This study describes a novel application of HLAMatchmaker to determine platelet compatibility in 16 alloimmunized patients with aplastic anemia refractory to random donor platelet transfusions. HLAMatchmaker is a software algorithm that predicts HLA compatibility by identifying immunogenic epitopes represented by amino acid triplets in antibody-accessible regions of human leukocyte antigen (HLA) molecules and determines the number of triplet mismatches (TMMs) and highly immunogenic triplet mismatches (HIMMs). Corrected count increments (CCIs) and molecular HLA typing were available for 523 transfusions. Conventional compatibility assessment based on cross-reactive group (CREG) determination was not predictive of transfusion outcome. Low HIMMs and TMMs numbers were associated with a higher likelihood of satisfactory (CCIs ≥ 8) compared with unsatisfactory (CCIs < 8) outcomes (median HIMMs = 4 vs 6, p2 value < .001; median TMMs = 11 vs 13, p2 value < .001). Although receiver operator characteristic curves revealed that HIMMs or TMMs number are not powerful predictors of individual transfusion outcome, a threshold of at least 3 HIMMs or at least 9 TMMs appeared to be associated with successful transfusions. Triplet-matched transfusions were successful, regardless of CREG matching. Our data validate HLAMatchmaker for platelet transfusions and demonstrate its potential to refine and expand donor selection for HLA-alloimmunized patients.
Introduction
Protracted platelet transfusion therapy is frequently complicated by refractoriness. Poor platelet increments are usually secondary to coexisting nonimmune causes such as fever, splenomegaly, sepsis, disseminated intravascular coagulation, or medications. However, immune refractoriness occurs in multiparous women and patients receiving prolonged blood component support. Immunologically mediated unresponsiveness to platelet transfusions typically results from the development of alloantibodies against HLA-A and -B locus antigens or, less frequently, antibodies to human platelet antigens. Megakaryocytes synthesize HLA-A and -B antigens, and those expressed on the platelet surface are primarily of intrinsic origin.1 The determination of donor-recipient compatibility for platelet transfusions is limited to comparisons of HLA-A and -B loci. Matching for HLA-C locus antigens is deemed unnecessary, as HLA-C antigens are expressed at very low levels on platelets and are not considered to play a role in immunologic platelet refractoriness.2
Guidelines for selecting platelets for HLA-alloimmunized patients have been described in excellent reviews by McFarland,3 Slichter,4 Schiffer,5 and Sacher et al.6 Selection of HLA antigen-compatible donors is essential for ensuring satisfactory posttransfusion platelet survivals; donors should lack the antigens against which the patient has made antibodies.3-6 When HLA-identical platelet donors are unavailable, the selection of HLA-matched or selectively mismatched platelet donors is based on the segregation of HLA antigens into serologically cross-reactive groups (CREGs).7 Attention to linked HLA specificities (Bw4/Bw6) is beneficial in some patients unresponsive to cross-reactive platelets, while mismatching for poorly expressed HLA-B locus antigens (eg, HLA-B44, B45) may be acceptable for certain donor-recipient pairs.8,9 These broad strategies have been implemented to decrease the size of the donor pool required to support all alloimmunized patients with matched products. However, this strategy is not always successful; previous studies report unsatisfactory increments in up to 40% of CREG-matched transfusions.7-10 The probability of finding 4 antigen-matched or CREG-matched products for patients with uncommon HLA phenotypes is low. Also, CREG matching gives more importance to public epitopes over private epitopes and is based on serologic data that has been largely supplanted by our current understanding of HLA polymorphisms at the structural level. Given the limitations of CREG matching, a better tool is required for the selection of HLA-compatible donors. Pretransfusion testing, such as platelet crossmatching assays, may help predict some transfusion failures, and it has been used by some centers to provide compatible platelets for patients with immune refractoriness.10-14 However, the reliability and practicability of crossmatching remains to be elucidated.
HLAMatchmaker is a computer program developed to determine HLA compatibility at the molecular level independently of identical or CREG-compatible HLA antigen matching, with the purpose of increasing the pool of donors whose products may be compatible for a given recipient.15 This is done by matching donor and recipient with the understanding that immunogenic epitopes are represented by amino acid triplets on exposed parts (ie, antibody-accessible positions) of protein sequences of HLA chains. The program computes the total number of triplet mismatches (TMMs) between the donor and recipient HLA repertoire. Analysis of serologic reactivity patterns of highly allosensitized patients using the triplet algorithm has helped to define the relative immunogenicity of mismatched triplets; the program can now identify the subset of highly immunogenic mismatches (HIMMs).16 This algorithm previously has been validated for the prediction of kidney transplant survival.17
Platelet refractoriness is a frequent complication in patients with severe aplastic anemia. HLA alloantibodies can be demonstrated in 30% to 40% of these patients. These patients represent an ideal population to study the effect of HLA matching on transfusion outcome since other confounding factors such as fever, splenomegaly, sepsis, or disseminated intravascular coagulation are rarely present in the natural course of the disease. In this retrospective study, we describe the novel application of HLAMatchmaker to determine platelet compatibility in 16 alloimmunized patients with aplastic anemia refractory to random donor platelet transfusion. All patients had received long-term support with apheresis platelets from donors selected according to the standard practice of enumerating the number of mismatched HLA antigens or CREGs. Since donor-recipient pairing had been based on classic serologic criteria of HLA matching, this database provided a logical platform to assess whether the application of a structurally based algorithm for donor selection could improve the prediction of transfusion outcome by segregating donors with structurally compatible, albeit distinct, HLA-antigen phenotypes. The clinical response to platelet transfusion was assessed by calculating the corrected count increments (CCIs), which normalize posttransfusion platelet counts according to patient's blood volume and platelet dose.18,19 HLAMatchmaker defined donor-recipient HLA-A and -B compatibility at the triplet level and determined the number of TMMs and HIMMs for all donor-recipient pairs. We then verified the HLAMatchmaker algorithm by correlating TMMs and HIMMs with CCIs and compared the results with matching, using conventional CREG criteria.
Patients, materials, and methods
Patient characteristics
HLA-alloimmunized patients were identified from among the severe aplastic anemia patients who received long-term platelet transfusion support at the National Institutes of Health Clinical Center from January 1999 through December 2002. All patients were molecularly HLA typed by the polymerase chain reaction-sequence specific primer (PCR-SSP) method.20 Approval was obtained from the National Heart, Lung, and Blood Institute institutional review board for these studies; informed consent was provided according to the Declaration of Helsinki.
Platelet transfusions
The selection of platelet donors for these alloimmunized patients was based on conventional criteria, that is, those with the least number of mismatches for HLA antigens and/or CREG were preferred. Transfusion responses were assessed calculating the 15 minutes-to-1-hour CCIs using the following formula: CCI = ([Posttransfusion platelet count/μL - pretransfusion platelet count/μL] × BSA) ÷ (Number of platelets transfused × 1011). The body surface area (BSA) was calculated according to George and Gehan.21
HLAMatchmaker
HLAMatchmaker considers each HLA antigen as a distinct string of polymorphic triplets that can induce specific alloantibodies. Sensitized patients do not produce alloantibodies against triplets present on their own HLA molecules. The algorithm assesses donor-recipient compatibility through intralocus and interlocus comparisons and determines what triplets in antibody-accessible positions on mismatched HLA molecules are different or shared between donor and patient. As an example, Table 1 shows the triplets in sequence positions for patient 15 with the HLA-A*0207, A*2402; B*0705/6, B*1502 phenotype. These constitute the self-triplet repertoire. This patient types serologically as HLA-B7, although molecular typing identifies the patient as having HLA-B*0705/6. In this example, we show how representative antigens of the B7 CREG are matched at the structural level. The HLA-B22 splits are well matched for this patient: B*5501 has no mismatched triplets, B*5601 has one mismatched triplet (151aRv), and B*5401 is mismatched for 45Ge and 151aRv. Several B7 cross-reactive antigens have more structural differences for this patient: the HLA-B40 splits B*4001 (equivalent to HLA-B60) and B*4002 (equivalent to HLA-B61) have 7 and 5 mismatched triplets, respectively. Moreover, B*2705 (equivalent to HLA-B27) has 8 mismatched triplets. Recent studies have shown that patients' antibody reactivity correlates with the number of mismatched triplets against donor HLA antigens.22
The molecular identification and enumeration of amino acid triplets among HLA alleles belonging to the same serologic family and cross-reactive groups for patient 15
. | Position: . | 9 . | 11 . | 14 . | 16 . | 41 . | 45 . | 56 . | 59 . | 62 . | 66 . | 70 . | 74 . | 76 . | 80 . | 82 . | 90 . | 105 . | 107 . | 127 . | 131 . | 138 . | 142 . | 144 . | 147 . | 149 . | 151 . | 156 . | 158 . | 163 . | 166 . | 171 . | 177 . | 180 . | 183 . | 186 . | 193 . | 199 . | Trp . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | A*0207 | F | Sv | R | Gr | A | Me | G | Y | Ge | rKv | aHs | H | Vd | gTI | IRg | A | S | W | K | R | M | mT | tKh | W | aAh | aHv | L | A | T | Ew | Y | Et | Q | A | K | Av | A | — |
| Patient | A*2402 | S | Sv | R | Gr | A | Me | G | Y | Ee | gKv | aHs | D | En | RIa | aLr | A | S | G | K | R | M | mI | tKr | W | aAh | aHv | Q | A | T | Dg | Y | Et | Q | P | K | Pi | A | — |
| Patient | B*0705 | Y | Sv | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | R | T | tI | tQr | W | aAr | aRe | R | A | E | Ew | Y | Dk | E | P | K | Pi | A | — |
| Patient | B*1502 | Y | Am | R | Gr | A | Ma | G | Y | Rn | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | L | Ew | Y | Et | Q | P | K | Pi | A | — |
| B7 CREG | B*5501 | Y | Am | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | T | Ew | Y | Et | Q | P | K | Pi | A | 0 |
| B7 CREG | B*5601 | Y | Am | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | L | Ew | Y | Et | Q | P | K | Pi | A | 1 |
| B7 CREG | B*5401 | Y | Am | R | Gr | A | Ge | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | T | Ew | Y | Et | Q | P | K | Pi | A | 2 |
| B7 CREG | B*4002 | H | Sv | R | Gr | T | Ke | G | Y | Re | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | R | T | tI | tQr | W | aAr | aRv | L | A | E | Ew | Y | Et | Q | P | K | Pi | A | 5 |
| B7 CREG | B*4001 | H | Am | R | Gr | T | Ke | G | Y | Re | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | R | T | tI | sQr | L | aAr | aRv | L | A | E | Ew | Y | Dk | E | P | K | Pi | A | 7 |
| B7 CREG | B*2705 | H | Sv | R | Gr | A | Ee | G | Y | Re | qIc | aKa | D | Ed | rTI | ILr | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | E | Ew | Y | Et | Q | P | K | Pi | A | 8 |
| Non Cross Reactive | B*1401 | Y | Sb | R | Gr | A | Ee | G | Y | Rn | qIc | tNt | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | T | Ew | H | Et | Q | P | K | Pi | A | 2 |
| Non Cross Reactive | B*3801 | Y | Sv | R | Gr | A | Ee | G | Y | Rn | qIc | tNt | Y | En | rla | aLr | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | T | T | Ew | Y | Et | Q | P | K | Pi | A | 3 |
. | Position: . | 9 . | 11 . | 14 . | 16 . | 41 . | 45 . | 56 . | 59 . | 62 . | 66 . | 70 . | 74 . | 76 . | 80 . | 82 . | 90 . | 105 . | 107 . | 127 . | 131 . | 138 . | 142 . | 144 . | 147 . | 149 . | 151 . | 156 . | 158 . | 163 . | 166 . | 171 . | 177 . | 180 . | 183 . | 186 . | 193 . | 199 . | Trp . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | A*0207 | F | Sv | R | Gr | A | Me | G | Y | Ge | rKv | aHs | H | Vd | gTI | IRg | A | S | W | K | R | M | mT | tKh | W | aAh | aHv | L | A | T | Ew | Y | Et | Q | A | K | Av | A | — |
| Patient | A*2402 | S | Sv | R | Gr | A | Me | G | Y | Ee | gKv | aHs | D | En | RIa | aLr | A | S | G | K | R | M | mI | tKr | W | aAh | aHv | Q | A | T | Dg | Y | Et | Q | P | K | Pi | A | — |
| Patient | B*0705 | Y | Sv | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | R | T | tI | tQr | W | aAr | aRe | R | A | E | Ew | Y | Dk | E | P | K | Pi | A | — |
| Patient | B*1502 | Y | Am | R | Gr | A | Ma | G | Y | Rn | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | L | Ew | Y | Et | Q | P | K | Pi | A | — |
| B7 CREG | B*5501 | Y | Am | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | T | Ew | Y | Et | Q | P | K | Pi | A | 0 |
| B7 CREG | B*5601 | Y | Am | R | Gr | A | Ee | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | L | Ew | Y | Et | Q | P | K | Pi | A | 1 |
| B7 CREG | B*5401 | Y | Am | R | Gr | A | Ge | G | Y | Rn | qIy | aQa | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | T | Ew | Y | Et | Q | P | K | Pi | A | 2 |
| B7 CREG | B*4002 | H | Sv | R | Gr | T | Ke | G | Y | Re | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | R | T | tI | tQr | W | aAr | aRv | L | A | E | Ew | Y | Et | Q | P | K | Pi | A | 5 |
| B7 CREG | B*4001 | H | Am | R | Gr | T | Ke | G | Y | Re | qIs | tNt | Y | Es | rNI | IRg | A | P | G | N | R | T | tI | sQr | L | aAr | aRv | L | A | E | Ew | Y | Dk | E | P | K | Pi | A | 7 |
| B7 CREG | B*2705 | H | Sv | R | Gr | A | Ee | G | Y | Re | qIc | aKa | D | Ed | rTI | ILr | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | A | E | Ew | Y | Et | Q | P | K | Pi | A | 8 |
| Non Cross Reactive | B*1401 | Y | Sb | R | Gr | A | Ee | G | Y | Rn | qIc | tNt | D | Es | rNI | IRg | A | P | G | N | S | T | tI | tQr | W | aAr | aRe | L | A | T | Ew | H | Et | Q | P | K | Pi | A | 2 |
| Non Cross Reactive | B*3801 | Y | Sv | R | Gr | A | Ee | G | Y | Rn | qIc | tNt | Y | En | rla | aLr | A | P | G | N | S | T | tI | tQr | W | aAr | aRv | L | T | T | Ew | Y | Et | Q | P | K | Pi | A | 3 |
The molecular identification and enumeration of amino acid triplets among HLA alleles belonging to the same serological family and cross-reactive groups for patient 15: the HLA matching algorithm is based on comparisons of linear sequences of amino acid triplets in antibody-accessible positions as motifs for potentially immunogenic epitopes. Each HLA antigen represents a distinct string of polymorphic triplets. The self-triplet repertoire for patient 15, with the HLA-A*0207, A*2402; B*0705/6, B*1502 phenotype is shown. Using HLA-B*0705 (serologically HLA-B7) as an example, this table also shows how representative antigens of the B7 CREG are matched at the structural level. While the HLA-B22 splits B*5501, B*5601, and B*5401 are well-matched, other B7-crossreactive antigens like B*4001, B*4002, and B*2705 have many mismatched triplets (see text for details). Last column: Trp indicates total number of triplet mismatches between HLA-B*0705 and each allele analyzed. The positions on top of the table represent polymorphic sites of HLA molecules accessible to antibodies. Each triplet is designated by its amino acid composition around a given position in the amino acid sequence. Amino acid residues are marked with the standard letter code; an uppercase corresponds to the residue in the numbered position, whereas the lowercase letters describe the nearest neighboring residues. For instance, the triplet 151 aRv represents an arginine (R) in position 151 with an alanine (a) in position 150 and a valine (v) in position 152. Many triplets are marked with one or two residues only, because their neighboring residues are the same on all HLA class I chains and they are, therefore, not shown.
This example illustrates that some CREG matches are structurally compatible, but others have considerable numbers of mismatched triplets. Conversely, certain antigens that are not considered cross-reacting for this patient's HLA antigens have fewer mismatched triplets. For instance, the non-cross-reacting B*1401 (equivalent to HLA-B64) and B*3801 are structurally much more compatible than the cross-reacting B*4001, B*4002, and B*2705 (Table 1).
The HLAMatchmaker (Ser 1.1) version used for this study is based on serologically defined HLA antigens. Program instructions, as well as newer molecular versions, can be downloaded from the website at http://tpis.upmc.edu.23 The serologic version assigns triplets from molecularly defined alleles corresponding to serologically defined antigens. Since the HLA types of our platelet donors and patients were molecularly defined, we used the 2001 HLA Dictionary to determine the serologic equivalents for all antigens.24
We compared the triplet repertoire of the patient with those of the donor and determined the number of mismatched triplets for each donor-patient pair. Previous studies have shown that HLA-A, -B antigen mismatched kidney transplants with no or few mismatched triplets have the same graft survival rates as the zero-antigen mismatches.17 HLA antigens of donor-recipient pairs also were compared using CREG-matching criteria. Transfusions were considered CREG compatible if the donor HLA antigens belonged to the same CREGs as any of patient's HLA antigens. Transfusions were considered CREG incompatible when one or more donor HLA antigens belonged to CREGs not represented by any of the patient's HLA antigens. Since many triplets are equivalent to known public epitopes, we have compared triplet matching with CREG matching as described by Duquesnoy et al.7
Statistical analysis
Median accompanied by the first and third quartile was calculated as measures of central tendency and dispersion unless otherwise specified. The association between TMMs and HIMMs with CCIs was analyzed using the χ2-test or Fisher exact test, as appropriate. Unpaired 2-tailed Student t test assuming unequal variance was applied unless otherwise stated to evaluate the level of statistical significance between 2 data sets, and 2-tailed P values (p2 value) are reported. Receiver operator characteristic (ROC) curves25 were constructed by computing the sensitivity and specificity using different thresholds of HIMMs and TMMs for predicting transfusion outcomes (ie, CCIs). Analyses were applied to the complete data set and to a limited data set in which data from patient 11, who received a disproportionably large number of transfusions (155), were removed to exclude an undue influence of this patient on the analysis.
Results
Patient characteristics and transfusions
Sixteen patients with aplastic anemia, a history of consistently poor increments following randomly selected platelet transfusions, and laboratory evidence of HLA alloimmunization were identified. Their characteristics are shown in Table 2. Although a total of 964 platelet products were transfused to this cohort of patients, pretransfusion and posttransfusion platelet counts necessary to calculate CCI values were available for only 523 (54%) transfusions from 314 donors with HLA types determined by DNA methods. Therefore, the analysis was limited to these platelet products. The number of transfusions received by individual patients varied remarkably, with one patient (patient 11) receiving 155 transfusions, corresponding to approximately 30% of the transfusions analyzed. The HLA-A and -B phenotype of each patient and “acceptable” donor HLA-A and -B antigens is shown in Table 3.
Details about patients and platelet transfusions
Characteristic . | Data . |
|---|---|
| No. patients with aplastic anemia | 16 |
| Age, y, median (range) | 40 (11-72) |
| No. female (%) | 10 (63) |
| Total transfusions | 964 |
| Transfusions with CCIs available | 523 |
| No. donors | 314 |
| Transfusion per patient, median (range) | 15 (6-155) |
Characteristic . | Data . |
|---|---|
| No. patients with aplastic anemia | 16 |
| Age, y, median (range) | 40 (11-72) |
| No. female (%) | 10 (63) |
| Total transfusions | 964 |
| Transfusions with CCIs available | 523 |
| No. donors | 314 |
| Transfusion per patient, median (range) | 15 (6-155) |
Identification of “acceptable” HLA-A and -B antigens by HLAMatchmaker
. | HLA type recipient . | . | . | . | HLA type donor . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | A . | A . | B . | B . | Acceptable HLA-A antigens . | Acceptable HLA-B antigens . | ||||
| 1 | A2 | A29 | B7 | B44 | A 69, 68, 74, 31, 32, 33 | B 55, 56, 70, 72, 42, 45, 50, 54, 61, 71, 75, 13, 41, 48, 49, 59, 62, 76, 77, 18, 35, 38, 47, 60, 67,* 39,* 64,* 8,* 65* | ||||
| 2 | A29 | A68 | B14 | A 69, 33, 34 | B 39, 65, 67, 55, 71 | |||||
| 3 | A2 | A11 | B7 | B51 | A 69, 68, 3, 32,* 74,* 25,* 34,* 66* | B 55, 78, 35, 53, 56, 71, 52, 54, 70, 72, 75, 76, 77, 18, 42, 58, 81, 59,* 64,* 65,* 67,* 38,* 39* | ||||
| 4 | A*0101 | A*0201 | B*0702 | A 36, 69, 68, 74* | B 55, 42, 81, 67* | |||||
| 5 | A24 | B7 | B62 | A 23 | B 76, 55, 70, 72, 75, 77, 56, 71, 42, 46, 54, 63, 59,* 64,* 67* | |||||
| 6 | A*0301 | B*1402 | B*4701 | B 39, 61, 65, 27, 55, 37, 54, 56, 67, 73, 70,* 71,* 72,* 18,* 50,* 75,* 62* | ||||||
| 7 | A*3101 | A*6901 | B*0702 | B*5501 | A 68, 29, 33, 74, 30 | B 54, 56, 42, 81, 67,* 39,* 64* | ||||
| 8 | A2 | A24 | B7 | B40 | A 23, 68, 69, 32,* 74* | B 48, 61, 81, 55, 41, 42, 54, 56, 49,* 50,* 59,* 67,* 70,* 72,* 18,* 38,* 39,* 64,* 71,* 75,* 77* | ||||
| 9 | A*2405 | A*2407 | B*3505 | B*5604 | A 23 | B 53, 54, 55, 51, 71, 78, 18, 42, 58, 70, 72, 75, 76, 77, 59,* 67,* 38,* 39,* 64* | ||||
| 10 | A2 | A30 | B7 | B40 | A 69, 68, 74, 31, 29, 32, 3* | B 48, 61, 81, 55, 41, 42, 54, 56, 50,* 67,* 70,* 72,* 18,* 39,* 64,* 71,* 75* | ||||
| 11 | A*2602 | A*3101 | B*1301 | B*5102 | A 25, 33, 34, 43, 66, 29, 74, 32 | B 52, 53, 77, 49, 63, 35, 57, 58, 62, 75, 78, 59,* 38* | ||||
| 12 | A*0201 | A*0301 | B*0702 | B*2705/13 | A 69, 68, 29,* 31,* 32,* 34,* 74* | B 55, 42, 54, 56, 73, 81, 67,* 39,* 64,* 65* | ||||
| 13 | A*3201 | A*7401/02 | B*1503 | B*5201 | B 70, 72, 75, 77, 18, 35, 51, 53, 63, 76, 78, 46, 49, 50, 57, 58, 71, 59,* 64* | |||||
| 14 | A2 | A11 | B51 | A 69, 68, 3, 74,* 32,* 34,* 66* | B 53, 52, 77, 58, 78, 59* | |||||
| 15 | A*0207 | A*2402 | B*0705/06 | B*1502 | A 23, 68, 69 | B 55, 70, 72, 75, 76, 77, 56, 71, 54, 63, 42, 46, 48, 49, 50, 52, 57, 81, 59,* 64,* 67,* 38,* 39,* 65* | ||||
| 16 | A1 | A3 | B7 | B8 | A 36, 11, 74* | B 42, 55, 54, 56, 67, 81, 39, 64 | ||||
. | HLA type recipient . | . | . | . | HLA type donor . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | A . | A . | B . | B . | Acceptable HLA-A antigens . | Acceptable HLA-B antigens . | ||||
| 1 | A2 | A29 | B7 | B44 | A 69, 68, 74, 31, 32, 33 | B 55, 56, 70, 72, 42, 45, 50, 54, 61, 71, 75, 13, 41, 48, 49, 59, 62, 76, 77, 18, 35, 38, 47, 60, 67,* 39,* 64,* 8,* 65* | ||||
| 2 | A29 | A68 | B14 | A 69, 33, 34 | B 39, 65, 67, 55, 71 | |||||
| 3 | A2 | A11 | B7 | B51 | A 69, 68, 3, 32,* 74,* 25,* 34,* 66* | B 55, 78, 35, 53, 56, 71, 52, 54, 70, 72, 75, 76, 77, 18, 42, 58, 81, 59,* 64,* 65,* 67,* 38,* 39* | ||||
| 4 | A*0101 | A*0201 | B*0702 | A 36, 69, 68, 74* | B 55, 42, 81, 67* | |||||
| 5 | A24 | B7 | B62 | A 23 | B 76, 55, 70, 72, 75, 77, 56, 71, 42, 46, 54, 63, 59,* 64,* 67* | |||||
| 6 | A*0301 | B*1402 | B*4701 | B 39, 61, 65, 27, 55, 37, 54, 56, 67, 73, 70,* 71,* 72,* 18,* 50,* 75,* 62* | ||||||
| 7 | A*3101 | A*6901 | B*0702 | B*5501 | A 68, 29, 33, 74, 30 | B 54, 56, 42, 81, 67,* 39,* 64* | ||||
| 8 | A2 | A24 | B7 | B40 | A 23, 68, 69, 32,* 74* | B 48, 61, 81, 55, 41, 42, 54, 56, 49,* 50,* 59,* 67,* 70,* 72,* 18,* 38,* 39,* 64,* 71,* 75,* 77* | ||||
| 9 | A*2405 | A*2407 | B*3505 | B*5604 | A 23 | B 53, 54, 55, 51, 71, 78, 18, 42, 58, 70, 72, 75, 76, 77, 59,* 67,* 38,* 39,* 64* | ||||
| 10 | A2 | A30 | B7 | B40 | A 69, 68, 74, 31, 29, 32, 3* | B 48, 61, 81, 55, 41, 42, 54, 56, 50,* 67,* 70,* 72,* 18,* 39,* 64,* 71,* 75* | ||||
| 11 | A*2602 | A*3101 | B*1301 | B*5102 | A 25, 33, 34, 43, 66, 29, 74, 32 | B 52, 53, 77, 49, 63, 35, 57, 58, 62, 75, 78, 59,* 38* | ||||
| 12 | A*0201 | A*0301 | B*0702 | B*2705/13 | A 69, 68, 29,* 31,* 32,* 34,* 74* | B 55, 42, 54, 56, 73, 81, 67,* 39,* 64,* 65* | ||||
| 13 | A*3201 | A*7401/02 | B*1503 | B*5201 | B 70, 72, 75, 77, 18, 35, 51, 53, 63, 76, 78, 46, 49, 50, 57, 58, 71, 59,* 64* | |||||
| 14 | A2 | A11 | B51 | A 69, 68, 3, 74,* 32,* 34,* 66* | B 53, 52, 77, 58, 78, 59* | |||||
| 15 | A*0207 | A*2402 | B*0705/06 | B*1502 | A 23, 68, 69 | B 55, 70, 72, 75, 76, 77, 56, 71, 54, 63, 42, 46, 48, 49, 50, 52, 57, 81, 59,* 64,* 67,* 38,* 39,* 65* | ||||
| 16 | A1 | A3 | B7 | B8 | A 36, 11, 74* | B 42, 55, 54, 56, 67, 81, 39, 64 | ||||
Although most acceptable mismatches also can be identified by CREG matching, many antigens (*) are considered incompatible by conventional CREG-matching criteria.
HLAMatchmaker-driven identification of donors with acceptable HLA-A and -B mismatches
Table 3 shows additional “acceptable” donor HLA-A and -B antigens identified by HLAMatchmaker. These antigens, mismatched for 0 to 3 triplets, are potentially HLA compatible based on triplotype repertoire, especially in the absence of corresponding antibodies in the patient's serum. Not surprisingly, most of these “acceptable” antigens belong to the same CREGs as the recipient antigens. Conversely, some CREG-mismatched antigens were fully compatible or minimally mismatched at the triplet level (marked with asterisks in Table 3). The patient's HLA phenotype influences the number of donor antigens with 0 to 3 triplet mismatches that can be identified. If patients have homozygous HLA phenotypes or closely cross-reacting antigens, few HLA antigens may be identified with 0 triplet mismatches. A median of 21 (first and third quartile, 18 and 26) “acceptable” antigens was identified for patients with heterozygous A and B alleles (n = 11), while those homozygous at either locus (n = 5) had 13 (first and third quartile, 8 and 16, t test p2 value < .01) “acceptable” antigens.
CREG-matching and transfusion responses
Only 170 (33%) of the 523 transfusions analyzed (Table 4) were fully CREG compatible (range, 7% to 83% of total transfusions for individual patients). The remaining transfusions had at least one donor antigen that belonged to a disparate CREG. The median platelet increments for the CREG-compatible and -incompatible transfusions were not significantly different (t test p2 value = .08), although a trend was noted toward better CCIs with CREG-compatible transfusions. Platelet transfusions yielding CCIs of at least 7.5 are generally considered satisfactory.6 Seventy-four percent of the fully CREG-matched transfusions and 71% of the CREG-incompatible transfusions examined in this study had CCIs greater than 8. Therefore, CREG-based pairing of donor and recipient was associated with minimal likelihood of clinical benefit when this assessment was done at the level of transfusion outcome, as determined by the 15-minute-to-1-hour CCIs.
Transfusions grouped by conventional CREG-matching criteria
. | N . | Median CCIs . |
|---|---|---|
| Transfusions analyzed | 523 | |
| CREG-compatible platelet transfusions | 170 | 12.4 (7.7-18.4) |
| CREG-incompatible platelet transfusions | 353 | 12.2 (7.2-16.6) |
. | N . | Median CCIs . |
|---|---|---|
| Transfusions analyzed | 523 | |
| CREG-compatible platelet transfusions | 170 | 12.4 (7.7-18.4) |
| CREG-incompatible platelet transfusions | 353 | 12.2 (7.2-16.6) |
HLA antigens of donor-recipient pairs were compared using cross-reactive groups. Transfusions were considered CREG incompatible when one or more donor HLA antigen fit in CREGs not represented by any of the patient's HLA antigens. Median CCIs are presented with the first and third quartile values in parentheses. The 2 cohorts were compared using unpaired, 2-tailed Student t test (p2 value = .08).
Triplet-matching and transfusion responses
The relationship between triplet mismatches and platelet increments was examined. The transfusion responses of fully matched donor-recipient pairs (ie, 0 triplet mismatches) were compared with the responses observed when donor HLA antigens were mismatched for no more than 3 or greater than 3 TMMs. The best outcomes (median CCIs = 18.4; first-third quartile, 11.3-23.4) were expectedly noted with fully matched transfusions. However, only 27 transfusions were fully matched (5% of all transfusions), underscoring the difficulty in finding fully matched donors using conventional matching techniques. Satisfactory CCIs (median = 13.3; first-third quartile, 9.2-19.2) were obtained for 72 transfusions from donors with no more than 3 TMMs, representing 14% of the transfusions. A much larger number (184, 35%) of transfusions were mismatched for up to 3 HIMMs (median CCIs, 14.0; first-third quartile, 9.8-19.2). Thus, acceptable mismatches allow the accrual of an incremental pool of donors likely to provide clinically useful products.
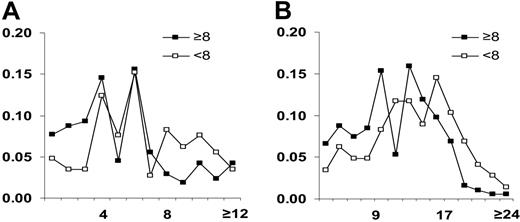
Transfusion outcomes were next defined as satisfactory (CCIs ≥ 8) or unsatisfactory (CCIs < 8) according to published guidelines.6 Seventy-two percent (n = 378) of the transfusions yielded CCIs of at least 8 (Tables 5 and 6). Transfusions yielding CCIs of at least 8 were associated with significantly less HIMMs (median = 4; first-third quartile, 2-6), compared to those associated with unsatisfactory increments (median HIMMs = 6; first-third quartile, 4-10, t test p2 value < .001). Similarly, TMMs were significantly decreased in satisfactory (median = 11; first-third quartile, 6-14), compared to unsatisfactory transfusions (median 13; first-third quartile, 9-18; p2 value < .001). Because of the exaggerated influence that patient 11, who received 155 transfusions, might have had on the analysis, we repeated the same analysis, excluding data from this patient. We then investigated the different ranges of mismatches that would be most predictive of good transfusion outcomes. A frequency analysis was entertained to evaluate the proportion of the total transfusions with acceptable (≥ 8 CCIs; black boxes, Figure 1) or not acceptable (< 8 CCIs; white boxes, Figure 1) outcomes, occurring in association with each number of HIMMs (Figure 1A) or TMMs (Figure 1B). We observed differences only when HIMMs were no more than 3; between 4 and 7 HIMMs the occurrence of satisfactory and unsatisfactory transfusion was similar, while at least 8 HIMMs were more frequently associated with poor outcomes (P value < .001 in χ2 test). Similarly, better outcomes were observed with no more than 9 TMMs; between 10 and 16 TMMs there was still a significant trend toward better outcomes but with some overlapping values (Figure 1B; Fisher exact test comparing ≤ 16 to > 16, p2 value < .001); at least 17 TMMs were consistently associated with a higher frequency of poor outcomes (χ2 test, P value < .001). Therefore, up to 16 TMMs may represent a reasonable, though less stringent, cutoff value for the selection of donors with a good likelihood of yielding satisfactory transfusion outcomes. There was no HLA locus-specific association with transfusion outcome, as mismatches at the HLA-A or -B locus bore the same weight. Thus, a defined threshold of acceptable mismatches may guide donor selection, as previously observed in renal transplantation.17
Relationship between triplet mismatches and corrected count increments
No. transfusions analyzed . | . | HIMMs . | . | . | TMMs . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCIs < 8 . | CCIs ≥ 8 . | Median when CCIs < 8 . | Median when CCIs ≥ 8 . | p2 value . | Median when CCIs < 8 . | Median when CCIs ≥ 8 . | p2 value . | |||||
| 145 | 378 | 6 (4-10) | 4 (2-6) | < .001 | 13 (9-18) | 11 (6-14) | < .001 | |||||
| 122* | 246* | 6 (4-9) | 4 (1-6) | < .001 | 13 (8-18) | 12 (4-15) | = .001 | |||||
No. transfusions analyzed . | . | HIMMs . | . | . | TMMs . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCIs < 8 . | CCIs ≥ 8 . | Median when CCIs < 8 . | Median when CCIs ≥ 8 . | p2 value . | Median when CCIs < 8 . | Median when CCIs ≥ 8 . | p2 value . | |||||
| 145 | 378 | 6 (4-10) | 4 (2-6) | < .001 | 13 (9-18) | 11 (6-14) | < .001 | |||||
| 122* | 246* | 6 (4-9) | 4 (1-6) | < .001 | 13 (8-18) | 12 (4-15) | = .001 | |||||
N = 523. CCIs were calculated for all platelet transfusions. HLAMatchmaker examined HLA compatibility at the triplet level for all donor-recipient pairs and determined the number of highly immunogenic triplet mismatches as well as the total number of triplet mismatches. Transfusions were divided into 2 groups: CCIs less than 8 or CCIs at least 8. The median number of HIMMs and TIMMs are presented, with the first and third quartile values (in parentheses). The 2 cohorts were compared using unpaired, 2-tailed Student t test. The median number of HIMMs and TIMMs are presented, with the first and third quartile values in parentheses.
Data presented for all transfusions with the exclusion of patient 11
Triplet mismatches subgroups for transfusions with CCI below 8 or at least 8
. | CCI . | . | P < .001 in χ2 test . | |
|---|---|---|---|---|
. | < 8 . | ≥ 8 . | . | |
| HIMMs | ||||
| No more than 3 | 30 (0.21) | 154 (0.41) | 184 | |
| 4 to 7 | 55 (0.38) | 152 (0.40) | 207 | |
| More than 8 | 60 (0.41) | 72 (0.19) | 132 | |
| Total | 145 | 378 | 523 | |
| TMMs | ||||
| No more than 9 | 40 (0.28) | 176 (0.47) | 216 | |
| 10 to 16 | 57 (0.39) | 150 (0.40) | 207 | |
| More than 17 | 48 (0.33) | 52 (0.14) | 100 | |
| Total | 145 | 378 | 523 | |
. | CCI . | . | P < .001 in χ2 test . | |
|---|---|---|---|---|
. | < 8 . | ≥ 8 . | . | |
| HIMMs | ||||
| No more than 3 | 30 (0.21) | 154 (0.41) | 184 | |
| 4 to 7 | 55 (0.38) | 152 (0.40) | 207 | |
| More than 8 | 60 (0.41) | 72 (0.19) | 132 | |
| Total | 145 | 378 | 523 | |
| TMMs | ||||
| No more than 9 | 40 (0.28) | 176 (0.47) | 216 | |
| 10 to 16 | 57 (0.39) | 150 (0.40) | 207 | |
| More than 17 | 48 (0.33) | 52 (0.14) | 100 | |
| Total | 145 | 378 | 523 | |
Values correspond to number (ratio in parentheses) of transfusions within each HIMM or TMM category.
Based on the previous observation, transfusions were segregated according to the number of HIMMs (≤ 3 and > 3) or TMMs (≤ 9 and > 9) (Table 7). The median CCIs for the HIMMs no more than 3 and greater than 3 groups were 14.0 (first and third quartile = 10 and 19, respectively) and 11.2 (first and third quartile = 6 and 16, respectively; t test p2 value < .001). Highly significant differences between HIMMs no more than 3 and greater than 3 subgroups also were noted in CREG-compatible transfusions and, to a lesser degree, in CREG-incompatible transfusions. Similarly, the TMM dichotomy, no more than 9 compared to greater than 9, resulted in significantly improved outcome (t test p2 value < .001). Similar results were seen when data from patient 11 were not used for the analysis (data not shown). This proposed threshold of no more than 3 HIMMs or greater than 9 TMMs is clinically relevant. It increased by 7-fold or greater the proportion of potential donors: from 5% of all transfusions when complete allelic match was used as a selection criterion to 35% and 41% when a threshold of no more than 3 HIMMs or no more than 9 TMMs, respectively, was applied. In addition, if the less stringent threshold of no more than 16 TMMs was applied, significant differences were still noted between transfusions (n = 100) with greater than 16 TMMs (median CCIs = 8.6; first-third quartile, 3-15), compared with transfusions (n = 423) with no more than 16 TMMs (median CCIs = 13.1; first-third quartile, 8-17, t test p2 value < .001). If this less stringent threshold was to be applied as a selection criterion, about 80% of transfusions could be considered acceptable.
Triplet mismatch subgroups and CCIs
. | CCIs when HIMMs ≤ 3 . | . | CCIs when HIMMs > 3 . | . | p2 value ≤3 vs > 3 . | CCIs when total MM ≤ 9 . | . | CCIs when total MM > 9 . | . | p2 value ≤ 9 vs > 9 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | No. . | Median . | No. . | Median . | . | No. . | Median . | No. . | Median . | . | ||||
| All transfusions | 184 | 14.0 (10-19) | 339 | 11.2 (6-16) | < .001 | 216 | 13.5 (10-18) | 307 | 11.2 (6-16) | < .001 | ||||
| CREG-compatible transfusions | 112 | 14.1 (10-20) | 58 | 8.5 (4-14) | < .001 | 120 | 13.2 (9-20) | 50 | 10.6 (4-15) | < .001 | ||||
| CREG-incompatible transfusions | 72 | 13.8 (10-17) | 281 | 11.7 (6-16) | .010 | 96 | 13.6 (10-17) | 257 | 11.2 (6-17) | .029 | ||||
. | CCIs when HIMMs ≤ 3 . | . | CCIs when HIMMs > 3 . | . | p2 value ≤3 vs > 3 . | CCIs when total MM ≤ 9 . | . | CCIs when total MM > 9 . | . | p2 value ≤ 9 vs > 9 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | No. . | Median . | No. . | Median . | . | No. . | Median . | No. . | Median . | . | ||||
| All transfusions | 184 | 14.0 (10-19) | 339 | 11.2 (6-16) | < .001 | 216 | 13.5 (10-18) | 307 | 11.2 (6-16) | < .001 | ||||
| CREG-compatible transfusions | 112 | 14.1 (10-20) | 58 | 8.5 (4-14) | < .001 | 120 | 13.2 (9-20) | 50 | 10.6 (4-15) | < .001 | ||||
| CREG-incompatible transfusions | 72 | 13.8 (10-17) | 281 | 11.7 (6-16) | .010 | 96 | 13.6 (10-17) | 257 | 11.2 (6-17) | .029 | ||||
HLAMatchmaker carried out HLA-A and -B compatibility determinations at triplet level and identified the number of highly immunogenic and total triplet mismatches for each donor-patient combination. Transfusions were sorted into subgroups based on the number of mismatches (≤ 3 and > 3 HIMMs; ≤ 9 and > 9 TMMs). The median CCIs for each group was determined, and comparisons between groups were done using the Student t test. The median number of HIMMs and TMMs are presented with the first and third quartile values (in parentheses). The 2 cohorts were compared using unpaired, 2-tailed Student t test.
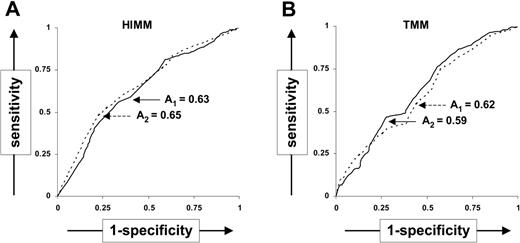
To measure the accuracy of TMMs and HIMMs as predictor of CCIs, we constructed ROC curves25 from the complete data set (Figure 2). While the specificity for TMMs no more than 9 or HIMMs no more than 3 was 0.76 and 0.83, respectively, the sensitivity (0.40 and 0.31, for TMMs no more than 9 or HIMMs no more than 3, respectively) was low. This suggests that while transfusions mismatched below these thresholds are very likely to produce favorable outcomes, exclusion of transfusions with mismatches above the same threshold may result in underuse of potentially useful products. The overall accuracy calculated as the area (A) under the ROC curve was 0.62 for TMMs and 0.63 for HIMMs, suggesting that neither TMMs nor HIMMs are powerful predictors of transfusion outcome. Similar results were obtained after the exclusion of patient 11 (A = 0.59 and 0.65 for TMMs and HIMMs, respectively). While the ROC curves suggest that TMMs and HIMMs are not accurate predictors of individual CCIs, donor-patient pairs with low TMMs and HIMMs are, in general, more likely to yield better transfusion outcomes.
Discussion
HLAMatchmaker is a tool to assess HLA compatibility at the amino acid level. Compared to CREG matching, it considers amino acid sequences of public and private epitopes and carries out both intralocus and interlocus comparisons. The basic premise of this program is that alloimmunized patients do not make antibodies against polymorphic amino acid triplets on mismatched HLA antigens, if these triplets are shared also with the patient's own HLA antigens. Duquesnoy et al7 verified this algorithm by carrying out a comprehensive serologic analysis of 127 well-characterized sera obtained from highly alloimmunized patients awaiting renal transplants. This study also identified triplets with different degrees of immunogenicity. Duquesnoy et al7 further verified the HLAMatchmaker algorithm in a retrospective study of renal transplant recipients, in which they analyzed the United Network for Organ Sharing (UNOS) and Eurotransplant databases of several thousand zero-HLA-DR-mismatched kidneys.17 They showed that HLA-A, -B mismatched kidneys that were compatible at the triplet level (UNOS, 0-2; Eurotransplant, 0-4 triplet mismatches) demonstrated almost identical survival rates as the zero-HLA-A, -B antigen mismatches defined by conventional criteria. Lobashevsky et al26 examined sera from highly sensitized renal transplant candidates and found that the number of highly immunogenic triplet mismatches between donor-recipient pairs reliably predicted final crossmatch results.26 Dankers et al22 showed a strong correlation between the numbers of triplet mismatches between donors and renal transplant recipients and the development of donor-specific antibodies in these patients, further validating the HLAMatchmaker algorithm.22
Frequency of HIMMs and TMMs in transfusions with at least 8 CCIs or less than 8 CCIs. HIMMs (A) and TMMs (B) in transfusions with at least 8 CCIs (▪) or less than 8 CCIs (□). The number of relevant HIMMs or TMMs is shown in the x-axes of the panels. TMMs were grouped by 2 (ie, 1-2, 3-4) to simplify the graphical presentation. Distribution of TMMs and HIMMs according to CCIs was analyzed using the χ2 test. NB: Fisher exact test comparing frequency of CCIs less than 8 or CCIs at least 8 in transfusions with no more than 16 compared to more than 16 TMMs demonstrated a p2 value of < .001.
Frequency of HIMMs and TMMs in transfusions with at least 8 CCIs or less than 8 CCIs. HIMMs (A) and TMMs (B) in transfusions with at least 8 CCIs (▪) or less than 8 CCIs (□). The number of relevant HIMMs or TMMs is shown in the x-axes of the panels. TMMs were grouped by 2 (ie, 1-2, 3-4) to simplify the graphical presentation. Distribution of TMMs and HIMMs according to CCIs was analyzed using the χ2 test. NB: Fisher exact test comparing frequency of CCIs less than 8 or CCIs at least 8 in transfusions with no more than 16 compared to more than 16 TMMs demonstrated a p2 value of < .001.
ROC curves to measure accuracy of triplet mismatches as CCI predictors. ROC curves show (1-specificity) versus sensitivity when using different thresholds of the number of HIMMs and TMMs to predict the transfusion outcomes (CCIs) with all patients (solid line) and with the exclusion of patient 11 (dashed line). The area beneath the ROC curves is indicated for the complete data set (A1) or for the data set exclusive of patient 11 (A2).
ROC curves to measure accuracy of triplet mismatches as CCI predictors. ROC curves show (1-specificity) versus sensitivity when using different thresholds of the number of HIMMs and TMMs to predict the transfusion outcomes (CCIs) with all patients (solid line) and with the exclusion of patient 11 (dashed line). The area beneath the ROC curves is indicated for the complete data set (A1) or for the data set exclusive of patient 11 (A2).
This is the first report of platelet transfusion outcomes when HLA-A and -B locus compatibility between donors and alloimmunized recipients is defined by the HLAMatchmaker program. Our analysis of 523 apheresis platelet transfusions demonstrated a direct relationship between posttransfusion platelet increments and HLA-A and -B matching at the triplet level. Donor-recipient pairs yielding good increments had significantly lower numbers of mismatched triplets (TMMs or HIMMs), compared to donor-recipient pairs associated with poor transfusion outcomes. However, when the mismatches exceeded 16 TMMs or 3 HIMMs, CCIs were unpredictable (Tables 5 and 6). When TMMs or HIMMs were more than 17 or 8, respectively, CCIs were predictably poor. ROC analysis demonstrated a weak association between the number of TMMs or HIMMs and CCIs. Overall, these results confirm previous studies in the context of solid organ transplantation. Our data suggest that the number of mismatches associated with good CCIs is defined by a relatively tight threshold for HIMMs and less well defined for TMMs, beyond which likelihood of successful transfusion approximates a random distribution.22,26 The proposed practical cutoff increased the proportion of potential donors from 5% of all transfusions when complete allelic match was used as a selection criterion to 35% and 80% when a threshold of no more than 3 HIMMs or no more than 16 TMMs, respectively, was applied. This threshold does not imply that HIMMs and TMMs are accurate predictors of individual CCIs. Rather, it suggests that the likelihood of good outcomes is in general higher if the number of triplet mismatches is below these limits.
We found that a higher proportion of CREG-matched donors were matched at the triplet level when compared with the CREG-mismatched donors. Interestingly, the transfusion outcomes were remarkably similar and independent of CREG status when the triplet mismatch data were correlated with CCIs. Conversely, a large number of triplet-mismatched, CREG-compatible transfusions had unfavorable outcomes. These data suggest that donor-recipient pairs that are comparably matched for triplets have similar transfusion outcomes, regardless of compatibility by CREG criteria. These observations support the HLAMatchmaker premise that donors with cross-reacting antigens, which are incompatible at the triplet level, are best avoided and suggest that CREG matching is redundant if HLAMatchmaker is used for determining HLA compatibility. The data also are consistent with the hypothesis that platelet donors matched at the triplet level must be considered compatible at the epitope level, even if donor HLA antigens appear mismatched by conventional criteria. Therefore, HLAMatchmaker should be applied at the time of HLA matching rather than after failure of HLA-matched platelet transfusion. Obviously, this hypothesis is best tested by a prospective follow-up of platelet increments and by the demonstration of a lack of alloreactivity to these mismatched antigens on subsequent antibody screening tests.
HLA alloimmunized patients may fail occasionally to respond adequately to HLA-matched or selectively mismatched platelets. Splenomegaly, which can seriously confound assessment of posttransfusion platelet increments, was absent in the study patients. However, no effort was made to exclude from this study transfusions given in the presence of other coexisting nonimmune causes of platelet refractoriness, such as fever, infection, bleeding, or administration of antibiotics or antifungal medications. These factors are for the most part unlikely to affect the results of this study, because measurement of 1-hour counts is believed to minimize their impact on CCIs.18 Platelet-specific antibodies must be ruled out in patients who repeatedly fail to respond to HLA-matched platelets. This was not a concern in our study, as all patients responded well to most platelet products administered. Although some older studies have demonstrated an inverse relationship between the age of stored platelet concentrates and platelet increments, a more recent study by Leach and AuBuchon27 involving apheresis platelets showed no effect of storage time on posttransfusion CCIs. Norol et al28 also showed equivalent responses to fresh and stored platelets in clinically stable patients. Infrequently, major ABO incompatibility (especially A1 platelets transfused into O recipients) may underlie a repeated failure to respond to HLA-matched platelets.29 However, the expression of A antigen is extremely variable on group A platelets. In fact, group A2 platelets express little, if any, A antigen.30 While our donors were typed for ABO blood group, they were not typed for A2 antigen. It is, therefore, not possible to precisely know if the group A platelets that were transfused to group O or B patients would be incompatible. The results of an analysis of platelet transfusion outcome based on ABO groups and HLAMatchmaker compatibility would be difficult to interpret or could lead to erroneous conclusions. Our study did not examine ABO compatibility and platelet storage time as independent variables for platelet increments. Finally, a limitation of the analysis was the use of identical donors for repeated transfusions in the same patient. This limits the power of the analysis, since several of the events were not truly independent assessments of histocompatibility, and future studies should address this point.
Our data show that 26% of the 170 fully CREG-matched transfusions were unsuccessful, suggesting that only 74% of available donations could be used successfully, according to these criteria. This is in agreement with the recognized inadequacy of serologic HLA typing for accurate characterization of all HLA polymorphisms.31 Since molecular HLA types can now be rapidly determined, all platelet donors should be retyped using molecular methods. Highly sensitized patients are likely to benefit from platelets that are closely HLA matched. However, from a platelet donor center's perspective, the routine provision of molecular-level matches for all alloimmunized patients would not be feasible.
HLAMatchmaker provides a tool to expand the number of compatible donors over conventional methods, which are based on the mere counting of numbers of mismatched HLA antigens or CREGs. As shown in this study, HLAMatchmaker is complementary to the identification of fully matched donors because it increases the pool of donors likely to yield good platelet increments. In addition, this algorithm is superior to CREG-based matching since platelet increments more significantly correlated with a threshold number of TMMs or HIMMs (Tables 5 and 6) than to the level of CREG compatibility (Table 4). In summary, centers that prefer HLA-matched platelets to crossmatched platelets can expand their pool of compatible donors by using HLAMatchmaker. This is best done when HLA-matched platelets are first provided to an alloimmunized patient; alternatively, this tool can be used to great advantage in severely alloimmunized recipients or those with rare HLA antigens.
Undetected HLA incompatibility has been suspected as a cause of refractoriness to HLA-matched platelet transfusions. The assignment of triplets to HLA antigens based solely on low-resolution molecular typing is a limitation of our study. High-resolution, DNA-based HLA typing methods can distinguish amino acid polymorphisms at the allele level. We are carrying out high-resolution, sequence-based HLA typing of patients and donors examined in this study. Analysis of this data with the molecular version of HLAMatchmaker should provide more sophisticated evidence of the validity of the HLAMatchmaker algorithm for HLA-matched platelet transfusion therapy. At present, we are developing tools for the resolution of cis-trans ambiguities for conclusive molecular definition at high resolution of the HLA-A and -B alleles studied.32
Prepublished online as Blood First Edition Paper, November 3, 2005; DOI 10.1182/blood-2004-10-4080.
Supported by grant RO1-AI-55933 from the National Institutes of Health (R.J.D.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This report is dedicated to the memory of Jim Reid, whose contributions to the HLA database and platelet donor search programs are acknowledged. We thank the National Institutes of Health Platelet Donor Center staff for data retrieval and the HLA Laboratory staff for technical help. We also thank Boyd Conley, Janet Browning, Sherry Sheldon, and Michele Hendery for database support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal