | . | Repopulation* . | . | |
|---|---|---|---|---|
| Group . | No. tested . | Secondary . | Tertiary . | |
| +++↑ | 6 | 6 | 4 | |
| +++↓ | 4 | 4 | 3 | |
| ++π↑ | 2 | 2 | 1 | |
| ++-↑ | 3 | 3 | 1 | |
| ++-↓ | 1 | 1 | 0 | |
| +-+↑ | 4 | 3 | 2 | |
| +-+↓ | 1 | 0 | ND | |
| +--↑ | 2 | 2 | 0 | |
| +--↓ | 2 | 2 | 0 | |
| --+ | 1 | 0 | ND | |
| --- | 1 | 0 | ND | |
ND indicates not done; tertiary transfers were not attempted if secondary hosts were not repopulated.
BM cells were isolated from primary and secondary hosts at least 7 months after transplantation for transfer into secondary and tertiary hosts, respectively. In each case, 5 × 106 BM cells were injected and the same methods and criteria were used to identify repopulated mice as for primary hosts (“Materials and methods”). Data are extended and modified from Muller-Sieburg et al.13
Discussion
We present here a systematic and complete analysis of HSC heterogeneity as assessed by clonal repopulation patterns obtained in rigorous long-term transplantation assays of a large number (97) of clonally derived HSCs. The results indicate that only a fraction of all possible patterns of repopulation is actually observed and that any additional patterns are likely to be extremely rare. Furthermore, we show a positive association between self-renewal and continuously increasing repopulation levels in the first 7 months after transplantation in the primary recipient. These findings are not consistent with a model in which HSCs are viewed as a homogeneous population of cells that respond to the conditions in vivo in a stochastic manner. Such models would be expected to give a functional continuum of clonal kinetic and self-renewal patterns. Such models also predict that each HSC clone should be able to regenerate the heterogeneity seen in the HSC compartment. However, as shown previously,4 when clonally derived HSCs self-renew in a primary host, they generate daughter HSCs that are remarkably similar to each other in repopulation kinetics as shown in multiple secondary hosts. Thus, self-renewal did perpetuate predetermined clonal patterns and did not generate heterogeneity.4 Overall, the data favor a model where the HSC compartment consists of a limited number of distinct types of HSCs, each with predictable, epigenetically fixed repopulation and self-renewal behavior.
The interpretation that HSCs show only a limited number of repopulation behaviors of course assumes that many repopulation patterns are realizable in the transplantation model. That this is the case is supported by a quick survey of repopulation kinetics found after transplantation with multiclonal HSC grafts. Just from data published from this laboratory,4,40,44 an additional 9 kinetics (not considering AUC) can readily be identified following transplantation of polyclonal grafts. Additional repopulation kinetics can be estimated from multiclonally repopulated hosts described by others.3,8,9,45 Thus, as expected (Figure 1B), combinations of clonal patterns can generate new kinetics seen after polyclonal transplants. This confirms that the transplantation assay can reveal many different patterns and emphasizes that individual HSCs realize only a subset of the possible kinetics.
HSC repopulation kinetics were compared here based on the 3 slopes of the repopulation kinetics and the AUC as an estimate of the clone size. The data suggest that these parameters are reasonably good predictors for self-renewal capacity and overall production of differentiated progeny by the HSC. These are arguably the most important functions of HSCs. Yet, it may be interesting to add more biologic parameters to the analysis, such as homing ability46 or the ability to respond to external stimuli,7,15 to refine the classification of the HSC compartment. It is also possible to add more parameters to the mathematic analysis. For example, some of the groups in Figure 2 appear to be less homogeneous than most others. This is mostly caused by differences in the steepness of the slopes (eg, Figure 2 the +–– ↑ group, initial segments). When additional parameters were added to classify the angle of the slopes, a visually more homogeneous picture emerges. Yet, each new classifier dramatically increases the number of possible groups with a concurrent increase in the probability that the HSC kinetics show only a limited subset of possible behaviors. Thus, adding more classifiers strengthens the interpretation.
An unexpected finding was that purified Lin–Rho–SP cells were selectively enriched for certain subsets of HSCs. There were few kinetics in the group of Lin–Rho–SP cells, and it is possible that an extended analysis would fill in the missing groups. Yet, it is likely that the kinetics published are the most prevalent in these populations and reflect the distribution of the HSCs in this population. This would support the idea that phenotype-based enrichment protocols deplete some and enrich other HSC subsets. It will be interesting to look at additional phenotypically defined populations that are enriched for HSCs. That a rigorous description of the composition of HSC populations is important was highlighted by the disparate results obtained by different groups that attempted to catalog the expressed gene program of HSCs.47-49 The (statistically) complete description of repopulation patterns of HSCs from adult B6 BM reported here now can serve as a standard against which the clonal composition of different populations of purified HSCs can be compared.
It is worth noting that the analysis shown here does not address how the diversity of HSC behaviors is generated or whether it changes during development and aging. It might be possible to isolate prospectively the different subsets of HSCs that define the different families. This would undoubtedly help in dissecting the molecular basis for these functional differences. Such functional differences would be of some interest for future applications of HSCs for therapeutic purposes. For example, HSCs that repopulate rapidly but for short time periods would be ideally suited for supportive therapy after myeloablation, whereas HSCs with long life spans would be the most desirable candidates for gene therapy.
Prepublished online as Blood First Edition Paper, November 15, 2005; DOI 10.1182/blood-2005-07-2970.
Supported by grants from the National Institutes of Health NIH-DK48015 and NIH-AG023197 (C.E.M.-S.) and grants from the National Cancer Institute of Canada (NCIC), with funds from the Terry Fox Run, the Stem Cell Network, and the NHLBI/NIH P01-55435 (C.J.E.). B.D. was supported by Stem Cell Network, the Canadian Institutes of Health Research (CIHR), and NCIC Studentships. N.U. was supported by CIHR and Kirin Fellowships.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal