Mammals have 2 distinct erythroid lineages. The primitive erythroid lineage originates in the yolk sac and generates a cohort of large erythroblasts that terminally differentiate in the bloodstream. The definitive erythroid lineage generates smaller enucleated erythrocytes that become the predominant cell in fetal and postnatal circulation. These lineages also have distinct globin expression patterns. Our studies in primary murine primitive erythroid cells indicate that βH1 is the predominant β-globin transcript in the early yolk sac. Thus, unlike the human, murine β-globin genes are not up-regulated in the order of their chromosomal arrangement. As primitive erythroblasts mature from proerythroblasts to reticulocytes, they undergo a βH1- to ϵy-globin switch, up-regulate adult β1- and β2-globins, and down-regulate ζ-globin. These changes in transcript levels correlate with changes in RNA polymerase II density at their promoters and transcribed regions. Furthermore, the ϵy- and βH1-globin genes in primitive erythroblasts reside within a single large hyperacetylated domain. These data suggest that this “maturational” βH1- to ϵy-globin switch is dynamically regulated at the transcriptional level. Globin switching during ontogeny is due not only to the sequential appearance of primitive and definitive lineages but also to changes in globin expression as primitive erythroblasts mature in the bloodstream.
Introduction
The first blood cells to circulate during mammalian embryogenesis consist of large primitive red cells. In the mouse embryo, primitive erythroid cells are generated from a transient wave of committed progenitors found in the yolk sac between embryonic day (E) 7.25 and E9.0 of gestation.1,2 Primitive proerythroblasts begin to emerge from blood islands beginning at E8.25, and enter the newly forming bloodstream,3 where they undergo terminal maturation. They transition as a semisynchronous cohort of cells from proerythroblasts to enucleated erythrocytes progressively undergoing a loss of proliferative capacity, a decrease in cell size, accumulation of hemoglobin, and nuclear condensation.4-6 Ultimately, they enucleate by E16.5 to become primitive erythrocytes that can circulate for several days after birth.7 After E11.5, primitive red cells are joined by increasing numbers of smaller definitive erythrocytes that are released from the fetal liver after enucleating. Over the ensuing 5 days, definitive erythrocytes become the predominant cell type in the fetal circulation and ultimately become the exclusive red-cell lineage in the adult.
The sine qua non of red cells is their accumulation of large amounts of hemoglobin composed of globin tetramers encoded by the α- and β-globin gene loci. Examination of globin expression in circulating human and mouse blood cells indicated that 5′ members of both globin loci are expressed in the embryo, while 3′ members are expressed in the adult, leading to the concept of globin switching during ontogeny.8 Since this switch in globin expression coincided temporally with the transition from primitive to definitive red cells, it was initially thought that primitive and definitive erythroid cells exclusively express embryonic and adult globins, respectively.9 However, this hypothesis did not explain the complexity of globin gene expression found in the mouse or the human. The human β-globin locus contains 5 functional genes that are expressed in the order of their arrangement within the locus (ϵ-Gγ-Aγ-δ-β). These genes undergo 2 major transitions in expression during ontogeny, the first from embryonic (ϵ) to fetal (Gγ,Aγ) globins and the second from fetal to adult (δ, β) globins. High-level globin gene expression at all developmental stages is driven by a large upstream sequence element called the locus control region (LCR). Considerable research over the past 3 decades has been focused on understanding the fetal to adult globin switch that occurs in definitive human red cells, and on understanding the role of the LCR in the activation of globin gene expression (reviewed by Stamatoyannopoulos10 ).
In the mouse, there are 4 functional β-globin genes (ϵy-βH1-β1-β2). The mouse βH1-globin and human γ-globin genes are thought to have evolved from a common ancestral γ-globin gene; however, the mouse βH1-globin has not been modified to be expressed in definitive fetal red cells as seen in humans. Thus, in the mouse, there is no fetal to adult hemoglobin switch and both ϵy- and βH1-globins are expressed in the primitive erythroid lineage.11,12 Embryonic βH1-globin transcripts are expressed in yolk sac blood islands as early as E7.5,13 but expression of ϵy-globin transcripts has not been investigated before E9.5.14 A relative transition in βH1- and ϵy-globin mRNA levels has been described in fetal blood between E10.5 and E13.5.14,15 The elucidation of globin gene ontogeny in the mouse is complicated by the progressive intermixing of primitive and definitive red cells in the bloodstream after E11.5.7,16 When nucleated primitive erythroid cells were separated from enucleated primitive and definitive erythrocytes at later stages of mouse gestation by unit gravity sedimentation, it became clear that low levels of adult hemoglobin also accumulate in primitive red cells.16 Taken together, these and other studies in mouse embryos17 suggested that all of the globin genes in the α- and β-globin clusters are expressed in primitive red cells, in contrast to definitive red cells that express only the adult (α1, α2, β1, β2) globin genes. This finding was confirmed by an analysis of globin transcription in single cells between E10.5 and E16.5 of mouse gestation.18
To systematically elucidate the spatial and temporal changes that occur in globin gene expression during differentiation of the primitive erythroid lineage, we analyzed mouse embryos by in situ hybridization and populations of primary primitive erythroid cells by quantitative polymerase chain reaction (qPCR). We also investigated the mechanisms that regulate globin gene expression by analyzing recruitment of polymerase and modifications of histones at the β-globin locus in primary primitive erythroid cells. Our results indicate that a βH1- to ϵy-globin switch occurs as primitive erythroid cells mature in the bloodstream. This “maturational” globin switch appears to be dynamically regulated at the transcriptional level.
Materials and methods
Collection of embryonic peripheral blood
Outbred Swiss Webster mice (Taconic, Germantown, NY) were mated overnight and vaginal plugs checked the following morning (E0.3). At specified times during gestation, mice were killed by cervical dislocation and peripheral blood was collected into PB-119 as previously described.7 We have determined by restriction fragment mapping and PCR analysis that these mice carry the S allele of the β-globin locus (data not shown). All animal experiments were approved by the University Committee on Animal Resources (University of Rochester, Rochester, NY).
Isolation of circulating primitive and definitive erythroid cells by FACS
Peripheral-blood cells from E12.5 and E15.5 mouse embryos were initially stained with Hoechst 33342 (Molecular Probes, Eugene, OR) at 1 μg/mL at 37°C for 30 minutes. After washing in PB-1, cells were stained in a cocktail containing 1 μg/mL thiazole orange (Sigma-Aldrich, St Louis, MO), and 0.2 μg/mL allophycocyanin anti-Ter119 (eBioscience, San Diego, CA) for 20 minutes on ice. Cells were washed and resuspended in PB-1 containing 5 μg/mL propidium iodine (Molecular Probes), and sorted on a FACSAria (BD Biosciences, San Jose, CA).
In situ hybridization
Gene-specific antisense probes were designed to murine Hbb-y globin (ϵy-globin, accession no. NM_008221) nucleotides 509 to 584, Hbb-bH1 globin (βH1-globin, accession no. NM_008219) nucleotides 187 to 288, Hbb-b1 globin (β1-globin, accession no. XM_489729) nucleotides 509 to 600, Hbb-b2 globin (β2-globin, accession no. NM_016956) nucleotides 479 to 582, Hba-a1 globin (α1-globin, accession no. NM_008218) nucleotides 38 to 120, and Hbb-x globin (ζ-globin, accession no. NM_010405) nucleotides 500 to 567. All accession numbers were obtained from the Entrez Nucleotide database.34 In situ hybridization was performed as previously described20 using in vitro–transcribed RNA probes internally labeled with 33P. Darkfield and brightfield images were obtained with an Optiphot microscope (Nikon, Melville, NY) using 10 × (NA 0.5), 20 × (NA 0.75), and 40 × (NA 0.85) objectives and a SPOT RT-Slider digital camera (Diagnostic Instruments, Sterling Heights, MI). Images were acquired, processed (including lens defect correction and brightness/contrast optimization), pseudocolored, and combined using Photoshop (Adobe, San Jose, CA) software with Fovea Pro (Reindeer Graphics, Asheville, NC) plugins.
Analysis of globin gene expression by qPCR
RNA was isolated from dissected yolk sac tissues and various red-cell populations with TRIzol (Invitrogen, Carlsbad, CA) per manufacturer's instructions and subsequently used to synthesize random primed cDNA via the SuperScript First-Strand Synthesis System for reverse transcription (RT)–PCR (Invitrogen). For every cDNA created, a reaction without RT was performed and used as a negative control for each primer pair. qPCR was performed using an iCycler with the MyiQ Single Color Real Time PCR detection system, running MyiQ System Software version 1.0 (Bio-Rad, Hercules, CA). Gene-specific primers were designed to regions of unique sequences listed previously for the different murine globin genes. We were unable to design primers to distinguish the α1- and α2-globins because of their high sequence homology, and therefore refer to the adult α-globin transcripts as α1/α2-globin. A dilution series of each cDNA was used with every primer pair to confirm range, linearity, and comparative amplification efficiencies. Primers to 18S21 were used as an internal control for cDNA quantity.
Analysis of histone acetylation and RNA polymerase II recruitment at the β-globin locus
Primitive erythrocytes were isolated from the bloodstream of E10.5 and E12.5 mouse embryos and subjected to analysis by chromatin immunoprecipitation. Formaldehyde crosslinking, sonication, immunoprecipitation, and DNA isolation were performed as described,22 with the following modifications. Cells were washed with phosphate-buffered saline (PBS) and cross-linked in 1% formaldehyde for 10 minutes at room temperature in a 10-mL volume. After quenching with 0.5 mL 2.5 M glycine and additional PBS washes, the cells were sonicated in a 3-mL volume for 80 seconds on a Misonix Sonicator 3000 (Farmingdale, NY) at a setting of 8 (approximate power output of 36-39 W). The sheared DNA was between 300 to 700 base pairs (bp) in length (Figure S1, available at the Blood website; see the Supplemental Materials link at the top of the online article). Antisera used were directed against (1) histone H3 acetylated at K9 and/or K16; (2) histone H4 acetylated at K5, K8, K12, and/or K16; and (3) histone H3 dimethylated at K4 (all from Upstate, Charlottesville, VA); or (4) RNA polymerase II (Santa Cruz sc-899; Santa Cruz Biotechnology, Santa Cruz, CA). Normal rabbit immunoglobulin G (IgG; Upstate) was also used as a control.
qPCR was performed as above (“Analysis of globin gene expression by qPCR”), except primers were designed to amplify regions corresponding to specific genomic sequences, as detailed in Figure 3 and listed in Table S1. Enrichments were calculated using the following formula:
E is the efficiency of the given primer set, as determined empirically, multiplied by 2 (such that for a primer set exhibiting 95% efficiency, E is 1.90); CtIgG and CtIP are the threshold cycles for fluorescence detection during real-time PCR determined for the normal rabbit IgG control immunoprecipitation (IP) and the test antibody (histone modification or RNA polymerase II [Pol II]), respectively. The “Control” sequence used was either the average of probes located within the inactive control genes pancreatic amylase, protamine 2, and T-cell receptor β (for histone modification chromatin immunoprecipitations [ChIPs]), or the value obtained for a probe located approximately 8.2 kb 3′ of the ϵy-globin transcription start site (for Pol II ChIPs). In the latter case, this probe was chosen because it consistently exhibited the lowest values for [E^(CtIgG - CtIP)] in the Pol II ChIP, even when compared with inactive gene loci.
Results
Changes in globin gene expression at the earliest stages of primitive erythroid maturation
Primitive erythroid cells originate in yolk sac blood islands that are derived from extraembryonic mesoderm cells. The onset of globin gene expression was examined by in situ hybridization, which allowed us to determine the spatial localization of globin transcripts. In E7.5 neural plate–stage embryos, βH1-globin transcripts were found exclusively in mesodermal cell masses (Figure 1A) localized between visceral endoderm and yolk sac mesothelium.13,23 ϵy-, β1-, and β2-globin transcripts were not detected by in situ analysis using the same conditions (Figure 1A). These findings suggest that βH1 transcripts are the first β-globin transcripts to accumulate in extraembryonic mesoderm cells as they commit to an erythroid fate during early gastrulation.
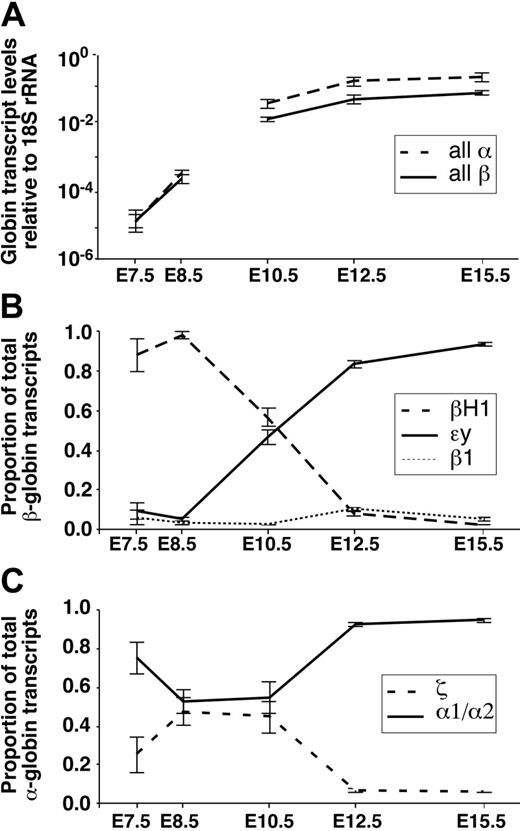
We also examined globin gene expression by qPCR using yolk sac RNAs from pooled, staged, E7.5 embryos. Consistent with the in situ results, the predominant β-globin gene expressed in neural plate–stage embryos was βH1-globin, which accounted for approximately 85% of the total β-globin transcripts (Figure 1A). Low levels of the remaining β-globin genes (ϵy, β1, and β2) were also detected in dissected yolk sac tissues from neural plate–stage embryos. qPCR analysis of the α-globin locus revealed ζ- and α1/α2-globin transcripts were also present in the yolk sac at E7.5. We found that α1/α2-globin transcripts accumulate at higher levels than ζ-globin transcripts, consistent with the qualitative results obtained in later headfold-stage mouse embryos.17 In aggregate, the qPCR analysis raises the possibility that transcription of all globin genes at both the α- and β-globin loci occurs at the onset of hematopoietic differentiation in the yolk sac. We cannot rule out that the trace amounts of β1- and β2-globin mRNAs detected by qPCR are due to maternal reticulocyte contamination. However, the majority of the α1/α2-globin transcripts are not due to maternal contamination because they are more than 10-fold higher than the combined β1- and β2-globin levels.
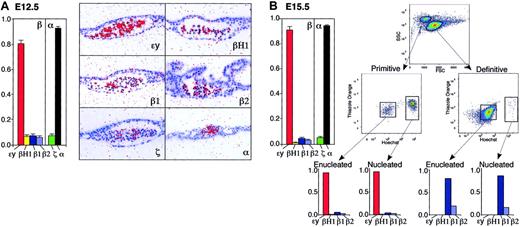
Changes in globin gene mRNA accumulation. (A) E7.5 yolk sac, (B) E8.5 yolk sac, and (C) E10.5 peripheral blood. Relative levels (mean ± SEM of at least 3 independent experiments) of the β-globin genes (ϵy, βH1, β1, and β2) and of the α-globin genes (ζ, α1/α2) quantified by qPCR are shown on the left. Total β- and total α-like globin transcript levels are each considered as 1. Expression of these same genes by in situ hybridization is shown in the right panels. Arrowheads indicate blood islands. For comparison, at each developmental time point, similar exposures of all of the globin genes are shown. In panel B, short exposures of the βH1- and ϵy-globins, marked by asterisks, highlight their differential transcript accumulation. ζ- and α1/α2-globin transcripts were not consistently detected at E7.5 (data not shown). Images were processed as described in “Materials and methods.”
Changes in globin gene mRNA accumulation. (A) E7.5 yolk sac, (B) E8.5 yolk sac, and (C) E10.5 peripheral blood. Relative levels (mean ± SEM of at least 3 independent experiments) of the β-globin genes (ϵy, βH1, β1, and β2) and of the α-globin genes (ζ, α1/α2) quantified by qPCR are shown on the left. Total β- and total α-like globin transcript levels are each considered as 1. Expression of these same genes by in situ hybridization is shown in the right panels. Arrowheads indicate blood islands. For comparison, at each developmental time point, similar exposures of all of the globin genes are shown. In panel B, short exposures of the βH1- and ϵy-globins, marked by asterisks, highlight their differential transcript accumulation. ζ- and α1/α2-globin transcripts were not consistently detected at E7.5 (data not shown). Images were processed as described in “Materials and methods.”
At E8.5, the onset of circulation finds the vast majority of primitive red cells still localized to yolk sac blood islands.3 We had previously determined by in situ hybridization that βH1-globin transcripts are markedly up-regulated between E7.5 and E8.5.13 Using 18S ribosomal RNA as a control for our qPCR analysis, we found an approximately 20-fold increase in globin transcript accumulation within the yolk sac at the onset of somitogenesis (Figure 5, data not shown). Consistent with our findings at E7.5, βH1 remains the predominant β-globin transcript in the yolk sac at E8.5 (Figure 1B). Lower levels of ϵy-globin transcripts are also present, as is evident in the shorter exposure in situ hybridization analysis (Figure 1B, βH1* and ϵy*). The detection of β1-globin transcripts in yolk sac blood islands by in situ hybridization confirms that transcription from the adult globin genes also occurs in primitive erythroblasts at these early stages of primitive erythroid maturation (Figure 1B and Palis et al2 ). Approximately equal amounts of ζ- and α1/α2-globin transcripts were detected by qPCR and evident qualitatively by in situ hybridization (Figure 1B).
By E10.5, a robust circulation of primitive erythroblasts is fully established.3 Analysis of globin expression in circulating red cells by in situ hybridization reveals relatively similar levels of βH1- and ϵy-globin transcripts in a homogeneous pattern (Figure 1C). Low levels of the β1- and β2-globin transcripts are also evident in primitive erythroblasts. Comparison of the ζ- and α1/α2-globin transcripts by in situ hybridization suggests that they both continue to accumulate at similar levels in maturing primitive erythroblasts. Since primitive erythroid cells constitute the only erythroid cells in the embryonic circulation at E9.5 and E10.5,7 we analyzed globin gene expression by qPCR using RNA isolated from peripheral-blood cells pooled from E10.5 embryos (Figure 1C). This analysis confirmed the in situ hybridization findings and indicates that the ϵy-globin gene expression is now up-regulated in circulating primitive erythroblasts to levels that are similar to those of the βH1-globin gene.
Changes in globin gene expression as primitive erythroid cells complete their maturation
As development proceeds, primitive erythroid cells continue to mature in the bloodstream as a semisynchronous cohort. In situ hybridization analysis of β-globin gene expression suggests that ϵy-globin becomes the predominant transcript in E12.5 mouse embryos (Figure 2A). In contrast, βH1-globin transcripts are expressed at lower levels when compared with previous developmental time points. Low, but similar, levels of β1- and β2-globin transcripts accumulate in circulating erythroblasts (Figure 2A). Analysis of the ζ- and α1/α2-globin genes also reveals a change from the relatively similar levels of expression at E8.5 and E10.5. Now, ζ-globin mRNAs are markedly lower than α1/α2-globin mRNAs (Figure 2A). At E12.5, more than 98% of the nucleated cells are primitive erythroblasts.7 Therefore, to accurately analyze globin gene expression in the primitive erythroid lineage by qPCR, we purified nucleated primitive erythroblasts from enucleated primitive and definitive erythrocytes by fluorescence-activated cell-sorting (FACS) relying on Ter119, Hoechst, and thiazole orange staining (Figure S2). This analysis confirms that ϵy-globin is the predominant β-globin transcript, accounting for 81% of the total (Figure 2A). These data indicate that a switch from βH1- to ϵy-globin expression occurs between E8.5 and E12.5 of mouse gestation. The qPCR analysis also reveals that β1- and β2-globin transcripts are up-regulated between E10.5 and E12.5 (Figure 2A). These results are in keeping with the detection of adult hemoglobin after E10.5 in primitive erythroblasts.16 Finally, the differential accumulation of the ζ- and α1/α2-globin mRNAs seen by in situ hybridization was also confirmed by the qPCR analysis (Figure 2A).
Changes in globin gene mRNA accumulation in primitive erythroid cells. (A) E12.5 and (B) E15.5 of mouse gestation. Relative levels (mean ± SEM of at least 3 independent experiments) of the β-globin genes (ϵy, βH1, β1, and β2) and of the α-globin genes (ζ, α1/α2) quantified by qPCR from nucleated primitive erythroid cells are shown on the left. Total β- and total α-like globin transcript levels are each considered as 1. Globin transcript accumulation in circulating blood cells at E12.5 visualized by in situ hybridization is shown in the right panels of A. Isolation by FACS of primitive and definitive orthochromatic erythroblasts and reticulocytes from E15.5 peripheral blood is shown on the right panels in B. These populations can be distinguished by forward-side scatter and further isolated as Ter119-positive cells (not shown). Orthochromatic erythroblasts can be distinguished from reticulocytes after staining RNA with thiazole orange and DNA with Hoechst. Also shown are the patterns of β-globin gene expression in each purified population quantified by qPCR. Late-stage primitive erythroid cells at E15.5 express predominantly ϵy-globin transcripts, while late stage definitive erythroid cells express only β1- and β2-globin transcripts. Images were processed as described in “Materials and methods.”
Changes in globin gene mRNA accumulation in primitive erythroid cells. (A) E12.5 and (B) E15.5 of mouse gestation. Relative levels (mean ± SEM of at least 3 independent experiments) of the β-globin genes (ϵy, βH1, β1, and β2) and of the α-globin genes (ζ, α1/α2) quantified by qPCR from nucleated primitive erythroid cells are shown on the left. Total β- and total α-like globin transcript levels are each considered as 1. Globin transcript accumulation in circulating blood cells at E12.5 visualized by in situ hybridization is shown in the right panels of A. Isolation by FACS of primitive and definitive orthochromatic erythroblasts and reticulocytes from E15.5 peripheral blood is shown on the right panels in B. These populations can be distinguished by forward-side scatter and further isolated as Ter119-positive cells (not shown). Orthochromatic erythroblasts can be distinguished from reticulocytes after staining RNA with thiazole orange and DNA with Hoechst. Also shown are the patterns of β-globin gene expression in each purified population quantified by qPCR. Late-stage primitive erythroid cells at E15.5 express predominantly ϵy-globin transcripts, while late stage definitive erythroid cells express only β1- and β2-globin transcripts. Images were processed as described in “Materials and methods.”
By E15.5, the rapid efflux of definitive erythrocytes from the liver has relegated primitive red cells to a minority of circulating cells.7 At this time point, primitive and definitive erythroid cells can be separated by FACS using forward and side scatter (Figure 2B). The incorporation of thiazole orange and Hoechst staining facilitated the separation of Ter119-positive orthochromatic erythroblasts and reticulocytes from both erythroid lineages (Figure 2B). Primitive and definitive red cells displayed a striking difference in globin gene expression (Figure 2B). Terminally differentiating primitive red cells contain predominantly ϵy-globin transcripts and almost no βH1-globin transcripts, while β1- and β2-globin transcripts continue to accumulate at low but relatively similar (< 2:1) levels (Figure 2B). In contrast, the cocirculating definitive orthochromatic erythroblasts and reticulocytes contain only the adult β1- and β2 globins, which accumulate at a 4:1 ratio. The differences in the amounts and relative levels of β1- and β2-globin transcripts indicate that these genes are differentially regulated in primitive and definitive erythroblasts.
Analysis of Pol II recruitment by ChIP
Our analysis of mRNA accumulation in primitive red cells, visualized by in situ hybridization or measured by qPCR, suggests a dynamic pattern of transcription within the β-globin locus as primitive erythroblasts mature. The degree to which transcriptional regulation contributes to the observed mRNA levels, however, can be obscured by regulation of RNA processing and stability. Therefore, to investigate whether changes in globin gene expression are mediated at the level of transcription, we examined Pol II occupancy at the gene promoters, as well as other regions within the locus, using the ChIP assay. Our analysis indicates that at E10.5, the ϵy- and βH1-globin promoters are substantially enriched in the anti–Pol II–bound fraction, while the β1- and β2-globin promoters do not appear enriched (Figure 3).
At E12.5, the ϵy-globin promoter is further enriched, Pol II binding to the βH1-globin promoter appears to decrease, and significant Pol II recruitment has occurred at the β1- and β2-globin promoters. Similarly, within the α-globin locus, a sharp decrease in Pol II binding is seen at the ζ promoter and gene between E10.5 and E12.5, while the α1/α2 globin gene(s) are further enriched in Pol II binding. The differences in Pol II occupancy at the various β-globin gene promoters are consistent with the changes that occur between E10.5 and E12.5 in mRNA levels of the different globin genes.
We also examined Pol II association at the 3′ ends of the genes, 1.2 to 1.4 kb from the promoters. At E10.5, Pol II association with the 3′ region of the βH1-globin gene is relatively low, while binding to the 3′ region of ϵy-globin is nearly as robust as at the promoter (Figure 3). No enrichment for Pol II association is seen at the 3′ ends of the β1- or β2-globin genes, consistent with the similar lack of Pol II binding at the promoters. At E12.5, association of Pol II with the 3′ end of ϵy-globin increases to the same degree as at the promoter, while there is little further change in Pol II binding to the 3′ end of βH1-globin. Pol II binding to the 3′ ends of the β1- and β2-globin genes increases, but not to the same degree as at the promoters. These data indicate that Pol II binding to the β-globin genes parallels their relative abundance during maturation of primitive erythroblasts, and suggests that this arises from a combination of regulatory mechanisms operative at initiation (Pol II at the promoter) or elongation (Pol II at the 3′ ends of the genes).
RNA polymerase II association with α- and β-globin gene sequences in primitive erythroid cells. Enrichments are shown in bar graph format relative to multiple control sequences elsewhere in the β-globin locus, of crosslinked chromatin samples immunoprecipitated using antiserum against RNA polymerase II, as determined by quantitative real-time PCR. ▪ represent enrichments at the indicated β-globin gene promoters, while □ represent enrichments at sequences located within the third exon or immediately downstream of the stop codon for each gene, approximately 1 kb 3′ of the promoters. The border between shaded and unshaded regions represents relative enrichment of 1.0-fold (ie, no enrichment [arrowhead]). The data shown represent averages of at least 3 PCRs for each sequence examined, derived from 2 (E10.5) or 3 (E12.5) separate immunoprecipitations. Error bars represent SEM.
RNA polymerase II association with α- and β-globin gene sequences in primitive erythroid cells. Enrichments are shown in bar graph format relative to multiple control sequences elsewhere in the β-globin locus, of crosslinked chromatin samples immunoprecipitated using antiserum against RNA polymerase II, as determined by quantitative real-time PCR. ▪ represent enrichments at the indicated β-globin gene promoters, while □ represent enrichments at sequences located within the third exon or immediately downstream of the stop codon for each gene, approximately 1 kb 3′ of the promoters. The border between shaded and unshaded regions represents relative enrichment of 1.0-fold (ie, no enrichment [arrowhead]). The data shown represent averages of at least 3 PCRs for each sequence examined, derived from 2 (E10.5) or 3 (E12.5) separate immunoprecipitations. Error bars represent SEM.
Histone modifications across the β-globin locus in primitive erythroblasts
In definitive erythroid cells, the active β-globin genes (β1 and β2) are associated with hyperacetylated domains, defined as large (10-12 kb) regions of DNA that are packaged into chromatin characterized by hyperacetylated core histones.24 Other modifications, such as dimethylation of histone H3 lysine 4, are also enriched within these domains. In contrast, the region containing the inactive globin genes (ϵy and βH1) is not associated with such high levels of histone hyperacetylation. The pattern of histone modifications suggests that active β-globin genes are encompassed within larger regions of modified, or “open,” chromatin. We therefore investigated whether globin gene activation is similarly associated with such broad patterns of histone modifications in primitive red cells and whether the dynamic pattern of globin gene expression during maturation of primitive erythroblasts correlates with histone modification patterns at the gene promoters or across larger regions of the locus.
We performed ChIP assays on peripheral-blood cells from E10.5 and E12.5 fetuses using antibodies against acetylated histone H3. ChIP assays were also performed using antibodies against acetylated histone H4, and histone H3 dimethylated at lysine 4 (Figure S3), but only with E12.5 blood, due to the small amount of sample obtained at E10.5. Histone hyperacetylation occurs over all of the sequences tested within the region encompassing the ϵy- and βH1-globin genes, including the βH1-globin promoter, which exhibits the highest enrichments for histone acetylation among all sequences tested. The data reveal a continuous domain of H3, H4, and histone H3 dimethylated at lysine 4 hyperacetylation stretching from the LCR, through both embryonic β-globin genes, and ending at an abrupt transition point located between 15.2 to 16.4 kb 3′ of the ϵy-globin transcription start site, or 5.2 to 6.4 kb 3′ of the βH1-globin transcription start site (Figure 4, Figure S3, and data not shown). This domain exhibits similar patterns of histone H3 acetylation at E10.5 (Figure 4), indicating that large-scale changes in chromatin structure do not accompany the switch from βH1- to ϵy-globin transcription that occurs at these time points. Finally, the β1- and β2-globin genes are each encompassed within a distinct domain of histone hyperacetylation (Figure 4), which is similar to the pattern of histone modifications seen in definitive erythroblasts.24 In this case, however, levels of acetylation in the population of peripheral-blood cells undergo a qualitative change between E10.5 and E12.5, with substantial increases occurring at most probes. Exceptions to this trend are the gene promoters and a sequence located 3′ of the β2-globin gene, at which acetylation levels are elevated at both time points.
Discussion
Primitive and definitive erythroid cells display multiple differences consistent with the notion that they constitute distinct erythroid lineages. The primitive erythroid lineage is transient, originating from a single wave of unique yolk sac–derived progenitors,2 while the definitive erythroid lineage continuously generates smaller erythrocytes in the fetus and adult. Furthermore, primitive and definitive erythroid cells have distinct patterns of globin gene expression. In the mouse, all of the globin genes of the α- and the β-globin loci are expressed in primitive erythroid cells. In contrast, only the adult (α1, α2, β1, β2) globin genes are expressed in definitive erythroid cells of the fetus (Figure 2B). Interestingly, at E15.5 of mouse gestation, the primitive and the definitive circulating red cells each consist primarily of orthochromatic erythroblasts and reticulocytes (Figure 2B and data not shown). Despite being at similar stages of maturation, the cells of these 2 erythroid lineages express strikingly different patterns of globin genes. Late-stage primitive erythroid cells contain predominantly ϵy-globin transcripts, while late-stage definitive erythroid cells contain only β1- and β2-globin transcripts. The marked differences in globin gene expression seen in similarly staged primitive and definitive erythroid cells indicate that the switch from embryonic to adult globins seen during development is due in a large part to the switch from primitive to definitive erythroid lineages.
Histone modifications within the β-globin locus in primary primitive erythroid cells at E10.5 and E12.5 indicate that the ϵy- and βH1-globin genes are contained within a single, large, hyperacetylated domain. The β-globin locus is represented to scale at the top, with the active globin genes indicated by arrows and exons of these genes by the thickest portions of the line. Unlabeled, thicker lines represent the positions of β-globin pseudogenes. The positions of PCR-amplified regions analyzed in the ChIP assay are indicated immediately below. Shown in bar graph format are enrichments, relative to multiple control sequences derived from loci containing genes inactive in erythroid cells, of cross-linked chromatin samples immunoprecipitated using antiserum against histone H3 acetylated at lysines 9 and/or 16. Chromatin was isolated from peripheral blood at E10.5 (□) and E12.5 (▪). The border between shaded and unshaded regions in each graph represents relative enrichment of 1.0-fold (arrowhead). Values for enrichment greater than 20-fold have been truncated in this representation. Error bars represent SEM.
Histone modifications within the β-globin locus in primary primitive erythroid cells at E10.5 and E12.5 indicate that the ϵy- and βH1-globin genes are contained within a single, large, hyperacetylated domain. The β-globin locus is represented to scale at the top, with the active globin genes indicated by arrows and exons of these genes by the thickest portions of the line. Unlabeled, thicker lines represent the positions of β-globin pseudogenes. The positions of PCR-amplified regions analyzed in the ChIP assay are indicated immediately below. Shown in bar graph format are enrichments, relative to multiple control sequences derived from loci containing genes inactive in erythroid cells, of cross-linked chromatin samples immunoprecipitated using antiserum against histone H3 acetylated at lysines 9 and/or 16. Chromatin was isolated from peripheral blood at E10.5 (□) and E12.5 (▪). The border between shaded and unshaded regions in each graph represents relative enrichment of 1.0-fold (arrowhead). Values for enrichment greater than 20-fold have been truncated in this representation. Error bars represent SEM.
However, we have also documented dramatic changes in globin gene expression within the primitive erythroid lineage as it matures as a semisynchronous wave from committed progenitors at E7.5 to reticulocytes at E15.5 of mouse gestation. β-globin gene expression has not been systematically examined in mouse embryos before E9.5.14,15 We found that βH1 is the first β-globin mRNA to significantly accumulate in immature primitive erythroid cells from E7.5 through E8.5, even though ϵy-globin is the closest gene to the LCR in the β-globin locus. Thus, unlike the human, the murine β-globin genes are not up-regulated in the order of their chromosomal arrangement. This nonlinear activation of the β-globin gene cluster in mice will need to be incorporated into models of globin gene regulation. A better understanding of the regulation of endogenous murine globin genes is necessary, particularly since mice serve as hosts for the study of exogenous human globin loci.
As primitive red cells continue to mature in the bloodstream, ϵy-globin transcripts are up-regulated and supersede βH1-globin mRNA levels (Figure 5). ϵy remains the predominant β-globin transcript as primitive orthochromatic erythroblasts enucleate between E12.5 and E15.5 and become reticulocytes. These changes in the relative levels of βH1- and ϵy-globin serve as a striking in vivo example of maturational globin switching. This concept was initially proposed by Papayannopoulou et al25 to describe the down-regulation of γ-globin expression as definitive erythroid cells mature in vitro in colonies derived from neonatal and adult human erythroid progenitors. The occurrence of maturational globin switching highlights the importance of cell context when studying globin gene regulation. Other, less dramatic, changes occur in the relative levels of β-globin genes as primitive erythroid cells mature in the bloodstream. There is a small increase both in β1- and in β2-globin transcript accumulation between E10.5 and E12.5 of gestation. It is likely that these transcripts are responsible for the progressive increase in adult hemoglobin found in primitive erythroblasts between E12.5 and E14.5.16
The down-regulation of βH1- and the up-regulation of ϵy-, β1-, and β2-globin mRNAs between E10.5 and E12.5 are paralleled by changes in RNA Pol II association at the respective globin gene promoters. These results support the concept that the βH1- to ϵy-globin switch in primitive erythroblasts is due primarily to changes in transcription at the β-globin locus.18 High-level expression of all of the β-globin genes is known to require the LCR, since mouse alleles harboring a deletion of the LCR exhibit β-globin mRNA levels only 1% to 4% of normal.26 ChIP-based studies of the ΔLCR allele have suggested that at least for the adult β1-globin gene, the primary deficiency is in transcriptional elongation. This was implied by the observation that Pol II recruitment to the β1-globin promoter is only 50% lower in the absence of the LCR, while association of Pol II to the 3′ portion of the gene is diminished approximately 10-fold.27 Our own data do not present such a clear difference, but as shown in Figure 3, while Pol II binding to the βH1-globin promoter decreases approximately 2-fold between E10.5 and E12.5, Pol II association at the 3′ end of the gene is already low at E10.5 and does not further decrease with time. Since βH1-globin mRNA levels appear to decrease before E10.5, these observations raise the possibility that a block of transcriptional elongation at βH1-globin precedes loss of Pol II binding to the promoter. It is therefore possible that the loss of βH1-globin expression reflects a loss of LCR activity at the βH1-globin promoter, perhaps due to the up-regulation of the ϵy-globin gene located between βH1-globin gene and the LCR. Similarly, while Pol II binding to the β1- and β2-globin promoters increases dramatically between E10.5 and E12.5, association with the 3′ regions of these genes is less robust. This suggests that while these genes and their associated domains are activated over this timespan, their expression remains low because the LCR interacts primarily with ϵy-globin.
Summary of changes in relative globin mRNA levels, normalized to 18S rRNA, as primitive erythroid cells mature from proerythroblasts at E7.5 to E8.5 to reticulocytes at E15.5 in the mouse. The E7.5 and E8.5 values are based on dissected yolk sac tissues, E10.5 on whole peripheral blood, and E12.5 and E15.5 values on sorted cell populations. (A) Levels of total α-globin and β-globin gene transcripts quantified by qPCR suggest that total α-globin transcripts accumulate in excess of β-globin transcripts. (B) Relative levels of ϵy-, βH1-, and β1-globin transcripts reveal the βH1- to ϵy-globin switch between E8.5 and E12.5. (C) Relative levels of the ζ- and α1/α2-globin transcripts reveal the predominance of α1/α2-globin gene from the earliest stages of primitive erythroid maturation. Error bars represent SEM.
Summary of changes in relative globin mRNA levels, normalized to 18S rRNA, as primitive erythroid cells mature from proerythroblasts at E7.5 to E8.5 to reticulocytes at E15.5 in the mouse. The E7.5 and E8.5 values are based on dissected yolk sac tissues, E10.5 on whole peripheral blood, and E12.5 and E15.5 values on sorted cell populations. (A) Levels of total α-globin and β-globin gene transcripts quantified by qPCR suggest that total α-globin transcripts accumulate in excess of β-globin transcripts. (B) Relative levels of ϵy-, βH1-, and β1-globin transcripts reveal the βH1- to ϵy-globin switch between E8.5 and E12.5. (C) Relative levels of the ζ- and α1/α2-globin transcripts reveal the predominance of α1/α2-globin gene from the earliest stages of primitive erythroid maturation. Error bars represent SEM.
In definitive erythroid cells, the inactive embryonic globin genes reside within a region of the locus that is not appreciably hyperacetylated, while the LCR and the regions containing the active β1- and β2-globin genes are extensively modified. These results suggest that differential expression of adult versus embryonic globin genes is regulated in part by chromatin modifications. Studies of histone modifications in primitive erythroid cells have to date been more limited. Forsberg et al examined E11.5 whole murine yolk sac tissues and found that the βH1- and β2-globin gene promoters were hyperacetylated, but they did not examine the other β-globin genes.28 When K562 cells were used as a surrogate for human primitive erythroid cells, H3 and H4 acetylation were found to be continuous between the LCR and the ϵy-globin gene. However, the fetal and adult globin genes were not examined.29 We therefore surveyed the entire β-globin locus in primary primitive erythroblasts at E10.5 and E12.5 of gestation by ChIP and found, in contrast to definitive erythroid cells, that the ϵy- and βH1-globin genes reside within a single large hyperacetylated domain that appears to be continuous with the LCR. However, we found no significant changes in histone modifications within this domain between E10.5 and E12.5 (Figure 4), indicating that the mechanism of down-regulation of βH1-globin transcription does not involve large-scale changes in chromatin structure, nor is it accompanied by a loss of histone acetylation at the promoter. These findings are consistent with the notion that embryonic globin gene regulation in primary primitive red cells involves changes in the interactions of local transcriptional complexes with the LCR. Across the region harboring the β1- and β2-globin genes, the pattern of histone modifications at E12.5 is similar to that observed in definitive cells, but at E10.5 levels of histone acetylation are significantly lower, although the promoter regions are extensively modified. This suggests that formation of the hyperacetylated domains encompassing these genes, as measured by ChIP, occurs simultaneously with their transcriptional activation, as measured by the accumulation of β1- and β2-globin transcripts between E10.5 and E12.5.
Little is known about the positive transcriptional regulation of the embryonic globin genes in primitive red cells, since most studies of embryonic globin genes have focused on the mechanism of their repression in definitive red cells (reviewed by Stamatoyannopoulos10 ). Members of the Kruppel-like family (KLF) of transcription factors are candidate regulators of embryonic globins,30,31 and KLF2 has recently been shown to regulate ϵy- and βH1-globin levels.32 However, the specific factors that regulate the βH1- to ϵy-globin switch in primitive red cells remain to be elucidated. Knowledge of the factors that regulate embryonic globin genes may have important implications for the therapy of hemoglobinopathies, as exemplified by the amelioration of anemia in a mouse model of sickle cell disease by the reactivation of embryonic ζ-globin gene.33
Prepublished online as Blood First Edition Paper, November 17, 2005; DOI 10.1182/blood-2005-08-3097.
Supported by funding from the Naitonal Institutes of Health (NIH) and by institutional funding from the University of Rochester School of Medicine and Dentistry.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to acknowledge the expert technical assistance of Anne Koniski for animal husbandry and Earl Walker for ChIP primer design. We thank Doug Higgs for helpful comments.


![Figure 3. RNA polymerase II association with α- and β-globin gene sequences in primitive erythroid cells. Enrichments are shown in bar graph format relative to multiple control sequences elsewhere in the β-globin locus, of crosslinked chromatin samples immunoprecipitated using antiserum against RNA polymerase II, as determined by quantitative real-time PCR. ▪ represent enrichments at the indicated β-globin gene promoters, while □ represent enrichments at sequences located within the third exon or immediately downstream of the stop codon for each gene, approximately 1 kb 3′ of the promoters. The border between shaded and unshaded regions represents relative enrichment of 1.0-fold (ie, no enrichment [arrowhead]). The data shown represent averages of at least 3 PCRs for each sequence examined, derived from 2 (E10.5) or 3 (E12.5) separate immunoprecipitations. Error bars represent SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/4/10.1182_blood-2005-08-3097/2/m_zh80040691000003.jpeg?Expires=1764962770&Signature=J97YjQ8nV0AixHOIZdSZmnyvoG8fcHe9vqq3s-GCgeRmMBxmRZz8bIjGAKfpIdWWmRuixpiQnwMgGoQ7nE4I0Dd~PMxL5XOYiw2xMpcVO1T3WmQ7XuUEyQLh2OIB6BJjhp34mitOSFwyVvfL0DpvhIPNcqdr1QnKeOgmO9seFrcLtHzTAum0AJqUKLn8~ebzi2qhctGeIRCbH1Bh0NHUJf~PLimLH6-wc9zJInZrbYkYPjpPg5sXPg7PP1wNgjNS-Pf7q78E0101BYfz5Vf1iZUXcDo9bDQfDDisCEWrewW0ZchfesqAzOFi1v3qUqkHF-jS8MbQO5GvmBEtd8fT7w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal