Abstract
Members of the Notch family of transmembrane receptors play an important role in cell fate determination. Over the past decade, a role for Notch in the pathogenesis of hematologic and solid malignancies has become apparent. Numerous cellular functions and microenvironmental cues associated with tumorigenesis are modulated by Notch signaling, including proliferation, apoptosis, adhesion, epithelial-to-mesenchymal transition, and angiogenesis. It is becoming increasingly evident that Notch signaling can be both oncogenic and tumor suppressive. This review highlights recent findings regarding the molecular and functional aspects of Notch-mediated neoplastic transformation. In addition, cellular mechanisms that potentially explain the complex role of Notch in tumorigenesis are discussed.
Introduction
In 1917, the first documented case of a strain of Drosophila characterized by notches at the ends of their wing blades was reported. These notches were caused by haploinsufficiency of an unknown gene, which was subsequently cloned in the mid-1980s and identified as the gene coding for the Drosophila transmembrane receptor Notch.1 Notch orthologues have since been identified in numerous organisms including mammals. A plethora of research has identified a role for Notch signaling in diverse cellular processes such as differentiation, proliferation, apoptosis, adhesion, and epithelial-to-mesenchymal transition (EMT). This review focuses on Notch signaling during the development and progression of hematologic and solid malignancies, with emphasis on recent findings in these fields.
The Notch receptor family
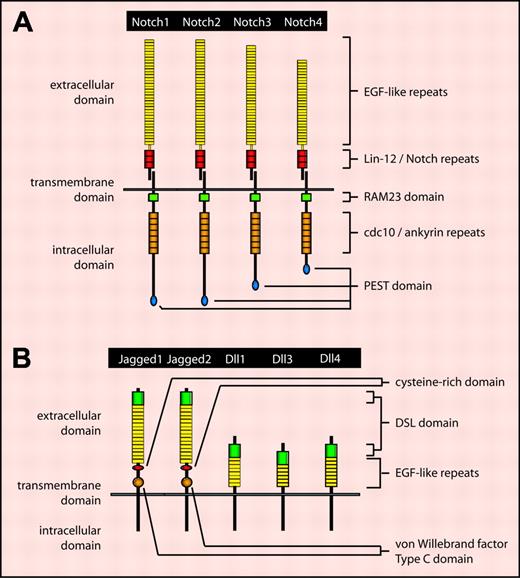
The mammalian family of Notch receptors consists of 4 members: Notch1 through Notch4 (Figure 1A). Notch proteins are synthesized as full-length unprocessed proteins composed of extracellular, transmembrane, and intracellular domains. Following transport through the secretory pathway to the trans-Golgi network, Notch is cleaved at a site referred to as the S1 cleavage site by a furinlike convertase to generate 2 subunits: one consisting of the majority of the extracellular domain and the other consisting of the remainder of the extracellular domain and the complete transmembrane and intracellular domains.2 These 2 subunits associate noncovalently, resulting in the cell-surface expression of a mature heterodimeric type I transmembrane receptor.
Notch activation and signaling
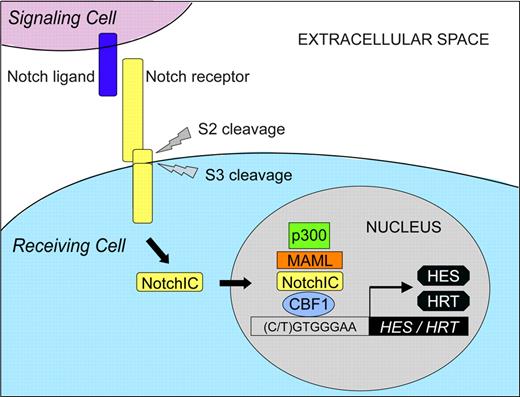
The Notch ligand family consists of 5 members in mammals: Jagged1/2 and Delta-like 1/3/4 (Dll1/3/4) (Figure 1B). Similar to Notch receptors, Notch ligands are type I transmembrane proteins. In the absence of ligand binding, heterodimeric Notch receptors are inactive. Within the extracellular domain of Notch receptors, numerous EGF-like repeats have been identified. Binding of Notch ligand to EGF-like repeats 11 and 12 on an adjacent Notch receptor induces a conformational change in the Notch extracellular domain, resulting in the exposure of an S2 cleavage site within the Notch extracellular domain (Figure 2).3 Following cleavage by the metalloprotease tumor necrosis factor-α converting enzyme, Notch receptors undergo regulated intramembranous proteolysis at a conserved S3 cleavage site located within the transmembrane domain. S3 cleavage is mediated by the γ-secretase complex comprising presenilin 1/2, nicastrin, Pen-2, and Aph-1.4 Hence, the Notch intracellular domain (NotchIC or NICD) is released into the cytoplasm, which subsequently translocates into the nucleus to effect Notch signaling.
Although Notch proteolytic processing at the S1 cleavage site does not result in Notch activation, processing at S2 and S3 cleavage sites is required for activation of the full-length receptor.3 Translocation of NotchIC to the nucleus is required for activation of Notch downstream signaling. In the absence of nuclear NotchIC, Notch target gene expression is repressed by the transcriptional repressor protein C protein binding factor 1 (CBF1; also known as RBP-Jκ or CSL for CBF1/Su(H)/Lag1). CBF1 has been shown to bind at least 2 corepressor complexes, individual components of which cooperate to inhibit transcriptional activation.3
When Notch signaling is activated, nuclear NotchIC binds to CBF1 and converts CBF1 from a transcriptional repressor to an activator.3 Mastermind-like (MAML), a family of transcriptional activator proteins comprising 3 members (MAML1-3), has been shown to be required for Notch signaling.5 MAML forms a ternary complex with CBF1-NotchIC via a direct interaction with NotchIC. In turn, MAML recruits the histone acetyltransferase p300/CBP that acetylates histones, thereby altering the structure of chromatin to a form amenable to active transcription. The ternary complex composed of CBF1-NotchIC-MAML thus functions as a transcriptional activator, resulting in Notch target gene transcription. Among the primary targets are several genes belonging to the basic helix-loop-helix (bHLH) family of proteins.6 In mammals, at least 2 families of bHLH proteins are induced following Notch activation: (1) the hairy/enhancer-of-split (HES) family and (2) the hairy-related transcription factor (HRT; also known as HEY, HESR, HERP, or CHF) family. Both the HES and HRT families function as transcriptional repressors.
The mammalian Notch receptor and ligand families. (A) The mammalian Notch receptor family comprises 4 members: Notch1, Notch2, Notch3, and Notch4. Notch proteins contain several conserved structural motifs. The extracellular domain contains a variable number of epidermal growth factor (EGF)–like repeats involved in ligand binding, and 3 Lin-12/Notch repeats involved in Notch heterodimerization. The intracellular domain contains a RAM23 motif involved in binding Notch downstream signaling proteins, 7 cdc10/ankyrin repeats also involved in mediating downstream signaling, and a PEST domain involved in Notch protein degradation. (B) The mammalian Notch ligand family consists of 5 members: Jagged1, Jagged2,Dll1, Dll3, and Dll4. Within the extracellular domain, Jagged family members contain a cysteine-rich region likely involved in the control of Notch receptor binding specificity, as well as a von Willebrand factor type C domain likely involved in ligand dimerization. These motifs are not present in Dll family members. Extracellular motifs common to all Notch ligands include a single Delta/Serrate/Lag-2 (DSL) domain involved in receptor binding, as well as a variable number of EGF-like repeats that may stabilize receptor binding.
The mammalian Notch receptor and ligand families. (A) The mammalian Notch receptor family comprises 4 members: Notch1, Notch2, Notch3, and Notch4. Notch proteins contain several conserved structural motifs. The extracellular domain contains a variable number of epidermal growth factor (EGF)–like repeats involved in ligand binding, and 3 Lin-12/Notch repeats involved in Notch heterodimerization. The intracellular domain contains a RAM23 motif involved in binding Notch downstream signaling proteins, 7 cdc10/ankyrin repeats also involved in mediating downstream signaling, and a PEST domain involved in Notch protein degradation. (B) The mammalian Notch ligand family consists of 5 members: Jagged1, Jagged2,Dll1, Dll3, and Dll4. Within the extracellular domain, Jagged family members contain a cysteine-rich region likely involved in the control of Notch receptor binding specificity, as well as a von Willebrand factor type C domain likely involved in ligand dimerization. These motifs are not present in Dll family members. Extracellular motifs common to all Notch ligands include a single Delta/Serrate/Lag-2 (DSL) domain involved in receptor binding, as well as a variable number of EGF-like repeats that may stabilize receptor binding.
Notch regulates cell differentiation and tissue development
Notch signaling plays an integral role in the differentiation of a variety of cell lineages (see Ohishi et al7 and Lai8 for recent in-depth reviews). Originally described as a mechanism for the inhibition of cell differentiation, the Notch signaling pathway was believed to maintain cells in an undifferentiated state, thus allowing cells to respond to inductive cues at appropriate times to facilitate cellular diversification.9 Indeed, Notch signaling has been shown to inhibit T-cell development,10 granulocytic differentiation,11 neurogenesis,12 and myogenesis.13 This view of Notch function, however, proved to be oversimplified, as Notch signaling has also been shown to direct cells toward alternate differentiation fates, for instance during gliogenesis.14 Thus Notch signaling can either block or promote cell differentiation, depending on the cell lineage.
Because of its ability to regulate the differentiation fate of individual cells, Notch signaling is intimately involved in the development of numerous tissues in multicellular organisms. In mammalian embryos, Notch receptors and ligands are widely expressed during organogenesis15 and indeed, play a role in the development of tissues derived from all 3 primary germ layers: endoderm (eg, pancreas16 ), mesoderm (eg, hematopoietic system,17 mammary gland,18 vasculature19 ), and ectoderm (eg, nervous system20 ). In addition to regulating embryonic tissue development, Notch signaling also controls fetal and postnatal tissue development, as well as the development and maturation of adult tissues.15 Hence precisely controlled Notch signaling is vital to the proper development of most tissues. Accordingly, perturbation of Notch signaling can manifest as tissue abnormalities ultimately leading to disease states such as cancer.
Notch activation can be oncogenic
Oncogenic Notch signaling in hematologic malignancies
It is well established that Notch signaling plays an important role in hematopoiesis.17 A recent study has identified Notch signaling as a critical factor involved in the maintenance of a pool of self-renewing hematopoietic stem cells (HSCs).21 Notch signaling is active in HSCs in vivo and is down-regulated as HSCs differentiate. Accordingly, inhibition of Notch signaling enhances HSC differentiation in vitro and in vivo. Importantly, the same study demonstrated a requirement for intact Notch signaling in Wnt-mediated HSC renewal, thus linking Notch and Wnt signaling in the process of HSC maintenance. In addition to maintaining an HSC population, Notch signaling is intimately involved in the maintenance of an uncommitted pool of lymphoid, myeloid, and erythroid precursor populations. Notch signaling also regulates the development and differentiation of multiple hematopoietic cell types, including T cells, B cells, monocytes, macrophages, dendritic cells, osteoclasts, and natural killer cells.22-28 Because of its important role in hematopoiesis, Notch signaling must be precisely regulated, as deregulated expression of Notch pathway elements can lead to the development of hematologic malignancies.
Ligand-mediated activation of Notch signaling. Notch expressed on the cell surface as a heterodimer is the mature, ligand-accessible form of the receptor. In the absence of ligand binding, heterodimeric Notch receptors are inactive. When Notch ligand binds to Notch receptor on an adjacent cell, a series of proteolytic cleavages occurs (referred to as S2 and S3 cleavages), resulting in release of the Notch intracellular domain (NotchIC) that subsequently translocates into the nucleus. In the absence of nuclear NotchIC, the transcription factor CBF1 binds to the DNA sequence 5′-(C/T)GTGGGAA-3′ within Notch target gene promoters and represses transcription. When Notch signaling is activated, nuclear NotchIC binds to CBF1 and, following recruitment of the nuclear protein MAML, results in the formation of a ternary complex that functions as a transcriptional activator. MAML recruits the histone acetyltransferase protein p300, resulting in histone acetylation and conversion of the local chromatin structure to a form amenable to active transcription, resulting in transcription of various Notch target genes including those belonging to the HES and HRT (HEY) families.
Ligand-mediated activation of Notch signaling. Notch expressed on the cell surface as a heterodimer is the mature, ligand-accessible form of the receptor. In the absence of ligand binding, heterodimeric Notch receptors are inactive. When Notch ligand binds to Notch receptor on an adjacent cell, a series of proteolytic cleavages occurs (referred to as S2 and S3 cleavages), resulting in release of the Notch intracellular domain (NotchIC) that subsequently translocates into the nucleus. In the absence of nuclear NotchIC, the transcription factor CBF1 binds to the DNA sequence 5′-(C/T)GTGGGAA-3′ within Notch target gene promoters and represses transcription. When Notch signaling is activated, nuclear NotchIC binds to CBF1 and, following recruitment of the nuclear protein MAML, results in the formation of a ternary complex that functions as a transcriptional activator. MAML recruits the histone acetyltransferase protein p300, resulting in histone acetylation and conversion of the local chromatin structure to a form amenable to active transcription, resulting in transcription of various Notch target genes including those belonging to the HES and HRT (HEY) families.
In humans, aberrant NOTCH1 expression has been identified as a causative factor in the development of T-cell acute lymphoblastic leukemia and lymphoma (T-ALL).29 Analysis of patient samples has revealed a chromosomal translocation, t(7;9)(q34:q34.3), that juxtaposes the coding region of the C-terminal domain of NOTCH1 adjacent to the T-cell receptor-β locus.29 This translocation results in the expression of a truncated constitutively active Notch1 protein, commonly referred to as translocation-associated Notch1 (TAN1).29 Lethally irradiated mice that received transplants of bone marrow cells transduced with Notch1IC develop T-ALL with 100% penetrance.30 Despite the ability of activated Notch1 to consistently induce T-cell leukemia in mice, less than 1% of human T-ALLs exhibit the t(7;9) translocation.31 However, activating mutations in NOTCH1 independent of t(7;9) have been identified in more than 50% of human T-ALL,32 with frequent occurrence in both childhood and adult cases of the disease.32,33 Activating mutations in Notch1 have since been reported in mouse models of T-ALL.34 Thus, deregulated Notch1 signaling plays a major role in the etiology of murine and human T-ALL. Activating mutations in Notch2 and Notch3 have also been shown to induce T-ALL.35,36 Similar to lethally irradiated mice that received transplants of Notch1IC-expressing bone marrow, transplantation of Dll4-expressing bone marrow results in the induction of T-ALL.37 Taken together, these studies suggest that aberrant Dll4-mediated activation of Notch signaling may promote leukemogenesis in T-ALL.
In contrast to T-cell malignancies, a definite role for activated Notch signaling in the development of pre-B-cell or B-cell malignancies is less clear. Lethally irradiated mice that received transplants of Notch1IC-expressing bone marrow exclusively develop T-cell malignancies, but exhibit disrupted B-cell development.38 Conversely, Notch1 deficiency results in the development of B cells at the expense of T cells.39 In human B-cell precursor ALL, Notch1 and HES1 mRNA expression is significantly lower than in T-ALL.40 Indeed, a majority of B-lymphoma cell lines exhibit intact Notch signaling that is precisely regulated.41 Taken together, these studies point against a role for aberrant Notch signaling in the development of B-cell malignancies. However, B-cell–derived Hodgkin lymphoma cells exhibit overexpression of Notch1 and Jagged1 protein.42 Moreover, nonmalignant cells adjacent to Notch1-positive Hodgkin cells express high levels of Jagged1 protein, raising the possibility that Jagged1-mediated Notch1 activation may be a contributing factor in the development of Hodgkin lymphoma.42 It is also of note that overexpression of Notch2 is observed in malignant cells from patients with B-cell chronic lymphocytic leukemia (B-CLL), and is associated with increased B-cell survival.43 Of interest, oncogenic viral proteins have been shown to use the Notch signaling pathway to induce B-cell immortalization.23 Thus, it is possible that in certain B-cell malignancies Notch activation plays an oncogenic role.
Deregulated Notch signaling is also involved in the pathogenesis of multiple myeloma. Primary human multiple myeloma samples exhibit overexpression of Jagged1/2 and Notch1/2 protein.44,45 Hence, both juxtacrine and paracrine mechanisms may contribute to myelomagenesis in vivo.44,45
Acute myelogenous leukemia (AML) is characterized by overexpression of Jagged1 mRNA/protein40,46 and Notch1 protein.46 Aberrant Jagged1-Notch1 signaling is posited to promote the development of AML by inducing excessive self-renewal with a concomitant block in cell differentiation.40 A recent study involving primary AML cells, however, has failed to demonstrate increased self-renewal in response to activated Notch signaling using recombinant Notch ligands.47 Instead, ligand-mediated Notch activation is reported to promote AML cell differentiation.47 Clearly, additional studies are needed to elucidate the role of Notch signaling in AML pathogenesis.
Oncogenic Notch signaling in solid tumors
Similar to hematologic malignancies, expression of Notch pathway elements has been observed in solid human tumors. Although the vast majority of these studies demonstrate correlative associations between aberrant Notch ligand/receptor expression and tumor development, they nonetheless suggest a possible causative role for deregulated Notch signaling in solid tumor pathogenesis. Up-regulation of Jagged1 mRNA and has been observed in human pancreatic cancer,48 and Jagged1 protein overexpression has been reported in human cancers of the prostate,49 cervix,50 and brain.51,52 Jagged2 mRNA is up-regulated in human cervical cancer,50 with Jagged2 protein overexpression reported in human pancreatic cancer.48 Dll1 expression is elevated in human cervical cancers at the mRNA level50 and in human brain cancers at both mRNA and protein levels.52
Deregulated Notch receptor expression has been reported in a growing number of solid human tumors. Increased Notch1 protein expression has been observed in human cancers of the cervix,50,53 colon,53 lung,53 pancreas,48 skin,54 and brain.51,52 Overexpression of Notch2 mRNA is observed in human brain cancer,55 with Notch2 protein overexpression detected in human cancers of the cervix,50,53 colon,53 pancreas,48 skin,54 and brain.51 Protein overexpression of Notch3 and Notch4 has been reported in human malignant melanoma54 and human pancreatic cancer.48 A recent report has identified Notch4 mRNA expression in human breast cancer.18 Among the many solid tumors exhibiting aberrant Notch expression, a causal role for Notch signaling in solid tumor pathogenesis has been best studied in breast cancer models in mice.
The first study to identify a role for Notch signaling in murine mammary gland tumorigenesis came from observations of Czech II mice infected with mouse mammary tumor virus (MMTV).56 Of the mammary tumors that developed in these mice, 18% (9 of 45 tumors) exhibited MMTV integration into the Notch4 locus. Of the tumors displaying MMTV integration into Notch4, all exhibited integration within a region of Notch4 that gives rise to a truncated protein consisting primarily of a constitutively active form encoding the transmembrane and intracellular domains. Similar to MMTV, intracisternal A particle integration into the Notch4 locus also results in expression of a constitutively active Notch4, and has been observed in spontaneous mammary tumors in both Czech II57 and Balb/c mouse strains.58 These studies clearly link aberrant Notch4 signaling to murine mammary gland tumorigenesis.
To directly demonstrate that activated Notch4 signaling is oncogenic in the murine mammary gland, activated Notch alleles have been introduced into mice. Transgenic mice expressing activated Notch4 under the control of the MMTV long-terminal repeat or the whey acidic protein promoter develop mammary carcinomas with subsequent metastasis to the lungs.59-61 A role for Notch1 in murine mammary tumorigenesis has also been reported. c-ErbB2 transgenic mice infected with MMTV develop mammary tumors, some of which exhibit MMTV integration into the Notch1 gene, resulting in expression of a constitutively activated Notch1 protein.62 Moreover, expression of either activated Notch1 or activated Notch4 has been shown to induce transformation of mouse mammary epithelial cells in vitro.63 Taken together, these studies demonstrate that activated Notch signaling plays a causal role in murine models of mammary tumorigenesis.
Recent studies have highlighted a potential role for Notch signaling in human breast cancer development. Overexpression of constitutively active Notch4 in normal human breast epithelial cells induces transformation in vitro.64 Among a panel of human breast cancer cell lines, 7 of 8 cell lines examined express elevated levels of a Notch4 RNA species corresponding to full-length Notch4.64 Furthermore, 2 of these cell lines express an additional Notch4 RNA species encoding a truncated constitutively active Notch4. In a study involving 7 breast cancer specimens, Notch1 protein expression was detected in all tumors examined, with normal breast tissue at the margins of tumor sections exhibiting little or no Notch1 protein expression.65 In a second study involving 25 specimens, mRNAs for all 4 Notch receptors were expressed at varying frequencies.18 A third study involving 97 specimens demonstrated that poorly differentiated breast tumors were associated with elevated levels of Notch1 protein and reduced patient survival.66 More importantly, a recent study has shown that more than 50% of human breast tumors express reduced protein levels of Numb, a negative regulator of Notch signaling, and a negative correlation exists between Numb expression and breast tumor grade.67 Taken together, these findings suggest an oncogenic role for Notch signaling in human breast cancer and a potential tumor-suppressive function for Numb in this disease.
The molecular and functional basis of Notch-induced oncogenesis
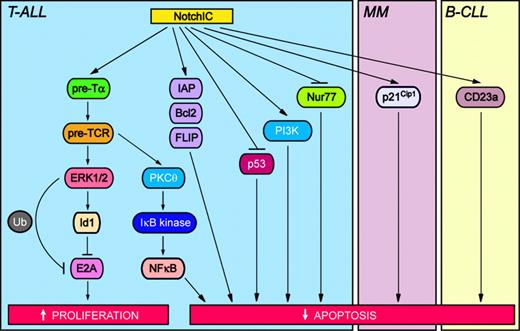
Two mechanisms involved in Notch-induced oncogenesis in hematologic malignancies include inhibition of apoptosis and induction of proliferation (Figure 3). In T cells, expression of activated Notch1 protects against T-cell receptor–mediated apoptosis by directly binding to and inhibiting the activity of the proapoptotic transcription factor Nur77.68 Activated Notch1 also protects T cells from apoptosis induced by dexamethasone, etoposide, or Fas receptor–mediated signaling via up-regulation of antiapoptotic proteins belonging to the inhibitor of apoptosis (IAP) and Bcl2 families, as well as FLICE-like inhibitor protein (FLIP).69,70 Recently, Notch signaling has been shown to promote the survival of pre-T cells through the maintenance of cell size, as well as through the promotion of glucose uptake and metabolism.71 Importantly, Notch promotes pre–T-cell viability by signaling through the phosphatidylinositol 3-kinase (PI3K) pathway.71 Notch can also protect against apoptosis by suppressing expression of the tumor suppressor p53, which in turn may promote T-cell lymphomagenesis.72
Oncogenic Notch signaling pathways in hematologic malignancies. Activation of Notch modulates signaling pathways that increase proliferation and inhibit apoptosis, thus contributing to neoplastic transformation. T-ALL indicates T-cell acute lymphoblastic leukemia; MM, multiple myeloma; and B-CLL, B-cell chronic lymphocytic leukemia. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced oncogenesis.”
Oncogenic Notch signaling pathways in hematologic malignancies. Activation of Notch modulates signaling pathways that increase proliferation and inhibit apoptosis, thus contributing to neoplastic transformation. T-ALL indicates T-cell acute lymphoblastic leukemia; MM, multiple myeloma; and B-CLL, B-cell chronic lymphocytic leukemia. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced oncogenesis.”
Several other players involved in Notch-induced T-cell neoplastic transformation have been identified. Induction of T-ALL by activated Notch1/3 requires signaling through the pre–T-cell receptor (pre-TCR), which promotes pre–T-cell proliferation and survival.73,74 In the absence of genes required for pre-TCR assembly, such as pre-Tα, Notch1IC-induced T-ALL is prevented.73 In fact, pre-Tα may be a direct target gene of Notch.75 Notch-induced pre-TCR signaling contributes to T-cell lymphomagenesis by activating extracellular signal-regulated kinase 1/2 (ERK1/2), which in turn up-regulates the expression of Id1, an inhibitor of the tumor suppressor protein E2A.76 Notch-mediated inhibition of E2A activity may also occur via ERK1/2-induced ubiquitination and degradation of E2A.77 Hence, activated Notch1/3 can inhibit DNA binding and the transcriptional activity of E2A,76,78 resulting in enhanced T-cell proliferation and inhibited apoptosis. This in turn may lead to T-cell neoplastic transformation.
Transgenic mice expressing activated Notch3 develop T-cell lymphomas characterized by constitutive activation of nuclear factor κB (NFκB) and thus enhanced NFκB-mediated cell survival.36 Recently, a mechanism linking Notch-induced pre-TCR signaling with NFκB activation has been demonstrated. Notch-induced pre-TCR signaling enhances the activation and membrane translocation of protein kinase Cθ (PKCθ), which in turn activates the IκB kinase complex resulting in increased NFκB activity.79
Molecules involved in the development of hematologic malignancies other than T-ALL are only now being elucidated. In B-CLL, expression of the transmembrane glycoprotein CD23a is associated with enhanced cell survival. Notch2 activation promotes B-CLL cell survival by up-regulating CD23a expression.80 In multiple myeloma, overexpression of activated Notch1 up-regulates the cyclin-dependent kinase inhibitor p21Cip1, resulting in cell cycle arrest and a concomitant protection from drug-induced apoptosis.81 Evidence suggests that Jagged2 up-regulation may play a role in the early pathogenesis of multiple myeloma. Overexpression of Jagged2 in myeloma cells is thought to induce Notch activation in adjacent bone marrow stromal cells via direct cell-cell contact.44 These activated stromal cells subsequently secrete IL6, resulting in paracrine stimulation of myeloma cells.44 Skeletrophin, a ubiquitin ligase expressed in myeloma cells, may increase ligand-dependent Notch activation in bone marrow stromal cells.82 Skeletrophin binds specifically to the intracellular domain of Jagged2 and is postulated to enhance endocytosis of Jagged2 into the myeloma cells. Because Notch activation involves transendocytosis of the Notch extracellular domain in conjunction with bound ligand into ligand-expressing cells,83 skeletrophin may stimulate Notch activation in stromal cells that are in direct cell-cell contact with myeloma cells.82
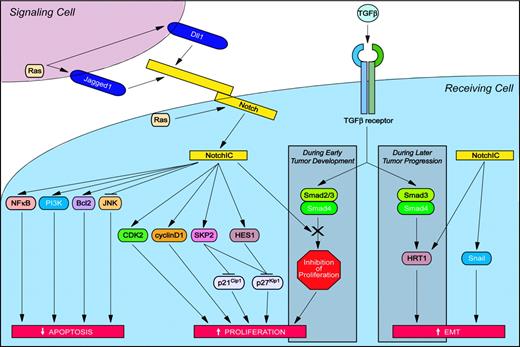
Numerous mechanisms have been suggested to play a role in Notch-induced solid tumor pathogenesis (Figure 4). Antiapoptotic effects of activated Notch proteins have been linked to the induction of Bcl2,84 as well as increased signaling through both the PI3K85 and NFκB86 signaling pathways. A positive feedback loop involving Jagged1-induced activation of NFκB signaling87 and NFκB-induced transcription of Jagged188 may further enhance the protective effects of Notch signaling. Activated Notch signaling can also protect against apoptosis by inhibiting the activation of c-Jun N-terminal kinase (JNK).84 By physically interacting with and inhibiting the function of JNK-interacting protein 1 (JIP1), a scaffold protein that enhances kinase activation during JNK signaling, Notch1IC effectively prevents JIP1-mediated JNK activation.89
Oncogenic Notch signaling pathways in solid tumors. Activation of Notch modulates signaling pathways that increase proliferation and inhibit apoptosis, thus contributing to neoplastic transformation. In addition, Notch activation can induce epithelial-to-mesenchymal transition (EMT) and thus promote the invasion and dissemination of cancer cells. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced oncogenesis.”
Oncogenic Notch signaling pathways in solid tumors. Activation of Notch modulates signaling pathways that increase proliferation and inhibit apoptosis, thus contributing to neoplastic transformation. In addition, Notch activation can induce epithelial-to-mesenchymal transition (EMT) and thus promote the invasion and dissemination of cancer cells. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced oncogenesis.”
Increased cell proliferation in response to Notch activation may also promote tumorigenesis. In a kidney epithelial cell line, activated Notch1 promotes cell cycle entry by enhancing CDK2 and cyclin D1 activity, the latter of which may be a direct target of Notch.90 In HeLa cells, HES1 promotes cell proliferation by repressing transcription of the cyclin-dependent kinase inhibitor p27Kip1.91 In 3T3 fibroblasts, Notch signaling attenuates p27Kip1 expression by a mechanism independent of transcriptional repression. Specifically, activated Notch1 induces expression of the S phase kinase–associated protein 2 (SKP2), a subunit of the ubiquitin-ligase complex SCFSKP2, which in turn enhances proteasome-mediated degradation of p27Kip1.92 The fact that SKP2 also promotes degradation of p21Cip1 further enhances the ability of Notch to promote cell cycle entry.92
The transforming ability of Notch may also be mediated through activated Ras signaling. Cell lines derived from primary tumors of Notch4IC transgenic mice exhibit activated PI3K and ERK signaling, 2 downstream branches of the Ras pathway.93 Interestingly, a separate study has demonstrated that the transforming ability of Ras may be related to the activation of Notch signaling.65 In human foreskin fibroblasts and human kidney epithelial cells expressing hTERT and SV40 oncoproteins, overexpression of activated Ras increases Notch1, Notch4, and Dll1 protein expression.65 Importantly, maintenance of the Ras-induced neoplastic phenotype has been shown to require sustained Notch signaling.65 Hence Notch may function as a downstream target of Ras and in a positive feedback loop also act as an activator of the Ras pathway during tumorigenesis.
Another mechanism of Notch-induced tumor development and progression may involve modulation of the transforming growth factor β (TGFβ) signaling pathway. During the early stages of tumor development, TGFβ inhibits the growth of most epithelial cell types and thus functions as a tumor suppressor. Activated Notch1 suppresses the growth-inhibitory effects of TGFβ by sequestering the transcriptional coactivator p300 from Smad3, a downstream effector of TGFβ signaling.94 Moreover, activated Notch4 has been shown to directly bind to and inhibit the signaling activity of Smad2, Smad3, and Smad4, resulting in attenuated TGFβ signaling in MCF-7 breast cancer cells.95 Hence cells exhibiting activated Notch signaling are resistant to the growth-inhibitory effects of TGFβ, which in turn may promote tumor development.
During the later stages of tumor progression, however, TGFβ promotes invasion and dissemination of cancer cells through the process of EMT. The tumor suppressor protein E–cadherin plays a role in cell-cell adhesion and maintenance of the differentiated epithelial phenotype. Human keratinocytes stimulated with TGFβ undergo EMT with an associated down-regulation of E-cadherin protein expression.96 Treatment with antisense HRT1 or functional inactivation of Jagged1, however, inhibits TGFβ-induced down-regulation of E-cadherin expression.96 Thus, the ability of TGFβ to induce EMT may be dependent upon Notch signaling. Indeed, TGFβ has been shown to induce the expression of Jagged1 protein,96 as well as the Notch target genes HES197 and HRT1.96 Despite these findings, and the fact that Notch target genes are expressed at sites of epithelial-mesenchymal cell-cell interactions during embryogenesis,98,99 a direct role for activated Notch signaling in the induction of EMT remains to be demonstrated. Of interest, recent studies have identified a role for Notch signaling in a specialized type of EMT, endothelial-to-mesenchymal transition.100,101 Jagged1 activation of endogenous Notch receptors in human endothelial cells promotes endothelial-to-mesenchymal transition,101 and endothelial cells expressing activated Notch1 or Notch4 repress VE-cadherin expression, potentially mediated by the transcriptional repressor Snail.100,101
Is activation of Notch signaling alone sufficient to induce transformation?
As a general rule, oncoproteins need to collaborate with other oncoproteins to cause cancer. Several reports have highlighted the importance of oncogenic cooperation in Notch-induced hematologic malignancies. Transgenic mice expressing either the E2A-PBX or c-Myc oncogenes develop T-ALL after a lengthy latency period.102,103 When infected with Moloney murine leukemia virus (MLV), however, lymphomas develop after a shortened latency period and exhibit proviral insertions into the Notch1 gene.102,103 Similarly, Notch1 transgenic mice develop T-ALL after a lengthy latency period, which is significantly shortened following MLV infection.104 The molecular basis of this shortened latency has recently been identified. These lymphomas exhibit viral integration into the Ikaros gene, which codes for a transcriptional repressor expressed during hematopoiesis.105 Wild-type Ikaros and CBF1 bind the same consensus DNA sequence to mediate transcriptional repression. Because viral integration results in the expression of a dominant-negative Ikaros isoform that prevents wild-type Ikaros from mediating transcriptional repression, genes normally regulated by Ikaros are derepressed and the promoters are thus free to be activated by Notch1IC/CBF1.
Oncogenic cooperation during Notch-induced solid tumorigenesis has also been reported. MMTV infection of c-erbB2 transgenic mice results in the development of mammary tumors with retroviral integration into the Notch1 gene, suggesting that Notch1 synergizes with ErbB2 to induce tumor formation.62 Interestingly, Notch activation can enhance ErbB2 promoter activity,106 raising the possibility that activated Notch signaling contributes to tumorigenesis by up-regulating ErbB2 expression. Cooperation between Notch and viral oncoproteins, such as simian virus 40 (SV40) large T, adenovirus E1A, and human papilloma virus (HPV) E6/E7 can also contribute to tumorigenesis.107 Taken together, these studies demonstrate that Notch signaling cooperates with numerous oncogenes to accelerate the development of hematologic and solid malignancies, consistent with a multistep model for tumorigenesis.
Notch activation can be tumor suppressive
Despite mounting evidence demonstrating an oncogenic role for activated Notch signaling in hematologic and solid malignancies, a re-evaluation of the tumor-promoting function of Notch was necessary with the discovery that Notch signaling could be tumor suppressive.
Tumor-suppressive Notch signaling in hematologic malignancies
Simultaneous activation of Notch1 and inactivation of Notch2 may play a role in T-cell lymphomagenesis. Specifically, γ-radiation–induced mouse thymic lymphomas exhibit overexpression of Notch1 mRNA and reduced Notch2 mRNA.108 Because activating mutations in Notch2 can induce T-ALL,35 additional studies are needed to clarify the role of Notch2 in T-cell malignancies.
A tumor-suppressive role for Notch signaling in B-cell malignancies has been suggested. In 4 separate studies, constitutive Notch signaling has been shown to inhibit proliferation and/or induce apoptosis in malignant B cells.81,109-111 Importantly, constitutively active forms of all 4 Notch receptors function as potent inducers of growth arrest and apoptosis in B-cell acute lymphoblastic leukemia cells, with Notch1IC also capable of inducing growth arrest/apoptosis in Hodgkin lymphoma and multiple myeloma cells.111 It should be noted, however, that Notch-induced growth arrest may actually promote malignant progression by protecting against drug-induced apoptosis, as has been suggested in the case of multiple myeloma.81 Ligand-induced Notch activation may also be tumor suppressive, as Dll1/4-mediated Notch activation suppresses the self-renewal capacity and long-term growth of AML cells.112
Tumor-suppressive Notch signaling in solid tumors
In human cervical cancers, early-disease–stage specimens exhibit enhanced Notch1 protein expression, whereas late-stage cervical cancers express reduced Notch1 protein levels.113 Because expression of activated Notch1 causes strong growth inhibition of HPV-positive cervical carcinoma cells, down-regulation of Notch1 in late-stage cervical cancer is posited to play a role in the maintenance of malignant transformation.113 Hence, Notch1 expression in early-stage cervical cancer may promote tumor formation, whereas expression in late-stage cervical cancer may be tumor suppressive.
A tumor-suppressive function for Notch1 has also been demonstrated in mouse skin. When Notch1 function is inactivated specifically in mouse skin, the epidermis undergoes hyperproliferation with subsequent development of skin tumors.114 These mice also have increased sensitivity to chemical-induced skin carcinogenesis.114 Recent findings suggest that Notch1 may function as a tumor suppressor in human skin as well. In normal differentiated human epidermis, Notch signaling is activated and functions to promote keratinocyte differentiation.115 Human basal cell carcinomas, in contrast, lack activated Notch1 signaling.54 A tumorsuppressive role for activated Notch signaling has also been suggested in the prostate,116 lung,117 brain,55 and liver.118
Preliminary studies of human breast cancer specimens suggest a possible tumor-suppressive role for Notch2. In contrast to poorly differentiated breast tumors, well-differentiated breast tumors are associated with elevated levels of Notch2 protein and increased patient survival.66 Interestingly, poorly differentiated breast tumors exhibit elevated levels of Notch1 protein and reduced patient survival.66 These findings suggest that in human breast cancer, Notch1 may be oncogenic, whereas Notch2 may be tumor suppressive.
The molecular and functional basis of Notch-induced tumor suppression
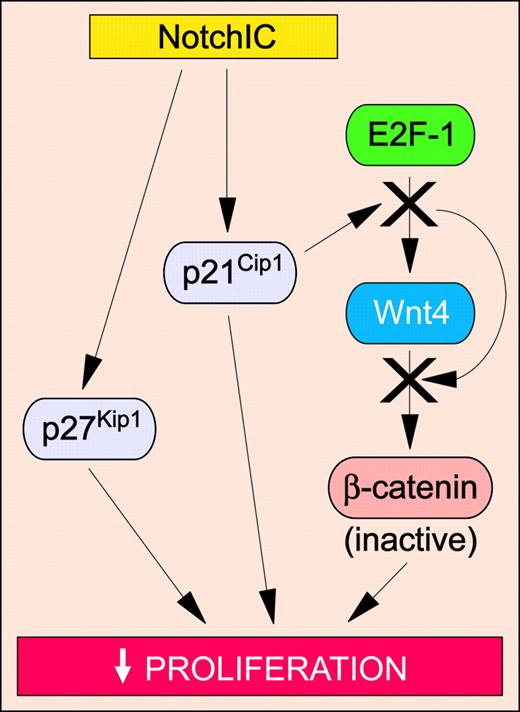
Because the tumor-suppressive function of Notch is a relatively new concept, the molecular mechanisms involved in this process have not been adequately elucidated. Nonetheless, several studies have highlighted antiproliferative effects of Notch signaling as an important mechanism (Figure 5). In small-cell lung cancer cell lines, expression of activated Notch1 or Notch2 has been shown to up-regulate the expression of p21Cip1 and p27Kip1, resulting in cell cycle arrest.117 In mouse keratinocytes, activated Notch1 signaling up-regulates p21Cip1, which in turn induces cell cycle arrest.119 This cessation of proliferation is necessary for terminal differentiation of the keratinocyte to occur. Accordingly, mice with targeted disruption of Notch1 in the epidermis develop skin tumors with decreased p21Cip1 levels.114 These mice also exhibit enhanced β-catenin signaling.114 Given that enforced expression of constitutively active Notch1 in mouse keratinocytes inhibits β-catenin signaling,114 these results suggest that Notch1 signaling prevents tumorigenesis in normal mouse skin by blocking proliferation through at least 2 distinct mechanisms: increasing p21Cip1 expression and repressing β-catenin–mediated Wnt signaling. A recent study has demonstrated that p21Cip1 expression is functionally linked to the Wnt signaling pathway in mouse keratinocytes. Specifically, Notch1 activation increases the expression of p21Cip1, which in turn associates with the E2F-1 transcription factor at the Wnt4 promoter.120 This association blocks the recruitment of c-Myc and p300, resulting in transcriptional repression of Wnt4 expression and hence attenuated Wnt signaling.120 Additional studies are needed to determine whether similar mechanisms occur during Notch-mediated tumor suppression in human keratinocytes.
Tumor-suppressive Notch signaling pathways in solid tumors. Activation of Notch modulates signaling pathways that inhibit proliferation, thus preventing neoplastic transformation. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced tumor suppression.”
Tumor-suppressive Notch signaling pathways in solid tumors. Activation of Notch modulates signaling pathways that inhibit proliferation, thus preventing neoplastic transformation. Specific signaling pathways and abbreviations of signaling components are outlined in “The molecular and functional basis of Notch-induced tumor suppression.”
The role of Notch in regulating angiogenesis
It is well known that signaling pathways involved in physiologic angiogenesis are commonly exploited during the induction of tumor angiogenesis. Evidence has emerged in support of a role for Notch signaling in modeling and maintaining the vasculature.19 Importantly, these findings highlight a potential role for deregulated Notch signaling in tumor angiogenesis and metastasis.
Notch activation regulates angiogenesis
The first evidence that activated Notch signaling could inhibit blood vessel development came from a study involving transgenic mice engineered to express activated Notch4 specifically in the mouse embryonic vasculature. These mice display vascular patterning defects consistent with attenuated vascular development.121 In an in vitro assay in which human endothelial cells coated on beads are stimulated to migrate into a 3-dimensional fibrin gel, expression of activated Notch4 inhibits spontaneous endothelial sprouting, as well as sprouting in response to fibroblast growth factor 2 (FGF2) and vascular endothelial growth factor (VEGF).122 In an in vivo chick chorioallantoic membrane (CAM) assay, expression of activated Notch4 in endothelial cells of the CAM is sufficient to inhibit VEGF-induced angiogenesis.122 These studies suggest that constitutive Notch activation inhibits endothelial sprouting in vitro and angiogenesis in vivo.
Gene targeting studies in the mouse have also highlighted the importance of the Notch pathway in angiogenesis. Mice in which the Notch1 allele is disrupted are embryonic lethal and exhibit defective vascular remodeling.123 Notch4 gene disruption, in contrast, results in viable mice with no apparent defects. Interestingly, Notch1/Notch4 double-deficient mice exhibit more severe vascular defects than Notch1-null mice. Taken together, these studies indicate that either constitutive activation or attenuation of Notch signaling results in a common vascular phenotype, disrupted blood vessel development. Hence precise regulation of Notch signaling is likely required for angiogenesis to occur. In accordance with this, human breast tumors exhibit Dll4 and Notch3 mRNA expression in some but not all tumor microvessels,18,124 suggesting that Notch activation may be required at specific stages of tumor angiogenesis.
The molecular and functional basis of Notch-induced inhibition of angiogenesis
Studies examining the effects of activated Notch signaling on endothelial cell phenotype and function have identified potential mechanisms involved in Notch-induced antiangiogenesis. Although the individual mechanisms target different stages of the angiogenic process, the net effect is common and involves establishment and maintenance of a stabilized endothelial cell network. Indeed, expression of activated Notch1 or overexpression of HES1 has been shown to stabilize endothelial tube formation in Matrigel.125 Interestingly, human umbilical vein endothelial cells (HUVECs) grown to confluence in 2-dimensional cultures in vitro exhibit increased expression of HRT1.126 Similarly, HUVECs cultured in 3-dimensional fibrin gels as endothelial tubes up-regulate Dll4, Notch1/4, and HRT1 mRNA expression.127
Up-regulated Notch signaling has been postulated to inhibit the formation of new endothelial sprouts and hence stabilize the vasculature by several distinct mechanisms. Activated Notch signaling can enhance endothelial cell adhesion to the extracellular matrix. Endothelial cells expressing activated Notch4 exhibit increased adhesion to collagen due to an enhanced affinity state of cell-surface β1 integrin receptors.122 A necessary role for the cdc10/ankyrin repeats of Notch4IC in the inhibition of endothelial sprouting has since been demonstrated.128 These findings suggest that Notch4 activation in endothelial cells, via a mechanism involving signaling mediated by the cdc10/ankyrin repeats, may inhibit angiogenesis by promoting β1 integrin–mediated adhesion to the underlying matrix, thereby preventing migration and effectively fixing the cells in place.
Attenuation of endothelial cell proliferation may also play a role in Notch-inhibited endothelial sprouting. It has been postulated that low or absent HRT1 expression may be permissive to endothelial proliferation, whereas overexpression of HRT1 may block this process, thus inhibiting the formation of new endothelial tubes.129 Recent evidence has confirmed an antiproliferative role for Notch signaling in endothelial cells. Expression of activated Notch1 or overexpression of HES1 induces endothelial cell cycle arrest in 2-dimensional cultures in vitro.125 Similarly, Jagged1-mediated Notch activation induces cell cycle arrest in primary human endothelial cells.126 The molecular mechanism of this inhibited proliferation is cell autonomous, and involves attenuation of retinoblastoma (Rb) protein phosphorylation.126 When stimulated with mitogens, endothelial cells begin cycling with accompanied up-regulation of p21Cip1 expression. This up-regulation of p21Cip1 is required for nuclear translocation of cyclin D–cdk4 complexes, which in turn phosphorylate Rb to permit S-phase entry. Activated Notch signaling inhibits mitogen-induced up-regulation of p21Cip1, thus inhibiting nuclear translocation of cyclin D–cdk4 complexes and Rb phosphorylation.126 Notch activation can also inhibit endothelial cell proliferation by down-regulating the expression of minichromosome maintenance proteins, which are essential for DNA replication.130
Another mechanism for Notch-inhibited sprouting involves down-regulation of endothelial cell responsiveness to VEGF. Specifically, overexpression of Notch4IC or HRT1 can down-regulate VEGF-R2 mRNA expression.128,129 Because the VEGFR2 (KDR) promoter contains E-box binding motifs, which are recognized by HRT proteins, activated Notch signaling may act via HRT1 to repress VEGF-R2 expression.
Notch activation has also been shown to inhibit endothelial cell apoptosis by inhibiting JNK activation and up-regulating Bcl2 expression through CBF1-dependent and -independent pathways, respectively.84 Thus by promoting adhesion and inhibiting proliferation and apoptosis, Notch appears to be important in stabilizing the vasculature, in particular the arteries and the microvasculature where Notch receptor/ligand expression is prominent. While Notch inhibition may promote proliferation, it would also result in loss of β1 integrin survival signals and susceptibility to apoptosis. Taken together with Notch gene disruption studies in mice,123 these findings suggest that selective activation or inhibition of Notch signaling within blood vessels of solid tumors may be a strategy with which to block tumor angiogenesis, and potentially elicit an antitumor response.
Discussion and conclusions
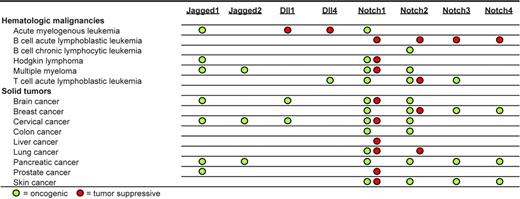
In efforts to elucidate the molecular mechanisms involved in cancer development, it is becoming increasingly important to identify the cellular factors that determine whether Notch signaling will be oncogenic or tumor suppressive. Several factors may play a role in determining whether Notch acts to promote or suppress tumorigenesis. One factor is the specific Notch paralog activated. In human breast cancer, Notch1 is often associated with oncogenesis, whereas Notch2 may be associated with tumor suppression.66 In contrast, in human brain tumors, Notch1 may be tumor suppressive with Notch2 playing an oncogenic role.55 Because most cancers express more than one type of Notch ligand and/or receptor, the overall expression profile of these ligands/receptors may ultimately determine whether Notch signaling will be oncogenic or oncosuppressive. Figure 6 summarizes our current understanding of the role of Notch ligands/receptors in the tumorigenesis of various human and animal neoplasias.
A second factor is cell type; that is, different cell types exhibit activation of different signaling pathways that may work in conjunction with Notch signaling to elicit a specific functional phenotype. For example, Notch activation in lung cancer cell lines up-regulates p21Cip1 and p27Kip1 expression to inhibit cell proliferation,117 but Notch activation in fibroblasts down-regulates p21Cip1 and p27Kip1 expression to promote cell cycling.92 Adding to the complexity is the fact that Notch activation in endothelial cells down-regulates p21Cip1 expression with an associated inhibition of proliferation.126 Notch activation can also inhibit or promote apoptosis, depending on the cell type. For example, Notch activation in Kaposi sarcoma cells inhibits apoptosis, whereas in hepatocellular carcinoma cells Notch promotes cell death.131,132
A third factor is the presence of specific cytokines/growth factors in the cellular microenvironment. Notch activation in the presence of macrophage colony-stimulating factor, but not granulocyte-macrophage colony-stimulating factor (GM-CSF), induces apoptosis in monocytes.24 Interestingly, Notch activation in the presence of GM-CSF inhibits the differentiation of monocytes into macrophages, but permits their differentiation into mature dendritic cells in the presence of GM-CSF, interleukin-4, and tumor necrosis factor-α.133 Moreover, Notch activation in the presence of both GM-CSF and TGFβ1 promotes monocyte differentiation into Langerhans cells.134
Neoplasias in which Notch receptors or ligands play potential oncogenic or tumor-suppressive roles.
Neoplasias in which Notch receptors or ligands play potential oncogenic or tumor-suppressive roles.
A fourth factor is the dosage of Notch signaling. In a leukemic T-cell line, low or high concentrations of Notch1IC promote or inhibit NFκB transactivation, respectively.135 In cord blood progenitor cells, ex vivo culture with low density of immobilized Dll1 promotes the survival of CD34+ cells, while a high concentration of Dll1-induced Notch activation inhibits the generation/survival of CD34+ cells.136 Similarly, the level of Notch signaling within the thymic microenvironment can determine the differentiation fate of hematopoietic precursor cells.137 Interestingly, microenvironmental factors may influence the level of Notch signaling. For example, hypoxia increases Notch1 protein expression in neuroblastoma cells,138 and estrogen induces Notch1 and Jagged1 mRNA expression in MCF7 breast cancer cells.139 Moreover, the relative expression of Notch signal modulators such as Deltex, Numb, and Fringe proteins also influences the intensity and ligand dependence of the Notch signal.140,141 These findings highlight the importance of precise Notch signal regulation during proper cellular development.
An intriguing concept is the possibility that activated Notch signaling within a single tumor may be both oncogenic and tumor suppressive. Because a single tumor is composed of a heterogenous population of cells, activation of Notch in different cell populations may have different effects on the growth of the tumor as a whole. For example within a single breast tumor, activated Notch signaling specifically in breast cancer cells may be oncogenic, whereas activated Notch signaling in endothelial cells within the breast tumor may block angiogenesis and thus be tumor suppressive. A recent study has found that Jagged1-transduced squamous cell carcinoma cells, when subcutaneously coimplanted with endothelial cells in a Matrigel-containing scaffold, exhibit enhanced tumor angiogenesis compared with untransduced controls.142 Although this finding suggests that Jagged1-expressing tumor cells can activate Notch signaling in neighboring endothelial cells to stimulate tumor angiogenesis, activation of the Notch pathway specifically in endothelial cells in this tumor model was not demonstrated.142 Additional studies are needed to elucidate whether endothelial-specific Notch activation is oncogenic or tumor suppressive in vivo.
Recent years have seen major advances in our understanding of the dual function of Notch signaling as both oncogene and tumor suppressor. Use of this knowledge in the near future will hopefully lead to the rational development of clinical therapeutics for the treatment of Notch-related malignancies.
Prepublished online as Blood First Edition Paper, November 15, 2005; DOI 10.1182/blood-2005-08-3329.
Supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society, the Heart and Stroke Foundation of British Columbia and the Yukon, the Canadian Institutes of Health Research, the Stem Cell Network, and the Canadian Breast Cancer Foundation (work in the laboratory of A.K.). K.G.L. was supported by a Doctoral Research Award from the Canadian Institutes of Health Research and a Predoctoral Fellowship Award from the Department of the Army (DAMD17-01-1-0164). The U.S. Army Medical Research Acquisition Activity, Fort Detrick, MD, is the awarding and administering acquisition office. A.K. is a Scholar of the Michael Smith Foundation for Health Research.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal