The BCL6 transcriptional repressor mediates survival, proliferation, and differentiation blockade of B cells during the germinal-center reaction and is frequently misregulated in B-cell non-Hodgkin lymphoma (BNHL). The p53 tumor-suppressor gene is central to tumorigenesis. Microarray analysis identified BCL6 as a primary target of p53. The BCL6 intron 1 contains a region in which 3 types of genetic alterations are frequent in BNHL: chromosomal translocations, point mutations, and internal deletions. We therefore defined it as TMDR (translocations, mutations, and deletions region). The BCL6 gene contains a p53 response element (p53RE) residing within the TMDR. This p53RE contains a motif known to be preferentially targeted by somatic hypermutation. This p53RE is evolutionarily conserved only in primates. The p53 protein binds to this RE in vitro and in vivo. Reporter assays revealed that the BCL6 p53RE can confer p53-dependent transcriptional activation. BCL6 mRNA and protein levels increased after chemotherapy/radiotherapy in human but not in murine tissues. The increase in BCL6 mRNA levels was attenuated by the p53 inhibitor PFT-α. Thus, we define the BCL6 gene as a new p53 target, regulated through a RE frequently disrupted in BNHL.
Introduction
The BCL6 transcriptional repressor mediates survival, proliferation, and differentiation blockade of B cells during the germinal-center (GC) reaction and is implicated in the pathogenesis of B-cell non-Hodgkin lymphoma (BNHL). BCL6 was originally identified by virtue of its involvement in 3q27 chromosomal translocations associated with 2 types of BNHL: diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL).1-3 BCL6 is selectively expressed in high levels in mature B cells in the GC, in which they undergo Ig gene somatic hypermutation (SHM), class switch recombination (CSR), and affinity maturation-based selection. BCL6 is required for GC formation and T-cell-dependent antibody responses.4 BCL6 was shown to suppress genes involved in lymphocyte activation, differentiation, cell-cycle arrest, and apoptosis.5 The balance between BCL6 and BLIMP-1, another transcriptional repressor, determines when and whether B cells will further differentiate into plasma cells. Introducing BCL6 into plasma cells caused them to “dedifferentiate” toward a B-cell state.6 There is growing evidence of BCL6's role in lymphomagenesis.7-10 Recently, BCL6 was shown to directly suppress p53 gene expression in GC B cells.11
TP53 (also called p53) is a key tumor-suppressor gene that is mutated or lost in approximately 50% of all human cancer cases worldwide.12 Downstream targets or upstream regulators of p53, such as p14ARF and MDM2, are altered in many tumors with intact TP53. p53 is activated in response to a variety of cellular and genotoxic stress conditions, leading to the induction of growth arrest, apoptosis, DNA repair, senescence, and differentiation.13 Many studies have shown that p53 exerts its various functions mainly by regulating gene expression of its target genes through a consensus DNA-binding site.14 Most p53 target genes are involved in mediating cell-cycle arrest and apoptosis.13
Three major types of genetic alterations affect the BCL6 gene in BNHL: chromosomal translocations, point mutations, and internal deletions. Chromosomal translocations involve chromosome 3q27 and occur in 30% to 40% of DLBCL cases and 6% to 15% of FL cases. These translocations occur in a highly conserved 4.0-kb regulatory region of the BCL6 gene, termed major translocation cluster (MTC), spanning the promoter, the first noncoding exon, and the 5′ region of the first intron.2,15 Within the MTC, a 110-bp breakpoint hyper-cluster region was defined.16 The second type of genetic alteration, point mutations, occur in the 5′ noncoding region of the gene, within which a major mutation cluster (MMC), approximately 730-bp long, was defined.17 These mutations are of somatic origin18 and serve as a marker of B-cell transit through the GC, as they occur in 30% to 50% of memory B cells. In addition, these mutations are found in approximately 50% of DLBCL cases.19 The third type of genetic alteration, internal deletions, was observed in 4 DLBCL patients and in a DLBCL cell line (Val), ranging from 1.5 to 2.4 kb in length. These deletions overlap over a 270-bp segment at the 5′ region of the BCL6 intron 1.20,21 All of the regions of genetic alterations specified earlier in this paragraph, the breakpoint hyper-cluster region within the MTC, the MMC, and the shared region of internal deletions, overlap over a 110-bp region located at the 5′ region of BCL6 intron 1. We therefore defined it as TMDR (translocations, mutations, and deletions region). Due to the high occurrence of genetic alterations found in this region in BNHL, it was hypothesized that disruption of binding of putative transcription factors to this region in the BCL6 gene may contribute to lymphomagenesis. Therefore, several attempts were made to identify such transcription factors,16,21,22 but no significant results were obtained so far.
In a previous study done in our laboratory, DNA microarrays were used to identify primary p53 target genes, one of which was BCL6.23 In the present study we show that the BCL6 gene contains a p53 response element (p53RE) residing within the TMDR. This p53RE contains a motif that is known to be preferentially targeted by SHM. This p53RE is evolutionarily conserved in primates but not in rodents. The p53 protein binds to this RE in vitro and in vivo. Luciferase reporter assays revealed that the BCL6 intron 1 can confer p53-dependent transcriptional activation through the p53RE. BCL6 mRNA levels increased after chemotherapy and/or γ-irradiation in human peripheral-blood lymphocytes (PBLs) and cell lines but not in murine tissues. The increase in BCL6 mRNA levels was attenuated by the p53 inhibitor PFT-α. BCL6 mRNA levels increased in response to activation of a temperature-sensitive p53 in human H1299 cells and not in the parental H1299 p53-null cells. Furthermore, BCL6 protein levels increased in response to chemotherapy in human lymphoblastoid B cells. Thus, we define the BCL6 gene as a new p53 target. In conjunction with the recent finding that BCL6 represses p53 gene expression,11 our data define a novel p53-BCL6 autoregulatory loop. Disruption of this loop may lead to deregulated BCL6 expression and promote BNHL.
Patients, materials, and methods
Patients
PBLs were obtained from patients treated at the Department of Hematology and Bone Marrow Transplantation, Chaim Sheba Medical Center, Israel. The patients were treated with 2 Gy twice a day ionizing radiation and/or 60 g/kg/d cyclophosphamide, as part of the treatment protocol. PBLs were taken for our research only when blood was drawn as part of the routine monitoring. This accounts for the fact that samples were taken neither consecutively nor in a uniform manner. Samples were taken always at 6 am. Control samples were obtained in the morning on the same day the treatment started. Details of the patients are as follows: (A) a 23-year-old female with prolymphocytic leukemia, (B) a 29-year-old male with T-cell acute lymphoblastic leukemia (T-ALL), (C) a 39-year-old female with Philadelphia-positive ALL. Chaim Sheba Medical Center institutional review board approval (Tel-Hashomer, Israel) and informed consent according to the Declaration of Helsinki were obtained in all cases. For each sample, 5 mL PB was drawn and white blood cells (WBCs) were separated. Total RNA was extracted from the WBCs and reverse transcribed.
Cell lines
An EBV-transformed lymphoblastoid B-cell line was established by EBV infection of peripheral blood of a healthy 30-year-old male and maintained at 37°C in RPMI-1640 medium containing 20% FCS. The human lung small-cell carcinoma cell line H1299 (lacking endogenous p53) expressing the mouse temperature-sensitive mutant p53Val135 (ts-p53Val135) was used in this study.24 The ts-p53Val135 assumes wild-type p53 conformation on temperature shift to 32°C. The control cell line used was the parental H1299 cells without ts-p53Val135. These 2 H1299 cell lines were maintained at 37°C in RPMI-1640 medium containing 10% FBS. U2OS human osteosarcoma cells and HCT116 human colon carcinoma cells, both p53 positive, were maintained at 37°C in DMEM and McCoy 5A medium, respectively, containing 10% FCS.
Mice
The Tp53+/- mice were obtained from Jackson Laboratories (Bar Harbor, ME) and maintained in the animal facilities of the Weizmann Institute of Science. Healthy 13- to 14-week-old male Tp53+/+ and Tp53-/- mice were subjected to total body irradiation using a 60Cobalt γ-source at a dose rate of 0.67 Gy/minute. At least 2 animals were used in each experimental group. Spleens were harvested at 0, 1, 2, 3, 4, 5, 6, and 24 hours after irradiation, and the thymuses were harvested at 0, 6, and 24 hours after irradiation. Total RNA was extracted from the tissues and reverse transcribed. The mice were humanely killed using an approved Institutional Animal Care and Use Committee Protocol. All animal experiments have been carried out with the approval of an ethics committee.
Sequencing of the Aotus nancymaae owl monkey Bcl6 intron 1
Sequencing was performed on DNA extracted from a PB sample taken from a 10-year-old male Aotus nancymaae, using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). A fragment of the Bcl6 intron 1 was amplified by polymerase chain reaction (PCR) using the following primers (5′ to 3′): forward, GATTAGTCCCACGTCTCTCC; reverse, GGAGCAAGGAAAGCAGTT.
Electrophoretic mobility-shift assay
Double-strand fragments were prepared from sense and antisense oligonucleotides spanning the p53RE derived from positions +696 to +728 (all positions are relative to the transcription start site) of the human BCL6 gene as follows (5′ to 3′; the p53-binding sites are underlined and the mutated nucleotides are denoted in lowercase): Oligo A, ACGAGACAGTGCTTGGGGGGTGATTCGGGCTAGTCTGGG; Oligo B, like oligo A without the extra guanosine in the middle of the first decamer, ACGAGACATGCTTGGGGGGTGATTCGGGCTAGTCTGGG; Oligo C, like oligo A with 2 mutations, ACGAGAaAGTGCTTGGGGGGTGATTCGGGaTAGTCTGGG. 32P end-labeled fragments were incubated with either 100 ng of baculovirus recombinant wild-type human p53 (Calbiochem, San Diego, CA) or mutant murine p53 (R270C) prepared in insect cells. Supershift assays were performed by adding 1 μL anti-p53 antibody (pAb421) to the reaction. Competition assays were performed by adding a 50-fold and 500-fold excess of unlabeled oligo A to the reaction.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis in the EBV-transformed lymphoblastoid B cells was performed using the EZ-ChIP kit, according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY). Immunoprecipitation was done with either anti-p53 antibody (Upstate Biotechnology; BP53-12), anti-GFP antibody (Santa Cruz Biotechnology, Santa Cruz, CA; catalog no. sc-8334), anti-RNA polymerase II antibody (Upstate Biotechnology), or normal mouse IgG (Upstate Biotechnology). The latter 2 antibodies, provided with the EZ-ChIP kit, served as positive and negative controls, respectively. Primers used for a sequence in the BCL6 intron 1 containing the p53RE were as follows (5′ to 3′): forward, GCGGCCGGAGCAGAGA; reverse, CACAAGCCGTACGCAAGCA (Figure 2B). Primers used for the CDKN1A (also called p21) promoter 100 bp upstream of the 5′ p53RE were as follows: forward, GCACTCTTGTCCCCCAG; reverse, TCTATGCCAGAGCTCAACAT. Primers used for a sequence from glyceraldehyde-3-phosphate dehydrogenase (GAPDH) exon 4 were as follows: forward, GTATTCCCCCAGGTTTACAT; reverse, TTCTGTCTTCCACTCACTCC. All PCR reactions were performed in 40 cycles, and the annealing temperatures for BCL6, p21, and GAPDH were 67°C, 53°C, and 53°C, respectively.
Plasmid constructs
To create a luciferase construct for BCL6, a 1.154-kb fragment spanning the 3′ end of the human BCL6 exon 1 and the 5′ region of intron 1 (nucleotides +135 to +1288; Figure 2A) was isolated from human placenta DNA, subcloned into the pGEM-T Easy vector (Promega, Madison, WI), and then cloned into the KpnI/HindIII site of the pGL2-Basic luciferase reporter vector (Promega). The primers for PCR of the fragment were as follows (5′ to 3′): forward, GGTACCGAGCTGACACCAAGTCCTC; reverse, AAGCTTCGACTCCCTCGACTACAAC. KpnI and HindIII sites were added to the 5′ end of the forward and reverse primers, respectively.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), according to the manufacturer's protocol. The wild-type BCL6 construct (see “Plasmid constructs”) served as a template.
The primers used for inserting the mutations in the BCL6 p53RE were as follows (5′ to 3′; the mutated nucleotides are denoted in lowercase): mut1-Sense, GCAGAGAGGACGAGAtAGTGCTTGGGGGGTG; antisense, CACCCCCCAAGCACTaTCTCGTCCTCTCTGC; and mut2-Sense, GGGGGGTGATTCGGGtTAGTCTGGGGGCTGTC; antisense, GACAGCCCCCAGACTAaCCCGAATCACCCCCC. All mutations were confirmed by DNA sequencing using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Transient transfections
U2OS cells were used for transient transfection assays with the JetPEI transfection reagent (Polyplus Transfection, Illkirch, France), according to the manufacturer's protocol. The plasmids added were 1 μg of either the wild-type or the mutated BCL6 luciferase reporter vectors (pGL2-basic; Promega) and either 100 to 1000 ng of wild-type p53 (pCDNA3; Promega) or 500 ng of mutant p53-R175H (pCDNA3; Promega). The dual luciferase reporter assay system (Promega) was used for measuring the level of Firefly luciferase activity and for normalizing the results to Renilla luciferase activity (CMV vector; Promega).
Doxorubicin and PFT-α treatments
Lymphoblastoid B cells were treated with 0.5 μg/mL doxorubicin and harvested after 6, 12, 18, and 24 hours. In the experiments with PFT-α, 40 μM of PFT-α or cyclic PFT-α (Alexis Biochemicals, San Diego, CA) were added 4 hours prior to treatment with 0.5 μg/mL doxorubicin, and cells were harvested after an additional 18 hours. U2OS cells were treated with 0.2 μg/mL doxorubicin and harvested after 6 and 24 hours.
Western blot analysis
Total protein was extracted from lymphoblastoid B cells and subjected to Western blot analysis using the following antibodies: anti-BCL6 (Imgenex, San Diego, CA; catalog no. IMG-582), anti-p53 (Santa Cruz Biotechnology; catalog no. sc-6243), and anti-β-actin (Santa Cruz Biotechnology; catalog no. sc-1616).
Quantitative real-time PCR analysis
Quantitative real-time PCR (QPCR) was performed using the TaqMan Universal PCR Master Mix and the SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's protocol, with the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Samples were normalized to either β2-microglobulin25 or β-actin in the human samples and to Gapdh in the murine samples. The primers used were as follows (5′ to 3′; unless noted, the final concentration was 500 nM): human BCL6 forward, CCTGCAGATGGAGCATGTTG; reverse, CATCAGCATCCGGCTGTTG; human p21 forward, GCAGACCAGCATGACAGATTTC; reverse, GCGCTTCCAGGACTGCAG; human β2-microglobulin forward, AGGCTATCCAGCGTACTCCAAAG; reverse, TCTGCTGGATGACGTGAGTAAAC; human β-actin forward, TGTGGCATCCACGAAACTACC; reverse, CTCAGGAGGAGCAATGATCTTGAT; murine Bcl6 forward, GGCCTCCTTCCGCTACAAG; reverse, CTGCGCTCCACAAATGTTACA; Taqman Probe (FAM), AAGACTGTCCACACGGG; murine Gapdh (final concentration 100 nM) forward, TGTGTCCGTCGTGGATCTGA; reverse, GATGCCTGCTTCACCACCTT; Taqman Probe (VIC)-CCTGGAGAAACCTGCCA; murine p21 forward, GGAACATCTCAGGGCCGAA; reverse, TGGGCACTTCAGGGTTTTCT.
Results
Overexpression of p53 increases BCL6 mRNA in human cells
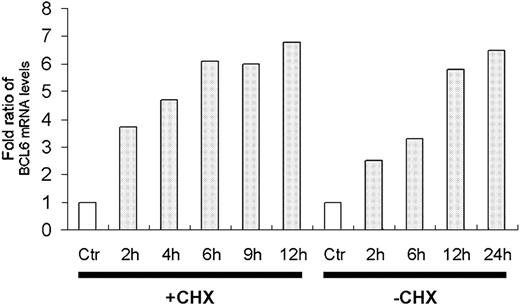
The transcriptional program regulated by p53 was analyzed using DNA microarrays. H1299 cells stably expressing the temperature-sensitive p53 were used to quantitate mRNA levels at different time points after shifting the temperature to 32°C, thus activating p53. Inhibition of protein synthesis by cycloheximide was employed to distinguish between primary and secondary target genes regulated by p53. BCL6 was up-regulated by p53 in the absence and presence of cycloheximide (Figure 1). Along with earlier data from our laboratory,23 these observations identify the BCL6 gene as a primary target of p53.
The TMDR contains a p53RE evolutionarily conserved in primates
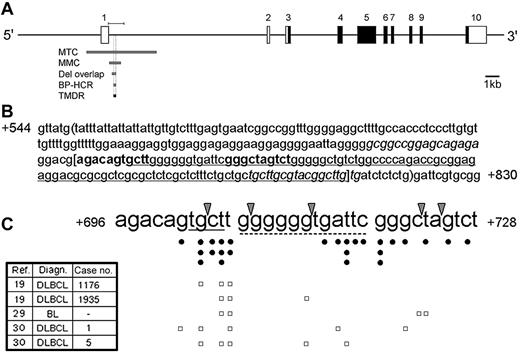
In this study we offer a new term, TMDR, to define a 110-bp region located at the 5′ region of the BCL6 intron 1 (Figure 2A). Its name is derived from the multiple genetic alterations frequently found in this region in BNHL.
p53 binds in a sequence-specific manner to p53REs within the DNA, typically composed of a repeat of the decamer consensus sequence 5′-RRRCWWGYYY-3′ (in which R denotes A or G; W denotes A or T, and Y denotes C or T) separated by 0 to 13 bp.14 The p53REs are not necessarily confined only to upstream regulatory regions. In fact, many p53-regulated genes (eg, GADD45, IGF-BP3, MDM2, and others)26,27 contain p53REs within their introns. The computer algorithm p53MH28 was used to search for p53REs in a region spanning from 5 kb upstream of the transcription start site to the end of intron 1 of the human and murine BCL6 genes. This analysis identified no potential p53RE whose score was above the cutoff defined by Hoh et al.28 The involvement of the 5′ region of the human BCL6 intron 1 in genetic alterations led us to focus our search on the TMDR. Indeed, we identified a putative p53RE at positions +696 to +728 of the human BCL6 gene (Figure 2B). Figure 2C depicts a compilation of documented genetic alterations in this p53RE in lymphomas, mainly in BNHL, gathered from previously published articles.16,19,22,29,30 Figure 2C also displays the specific mutational status of 5 patients from these studies who harbor at least 3 mutations in the p53RE, predicted to severely attenuate p53 binding to this site, out of whom 4 were diagnosed with DLBCL. Since some of the published reports do not describe the mutational status of each patient or the exact location of the mutations, but rather give an overall description of the mutations, it is likely that there are other patients who harbor multiple mutations in this p53RE. In addition, several studies showed that the RGYW motif (A/GGC/TA/T) and its reverse complement WRCY (A/TA/GCC/T) motif are preferred targets for SHM in Ig genes31 as well as in the BCL6 gene.19,32,33 One of these studies, addressing the BCL6 gene, defined the TGCT motif as the second most frequently targeted RGYW/WRCY motif by SHM.19 Indeed, the putative BCL6 p53RE contains a WRCY motif (ie, TGCT) but not an RGYW motif (Figure 2C). Furthermore, palindromic sequences were shown to be preferred targets of SHM,34 and the structure of the consensus p53RE is palindromic. Noteworthy in this context are the results of a recently published meta-analysis of BCL6 mutations in BNHL, according to which the rate of mutations in the BCL6 p53RE is similar to that in the entire MMC.33
BCL6 is a primary target of p53. H1299 cells expressing the ts-p53Val135 were used to identify p53 target genes. The mRNA levels were determined using DNA microarrays and the ratios were calculated by dividing the expression level at each time point by that of the respective control cells. Ctr indicates control cells of H1299 devoid of p53 after 2 hours at 32°C; h, number of hours after shifting the temperature to 32°C; +CHX/-CHX, presence or absence, respectively, of exposure to 10 μg/μL cycloheximide 30 minutes prior to temperature shift to 32°C. □ indicates control.
BCL6 is a primary target of p53. H1299 cells expressing the ts-p53Val135 were used to identify p53 target genes. The mRNA levels were determined using DNA microarrays and the ratios were calculated by dividing the expression level at each time point by that of the respective control cells. Ctr indicates control cells of H1299 devoid of p53 after 2 hours at 32°C; h, number of hours after shifting the temperature to 32°C; +CHX/-CHX, presence or absence, respectively, of exposure to 10 μg/μL cycloheximide 30 minutes prior to temperature shift to 32°C. □ indicates control.
The TMDR contains a p53RE evolutionarily conserved in primates. (A) Schematic representation of the human BCL6 gene. Coding and noncoding exons are indicated by ▪ and □, respectively. MTC indicates major translocation cluster; MMC, major mutation cluster; Del overlap, region of internal deletions overlap; BP-HCR, breakpoint hyper-cluster region; TMDR, translocations, mutations, and deletions region; and the bar above the BCL6 gene, fragment cloned for the luciferase assay. (B) Sequence of a segment of the human BCL6 intron 1 containing a p53RE. The nucleotides are numbered relative to the transcription start site. The 2 decamers of the p53RE are in bold; the TMDR is between brackets; the BP-HCR is underlined; the region of deletions overlap is between parentheses; the primers used for the BCL6 ChIP analysis are italicized. (C) Reported translocations and mutations in the BCL6 p53RE. Arrowheads indicate positions of breakpoints of translocations; •, point mutations; □, each row represents the point mutations of a single patient with at least 3 mutations in the p53RE; Ref, reference; Diagn, diagnosis; DLBCL, diffuse large B-cell lymphoma; BL, Burkitt lymphoma; and —, not available. The WRCY motif (tgct) is underlined with a solid line; the 12-bp spacer is underlined with a dashed line; the details of the patients appear in the left table, along with the case no. from the original article.
The TMDR contains a p53RE evolutionarily conserved in primates. (A) Schematic representation of the human BCL6 gene. Coding and noncoding exons are indicated by ▪ and □, respectively. MTC indicates major translocation cluster; MMC, major mutation cluster; Del overlap, region of internal deletions overlap; BP-HCR, breakpoint hyper-cluster region; TMDR, translocations, mutations, and deletions region; and the bar above the BCL6 gene, fragment cloned for the luciferase assay. (B) Sequence of a segment of the human BCL6 intron 1 containing a p53RE. The nucleotides are numbered relative to the transcription start site. The 2 decamers of the p53RE are in bold; the TMDR is between brackets; the BP-HCR is underlined; the region of deletions overlap is between parentheses; the primers used for the BCL6 ChIP analysis are italicized. (C) Reported translocations and mutations in the BCL6 p53RE. Arrowheads indicate positions of breakpoints of translocations; •, point mutations; □, each row represents the point mutations of a single patient with at least 3 mutations in the p53RE; Ref, reference; Diagn, diagnosis; DLBCL, diffuse large B-cell lymphoma; BL, Burkitt lymphoma; and —, not available. The WRCY motif (tgct) is underlined with a solid line; the 12-bp spacer is underlined with a dashed line; the details of the patients appear in the left table, along with the case no. from the original article.
The putative BCL6 p53RE was not identified by the p53MH algorithm, since it contains an extra nucleotide (ie, guanosine) between the 2 pentamers of the first decamer (Figure 2C). This phenomenon was also observed in the murine Mdm2 p53RE (GAGCTAAGTCCTGACATGTCT26,27 ) and in 1 of the 20 clones, namely “11A2,” by which the consensus p53RE was originally defined (AAACAATGCCCAGACTTGTCT14 ). Further analysis using the BLAST search,35 BLAT search,36 and sequencing of the relevant segment of the Aotus nancymaae owl monkey Bcl6 intron 1 revealed that the BCL6 p53RE is evolutionarily conserved in primates and only partially conserved in Canis familiaris (Table 1). However, this p53RE is not evolutionarily conserved in Mus musculus and Rattus norvegicus.
The BCL6 p53RE is evolutionarily conserved in primates
. | Consensus p53RE . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species . | RRR . | C . | W . | * . | W . | G . | YYY . | 0- to 13-bp spacer . | RRR . | C . | W . | W . | G . | YYY . | |||||||||||||
| Homo sapiens | aga | c | a | g | t | g | ctt | ggggggtgattc | ggg | c | t | a | g | tct | |||||||||||||
| Pan troglodytes | aga | c | a | g | t | g | ctt | ggggggtgattc | ggg | c | t | a | g | tct | |||||||||||||
| Macaca fascicularis | aga | c | a | c | t | g | ctt | gggggtgattc | ggg | c | a | a | g | tct | |||||||||||||
| Macaca mulatta | aga | c | a | c | t | g | ctt | gggggtgattc | ggg | c | a | a | g | tct | |||||||||||||
| Aotus nancymaae | aga | c | a | g | t | g | ctt | ggggggcgattc | ggg | c | a | g | g | tct | |||||||||||||
| Canis familiaris | aga | c | c | g | c | g | ctt | ggggtgctgc | ggg | c | c | a | g | gct | |||||||||||||
. | Consensus p53RE . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species . | RRR . | C . | W . | * . | W . | G . | YYY . | 0- to 13-bp spacer . | RRR . | C . | W . | W . | G . | YYY . | |||||||||||||
| Homo sapiens | aga | c | a | g | t | g | ctt | ggggggtgattc | ggg | c | t | a | g | tct | |||||||||||||
| Pan troglodytes | aga | c | a | g | t | g | ctt | ggggggtgattc | ggg | c | t | a | g | tct | |||||||||||||
| Macaca fascicularis | aga | c | a | c | t | g | ctt | gggggtgattc | ggg | c | a | a | g | tct | |||||||||||||
| Macaca mulatta | aga | c | a | c | t | g | ctt | gggggtgattc | ggg | c | a | a | g | tct | |||||||||||||
| Aotus nancymaae | aga | c | a | g | t | g | ctt | ggggggcgattc | ggg | c | a | g | g | tct | |||||||||||||
| Canis familiaris | aga | c | c | g | c | g | ctt | ggggtgctgc | ggg | c | c | a | g | gct | |||||||||||||
Mismatches to the consensus p53RE are italicized.
Not part of the consensus p53RE; added to enable convenient comparison with the discovered p53REs
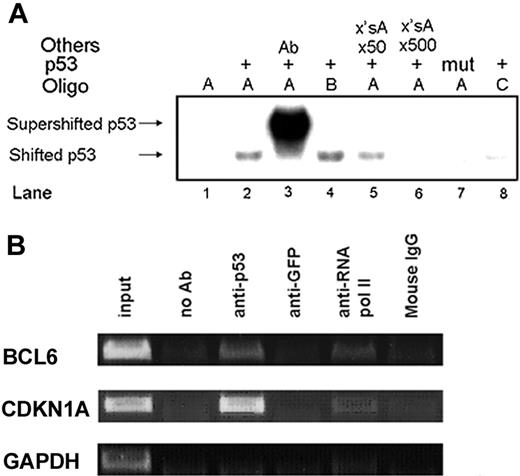
p53 binds to the BCL6 p53RE in vitro and in vivo. (A) p53 in vitro binding to the BCL6 p53RE was assayed by EMSA. Wild-type (lanes 2-6, 8) and mutant (lane 7) p53 proteins were incubated with the specified oligos. Oligo A, wild-type BCL6 p53RE (lanes 1-3, 5-7); Oligo B, like oligo A without the extra guanosine in the middle of the first decamer of the BCL6 p53RE (lane 4); Oligo C, mutated BCL6 p53RE (lane 8); supershift with anti-p53 antibody (lane 3); competition with a 50-fold or 500-fold excess of unlabeled oligo A (lanes 5 and 6, respectively). Ab indicates antibody. ×'sA indicates an excess of unlabeled Oligo A. (B) p53 in vivo binding to the BCL6 p53RE was assayed by ChIP in lymphoblastoid B cells. Immunoprecipitation of p53 protein-DNA complexes was done with anti-p53 antibody. Negative controls were chromatin immunoprecipitated with an irrelevant antibody (anti-GFP) and normal mouse IgG, as well as samples to which no antibody was added (no Ab). Positive control was chromatin immunoprecipitated with anti-RNA polymerase II antibody. Primers specific for the CDKN1A promoter and GAPDH exon 4 served as positive and negative controls, respectively. Input indicates 0.1% of the sonicated chromatin before immunoprecipitation.
p53 binds to the BCL6 p53RE in vitro and in vivo. (A) p53 in vitro binding to the BCL6 p53RE was assayed by EMSA. Wild-type (lanes 2-6, 8) and mutant (lane 7) p53 proteins were incubated with the specified oligos. Oligo A, wild-type BCL6 p53RE (lanes 1-3, 5-7); Oligo B, like oligo A without the extra guanosine in the middle of the first decamer of the BCL6 p53RE (lane 4); Oligo C, mutated BCL6 p53RE (lane 8); supershift with anti-p53 antibody (lane 3); competition with a 50-fold or 500-fold excess of unlabeled oligo A (lanes 5 and 6, respectively). Ab indicates antibody. ×'sA indicates an excess of unlabeled Oligo A. (B) p53 in vivo binding to the BCL6 p53RE was assayed by ChIP in lymphoblastoid B cells. Immunoprecipitation of p53 protein-DNA complexes was done with anti-p53 antibody. Negative controls were chromatin immunoprecipitated with an irrelevant antibody (anti-GFP) and normal mouse IgG, as well as samples to which no antibody was added (no Ab). Positive control was chromatin immunoprecipitated with anti-RNA polymerase II antibody. Primers specific for the CDKN1A promoter and GAPDH exon 4 served as positive and negative controls, respectively. Input indicates 0.1% of the sonicated chromatin before immunoprecipitation.
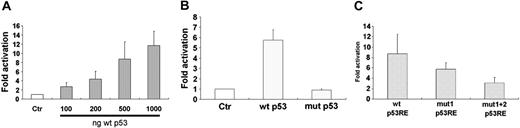
p53 activates transcription from the human BCL6 p53RE. (A) U2OS cells were cotransfected with 1 μg of wild-type BCL6 reporter vector and either an empty vector (Ctr) or wild-type p53-expressing vector in increasing concentrations. Results display mean ± SD of 4 independent experiments, each in triplicate. □ indicates control. (B) U2OS cells were cotransfected with 1 μg of wild-type BCL6 reporter vector and either an empty vector (Ctr) or 500 ng of either wild-type p53 (wt p53) or mutated p53-R175H (mut p53) expressing vectors. Results display mean ± SD of 3 independent experiments, each in triplicate. □ indicates control. (C) U2OS cells were cotransfected with 1 μgof either wild-type BCL6 reporter vector (wt p53RE) or BCL6 reporter vector with a mutation in the first decamer of the p53RE (mut1 p53RE) or in both decamers (mut1 + 2 p53RE) and 500 ng of either wild-type p53-expressing vector or an empty vector. Results displayed are fold activations of luciferase of each of the reporter vectors measured after addition of wild-type p53 divided by that measured after addition of an empty vector. Results display mean ± SD of 4 independent experiments, each in triplicate.
p53 activates transcription from the human BCL6 p53RE. (A) U2OS cells were cotransfected with 1 μg of wild-type BCL6 reporter vector and either an empty vector (Ctr) or wild-type p53-expressing vector in increasing concentrations. Results display mean ± SD of 4 independent experiments, each in triplicate. □ indicates control. (B) U2OS cells were cotransfected with 1 μg of wild-type BCL6 reporter vector and either an empty vector (Ctr) or 500 ng of either wild-type p53 (wt p53) or mutated p53-R175H (mut p53) expressing vectors. Results display mean ± SD of 3 independent experiments, each in triplicate. □ indicates control. (C) U2OS cells were cotransfected with 1 μgof either wild-type BCL6 reporter vector (wt p53RE) or BCL6 reporter vector with a mutation in the first decamer of the p53RE (mut1 p53RE) or in both decamers (mut1 + 2 p53RE) and 500 ng of either wild-type p53-expressing vector or an empty vector. Results displayed are fold activations of luciferase of each of the reporter vectors measured after addition of wild-type p53 divided by that measured after addition of an empty vector. Results display mean ± SD of 4 independent experiments, each in triplicate.
The p53 protein binds to the BCL6 p53RE in vitro
The ability of p53 to bind the putative BCL6 p53RE was determined by an electrophoretic mobility-shift assay (EMSA). Figure 3A demonstrates that a specific protein-DNA complex was formed when wild-type p53 (Figure 3A, lane 2), but not mutant p53 (Figure 3A, lane 7), was used in conjunction with a synthetic double-stranded oligonucleotide comprising the putative BCL6 p53RE. In addition, a p53-specific antibody (pAb421) was able to supershift the p53-DNA complex (Figure 3A, lane 3) along with its known ability to stabilize DNA binding of p53.37,38 The addition of a 50- or 500-fold excess of unlabeled p53 target site oligonucleotide reduced and diminished, respectively, p53 binding to the labeled probe (Figure 3A, lanes 5 and 6, respectively). Deletion of the extra guanosine in the middle of the first decamer slightly enhanced the binding of p53 (Figure 3A, lane 4). These results indicate that p53 binds specifically to the putative BCL6 p53RE in vitro.
The p53 protein binds to the BCL6 p53RE in vivo
ChIP analysis was used to determine whether the BCL6 p53RE is bound by p53 in vivo. Lymphoblastoid B cells were subjected to ChIP analysis with anti-p53 antibody. Since p53-DNA binding activity is not significantly increased in response to genotoxic stress,39 ChIP analysis was performed on untreated cells. Primers were designed to amplify a short segment of the BCL6 gene containing the p53RE (Figure 2B). As shown in Figure 3B, this segment was amplified by PCR when using an anti-p53 antibody or anti-RNA polymerase II antibody as a positive control and not when using the negative controls. HCT116 and U2OS cells were also subjected to ChIP analysis and provided similar results (data not shown). These results, together with the EMSA data, indicate that the putative BCL6 p53RE is functional.
The p53 protein activates transcription from the human BCL6 p53RE
To determine whether the putative BCL6 p53RE is transcriptionally regulated by p53, a 1.154-kb fragment of the human BCL6 gene, spanning the 3′ end of exon 1 and the 5′ region of intron 1, containing the TMDR with the p53RE in it (Figure 2A), was cloned into a luciferase reporter vector. U2OS cells were cotransfected with this BCL6 reporter vector and a wild-type or mutant p53-expressing vector. As shown in Figure 4A and 4B, wild-type p53, but not mutant p53, could activate transcription from the human BCL6 intron 1. To further investigate the role of the BCL6 p53RE in mediating activation of BCL6 transcription, we generated BCL6 reporter vectors bearing mutations in the p53RE in the most conserved residue, namely, the cytidine in the fourth position of the decamer. As shown in Figure 4C, introducing a mutation in one of the decamers of the p53RE reduced activation of BCL6 transcription, and introducing mutations in both decamers of the p53RE significantly reduced activation of BCL6 transcription.
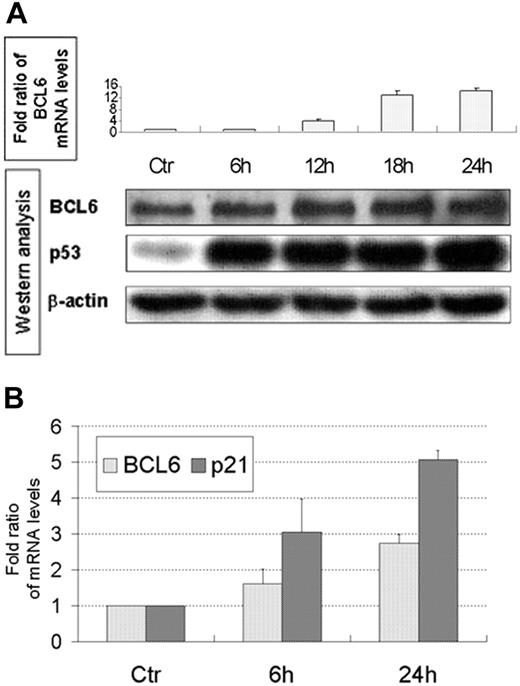
Doxorubicin induces BCL6 mRNA and protein in human cells
Chemotherapy induces p53 activation. Hence, we examined the levels of BCL6 mRNA, using QPCR after treating lymphoblastoid B cells and U2OS cells with doxorubicin. Indeed, an increase in BCL6 mRNA levels was observed in both cell lines (Figure 5A, top; 5B) and in p21 mRNA levels in U2OS cells (Figure 5B). The increase in p21 mRNA levels, a major transcriptional target of p53, is directly related to p53 activity and suggests that the increase in BCL6 mRNA levels is also due to p53 activity. Furthermore, an increase in BCL6 protein levels was observed in lymphoblastoid B cells (Figure 5A, bottom).
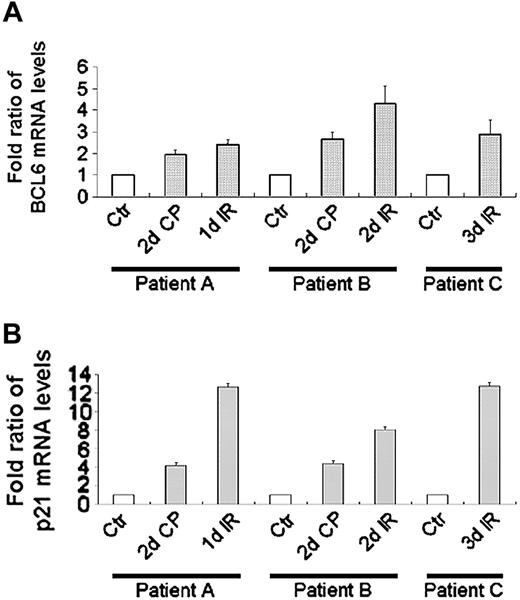
BCL6 mRNA increases after chemotherapy/radiotherapy in human patients
In view of the high levels of BCL6 mRNA detected in whole blood, as reported by Genomics Institute of the Novartis Research Foundation (GNF),40 we examined PBLs taken from human patients subjected to chemotherapy and/or whole body γ-irradiation as part of their treatment. QPCR analysis revealed that chemotherapy/radiotherapy resulted in an increase in BCL6 mRNA (Figure 6A) as well as in p21 mRNA (Figure 6B). These results, together with the doxorubicin-induced increase in BCL6 mRNA and protein in lymphoblastoid B cells and U2OS cells, indicate that BCL6 is up-regulated in response to DNA damage under conditions that lead to p53 activation.
Doxorubicin induces BCL6 mRNA and protein in lymphoblastoid B cells and mRNA in U2OS cells. (A) (Top) Lymphoblastoid B cells were treated with doxorubicin and harvested at the indicated times. BCL6 mRNA levels were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications repeated 3 times, each in duplicate. Ctr indicates untreated cells. (Bottom) Western blot analysis of BCL6, p53, and β-actin in the lymphoblastoid B cells after the same treatment with doxorubicin. Results displayed are representative of 2 independent experiments. (B) U2OS cells were treated with doxorubicin and harvested at the indicated times. BCL6 mRNA levels were determined using QPCR and normalized to β-actin. p21 served as positive control. Results display mean ± SD of quantifications of 3 independent experiments, each repeated 3 times, each in duplicate. Ctr indicates untreated cells.
Doxorubicin induces BCL6 mRNA and protein in lymphoblastoid B cells and mRNA in U2OS cells. (A) (Top) Lymphoblastoid B cells were treated with doxorubicin and harvested at the indicated times. BCL6 mRNA levels were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications repeated 3 times, each in duplicate. Ctr indicates untreated cells. (Bottom) Western blot analysis of BCL6, p53, and β-actin in the lymphoblastoid B cells after the same treatment with doxorubicin. Results displayed are representative of 2 independent experiments. (B) U2OS cells were treated with doxorubicin and harvested at the indicated times. BCL6 mRNA levels were determined using QPCR and normalized to β-actin. p21 served as positive control. Results display mean ± SD of quantifications of 3 independent experiments, each repeated 3 times, each in duplicate. Ctr indicates untreated cells.
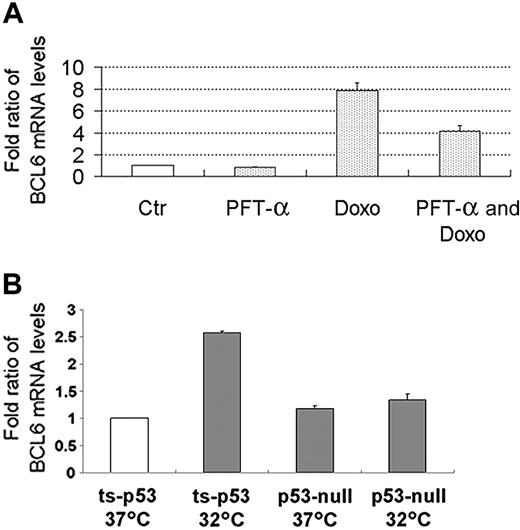
The increase in BCL6 mRNA is p53 dependent
To determine whether the induction of BCL6 by DNA damage was p53 dependent, we examined the effects of pretreatment with the p53 inhibitor PFT-α on the increase in BCL6 mRNA upon doxorubicin treatment. Indeed, pretreatment with PFT-α attenuated the increase in BCL6 mRNA (Figure 7A); similar results were obtained with pretreatment with cyclic PFT-α (data not shown). Thus, the increase in BCL6 mRNA levels after treatment with doxorubicin is due, at least in part, to the activity of p53. The incomplete abrogation of this increase may be due to the contribution of additional regulators of BCL6 expression or to incomplete inhibition of p53 by the drug. In order to validate that BCL6 is directly activated by p53, we used H1299 cells stably expressing the ts-p53Val135 and the parental H1299 p53-null cells. Indeed, expression of BCL6 was induced in H1299 cells with ts-p53Val135 and not in H1299 p53-null cells (Figure 7B).
Bcl6 mRNA is not induced in irradiated mice
To determine whether Bcl6 is induced by irradiation in mice, Tp53+/+ and age-matched Tp53-/- mice were treated with 5 Gy ionizing radiation, and the spleens and thymuses were harvested at 0, 6, and 24 hours after irradiation. Bcl6 and p21 mRNA levels were monitored by QPCR. In contrast to the p53-dependent induction of p21, there was no significant change in Bcl6 mRNA in response to γ-irradiation in either Tp53+/+ or Tp53-/- mice (Supplemental Figure S1, available at the Blood website; click on the Supplemental Figure link at the top of the online article). In view of the negative autoregulation of Bcl6,41 spleens were harvested also at 1-hour intervals between 0 and 6 hours in order to detect any transient increase in Bcl6 mRNA, which was not observed. As expected,42 basal Bcl6 mRNA levels were similar in both Tp53+/+ and Tp53-/- mice (data not shown). Thus, as predicted from the lack of a p53RE in the Bcl6 gene in rodents, p53 does not appear to activate Bcl6 in mice.
Discussion
In this study we identified BCL6 as a new target gene of p53. We propose that in B cells undergoing maturation in the GC, p53 is activated in response to the breaks formed in the genomic DNA due to SHM and CSR.34,43 Indeed, double-strand breaks generated by V(D)J recombination were shown to activate a p53-dependent DNA damage checkpoint in scid lymphocyte precursors, limiting the oncogenic potential of these breaks.44 Thus, in B cells undergoing maturation in the GC, extended activity of p53 is likely to lead to apoptosis or permanent cell-cycle exit. This undesirable outcome is avoided through the induction of BCL6 expression by p53. The ability of BCL6 to affect cell-cycle control was demonstrated in several studies, including DNA microarray screening for BCL6 target genes,5 the discovery of BCL6's ability to inhibit cellular senescence induced by p53,45 the identification of the human programmed cell-death 2 gene (PCDC2) as a BCL6 target,46 and the ability of BCL6 to immortalize primary B cells in the absence of p53 function.7 Interference with BCL6 function by a specific peptide revealed that BCL6 is essential for survival and proliferation of B-cell lymphoma cells.10 Importantly, BCL6 can directly suppress p53 gene expression and p53-mediated apoptotic responses to DNA damage in B cells.11 Combined with our findings, this defines a novel autoregulatory loop in which p53 is activated in response to genomic alterations that occur normally during B-cell maturation in the GC. In the correct physiologic context, this subsequently leads to induction of BCL6, which down-regulates p53 mRNA and spares the cell from the apoptotic effect of extended p53 activation. This down-regulation of p53 is predicted, in turn, to attenuate the induction of BCL6, allowing it to return to baseline levels and thereby defining a tightly controlled time window for completion of B-cell maturation.
BCL6 mRNA increases after chemotherapy/radiotherapy in human patients. BCL6 (A) and p21 (B) mRNA levels in WBCs taken from patients treated with chemotherapy/radiotherapy were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications repeated 3 times, each in triplicate. Labels adjacent to the x-axis indicate the time in which the PB was taken. Fold ratios of mRNA levels were calculated in comparison to the control samples for each patient. Ctr indicates control; d, number of days of the detailed treatment; CP, cyclophosphamide; IR, whole body irradiation; patient A, a 23-year-old female with prolymphocytic leukemia; patient B, a 29-year-old male with T-ALL; patient C, a 39-year-old female with Philadelphia-positive ALL. □ indicates control.
BCL6 mRNA increases after chemotherapy/radiotherapy in human patients. BCL6 (A) and p21 (B) mRNA levels in WBCs taken from patients treated with chemotherapy/radiotherapy were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications repeated 3 times, each in triplicate. Labels adjacent to the x-axis indicate the time in which the PB was taken. Fold ratios of mRNA levels were calculated in comparison to the control samples for each patient. Ctr indicates control; d, number of days of the detailed treatment; CP, cyclophosphamide; IR, whole body irradiation; patient A, a 23-year-old female with prolymphocytic leukemia; patient B, a 29-year-old male with T-ALL; patient C, a 39-year-old female with Philadelphia-positive ALL. □ indicates control.
The increase in BCL6 mRNA is p53 dependent. (A) The increase in BCL6 mRNA levels after treatment with doxorubicin in lymphoblastoid B cells is attenuated as a result of pretreatment with PFT-α. Lymphoblastoid B cells were subjected to the detailed treatments. BCL6 mRNA levels were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications of 3 independent experiments, each repeated 2 times, each in duplicate. Ctr indicates untreated cells. PFT-α was added at -4 hours; doxorubicin (Doxo) was added at 0 hours; all samples were harvested at +18 hours. □ indicates control. (B) H1299 cells with ts-p53Val135 (ts-p53) or the parental H1299 cells (p53-null) were maintained either at 37°C (p53 inactive) or placed overnight at 32°C (p53 active). BCL6 mRNA levels were determined using QPCR and normalized to β-actin. Fold ratios of mRNA levels were calculated in comparison to that measured in the H1299 cells with ts-p53Val135 maintained at 37°C. Results display mean ± SD of quantifications repeated 3 times, each in triplicate. □ indicates control.
The increase in BCL6 mRNA is p53 dependent. (A) The increase in BCL6 mRNA levels after treatment with doxorubicin in lymphoblastoid B cells is attenuated as a result of pretreatment with PFT-α. Lymphoblastoid B cells were subjected to the detailed treatments. BCL6 mRNA levels were determined using QPCR and normalized to β2-microglobulin. Results display mean ± SD of quantifications of 3 independent experiments, each repeated 2 times, each in duplicate. Ctr indicates untreated cells. PFT-α was added at -4 hours; doxorubicin (Doxo) was added at 0 hours; all samples were harvested at +18 hours. □ indicates control. (B) H1299 cells with ts-p53Val135 (ts-p53) or the parental H1299 cells (p53-null) were maintained either at 37°C (p53 inactive) or placed overnight at 32°C (p53 active). BCL6 mRNA levels were determined using QPCR and normalized to β-actin. Fold ratios of mRNA levels were calculated in comparison to that measured in the H1299 cells with ts-p53Val135 maintained at 37°C. Results display mean ± SD of quantifications repeated 3 times, each in triplicate. □ indicates control.
The p53-BCL6 autoregulatory loop is not unique for either of these proteins. A similar loop exists for p53 and its negative regulator MDM2.47 In addition, a double-negative loop, in which each partner suppresses the other, is already defined for BCL6 and BLIMP-1.5,6 Thus, BCL6 might reside at the junction between 2 feedback loops.
Although delineation of the p53 pathways has become clearer, as many target genes related to apoptosis and cell-cycle arrest have been identified, the molecular mechanisms behind p53-mediated differentiation are still poorly understood. Here we identify a new target gene of p53, which is closely related to regulation of differentiation.
The novel BCL6 p53RE is evolutionarily conserved only in primates. In order to confirm the lack of BCL6 activation by p53 in rodents, we performed experiments in Tp53+/+ and Tp53-/- mice, which indeed showed that BCL6 is not induced by p53 in mice. Therefore, we hypothesize that BCL6 induction by p53, and hence the p53-BCL6 loop, exists only in primates.
Our results are apparently different from those reported by others.11,48 We show that under conditions that elevate p53 levels (Figures 5, 6) or activation of p53 (Figure 7B), BCL6 levels are increased. On the other hand, it was shown that under conditions that activate p53 (etoposide treatment), BCL6 is down-regulated in the Burkitt lymphoma cell lines Daudi, Raji, and Ramos and in DLBCL Val cells.11,48 The reason for this discrepancy may depend on the functionality of p53 and the status of the BCL6 gene in the particular cell line used and on the type of insult used for p53 activation. While the EBV-transformed lymphoblastoid B-cell line used by us has a functional p53, it was shown that the Burkitt lymphoma cell lines specified earlier in this paragraph neither carry a normal TP53 gene nor display normal p53 responses to various insults, including etoposide treatment.49-51 In addition, although DLBCL Val cells do possess a functional p53, the BCL6 gene in these cells contains a deletion in one allele (see “Introduction”) and a translocation in the second allele,52 resulting in the elimination of the p53RE in both alleles. Hence, in those Burkitt lymphoma and DLBCL Val cells the regulation of BCL6 by p53 is expected to be impaired.
Compilation of previous studies was used to identify the TMDR, a region commonly affected by genetic alterations in BNHL. Therefore, it is plausible to assume that the TMDR mediates the interaction with various transcription and regulatory factors that normally regulate BCL6 expression and whose disruption by genetic alterations contributes to deregulated BCL6 expression in BNHL. Several attempts were made to identify binding sites of transcription factors within the TMDR using EMSA probes within the breakpoint hyper-cluster region16,22 and within the region of deletion overlap,21 but no significant results were hitherto obtained. In this study, we identified a p53RE, containing an extra nucleotide, located in the TMDR. p53 interacts with DNA as a tetramer (ie, each p53 monomer binds 1 of the 4 pentamers comprising the 2 decamers of the p53RE).14 We hypothesize that the addition of a single base between the 2 pentamers, as observed in the BCL6 p53RE, does not interfere severely with p53 binding. Assuming that p53 binding to the RE is abrogated in those BNHL cases harboring either translocations located in the RE or downstream to the RE, multiple mutations in the RE, or deletions of the RE, the predicted result is deregulation of BCL6 expression. The clinical data (ie, genetic alterations in the BCL6 p53RE found in BNHL) indicate that there is a specific phase in lymphomagenesis in which the loss of BCL6 activation by p53 is related to formation of BNHL. This deregulation may not be evident as a high/low level of BCL6 in the full form of BNHL because the underlying reason for BNHL formation may be the untimely activation of BCL6 during GC reaction rather than the absolute level of expression.
So far, the linkage between lymphomagenesis and genetic alterations in BCL6 is unclear. Assuming that the frequent presence of translocations in the BCL6 gene in BNHL is related to deregulation of BCL6 expression and lymphomagenesis, studies were aimed at finding a correlation between these manifestations. The promoter substitution phenomenon caused by translocations was reported to trigger persistent expression of BCL6.53,54 However, several other studies showed no significant correlation between rearrangements of the BCL6 gene and deregulation of its expression.55-58 Therefore, it was concluded that BCL6 gene rearrangements may not necessarily cause elevation of BCL6 expression and that these rearrangements are not a sine qua non for increased BCL6 expression.58 In addition, there is no consensus over the effect of BCL6 translocations on prognosis. Previous studies have reported that translocations may indicate a favorable prognosis,59 an unfavorable prognosis,60 or no effect.15,61
Point mutations in the BCL6 gene were considered a histogenetic marker for the normal physiologic transit of B cells through the GC.18 Thus, the high proportion of BNHL cases with mutations in BCL6 reflects the frequent GC or post-GC origin of these tumors rather than their malignant nature. Studies showed that mutations are correlated with favorable prognosis in DLBCL,29,62 unfavorable prognosis in FL63 and chronic lymphocytic leukemia,64 or no effect in DLBCL19 and classic Hodgkin disease.65 These contradicting conclusions may imply that the location of the mutations determines their effect. Thus, stratification of the mutations could refine the correlation with prognosis. This refinement may also explain the insignificant difference between the frequency of mutations reported in normal and malignant B cells.18 Indeed, it was shown that a subset of specific mutations in the first noncoding exon of BCL6 causes disruption of its negative autoregulation in DLBCL, thus deregulating BCL6 expression.41 As described in “Introduction,” BCL6 mediates survival, proliferation, and differentiation blockade. Thus, aberrant high expression of BCL6, caused by disruption of its negative autoregulation, may lead to lymphomagenesis. It is noteworthy that elevated BCL6 mRNA and protein levels were demonstrated to predict favorable prognosis in several studies of DLBCL,66 T-ALL,67 and primary central nervous system lymphoma.68
Further studies are needed to define a subgroup of BNHL patients in which the BCL6 p53RE is not functional and to search for clinical implications.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-04-1629.
G.R. holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel-Aviv University, Israel.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the Arison family for their donation to Pediatric Oncology. We thank Hanna Ben-Bassat for establishing the EBV-transformed lymphoblastoid B-cell line. This work was performed in partial fulfillment of the requirements for a PhD degree of Ofer Margalit, and Hila Amram, Sackler Faculty of Medicine, Tel-Aviv University, Israel.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal