The BCR-ABL oncoprotein of chronic myelogenous leukemia (CML) localizes to the cell cytoplasm, where it activates proliferative and antiapoptotic signaling pathways. We previously reported that the combination of the ABL kinase inhibitor imatinib mesylate (IM) and the nuclear export inhibitor leptomycin B (LMB) traps BCR-ABL inside the nucleus, triggering the death of the leukemic cells. To evaluate the efficacy of the combination of IM and LMB on human cells we collected CD34-positive cells from 6 healthy donors and myeloid progenitors from 35 patients with CML. The sequential addition of IM and LMB generated the strongest reduction in the proliferative potential of the leukemic cells, with limited toxicity to normal myeloid precursors. Furthermore, nested reverse transcriptase-polymerase chain reaction (RT-PCR) analysis on colonies representative of each experimental condition demonstrated that the combination of IM and LMB was the most effective regimen in reducing the number of BCR-ABL-positive colonies. The efficacy of the 2-drug association was independent of the clinical characteristics of the patients. Our results indicate that strategies aimed at the nuclear entrapment of BCR-ABL efficiently kill human leukemic cells, suggesting that the clinical development of this approach could be of significant therapeutic value for newly diagnosed and IM-resistant CML patients.
Introduction
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder characterized by a shortened chromosome 22 indicated as the Philadelphia chromosome (Ph).1,2 This cytogenetic alteration—produced by a balanced translocation involving chromosomes 9 and 22—juxtaposes the first exons of the BCR gene with most of the ABL sequence, thus generating the BCR-ABL chimeric oncogene.3,4 The resulting BCR-ABL oncoprotein is a constitutively active tyrosine kinase that activates multiple proliferative and antiapoptotic signaling pathways, thereby facilitating the expansion of the leukemic myeloid progenitors.5
The development of imatinib mesylate (IM), a semispecific inhibitor of BCR-ABL kinase activity, has radically changed the therapeutic approach to CML.6 IM is currently considered the first line of treatment for patients with CML in the chronic phase of the disease and is the drug of choice to induce a rapid—although usually short-lived—remission in the more advanced stages of CML.7-11 However, about one-third of chronic-phase patients treated with IM alone will develop resistance to the drug. This phenomenon is usually due to the reactivation of the BCR-ABL tyrosine kinase that can be explained by 2 alternative mechanisms: (1) expansion of pre-existing BCR-ABL mutant clones that have a lower affinity for IM or have lost their ability to bind the compound caused by the selective pressure exerted by IM treatment; (2) amplification or overexpression of the BCR-ABL gene, leading to an increase in the number of BCR-ABL protein molecules expressed by each leukemic cell. Under these circumstances, the amount of drug that penetrates inside the cells is insufficient to inactivate all copies of BCR-ABL, and the patients lose their response to the drug.12-16 Afurther concern is the increasing evidence suggesting that treatment with IM alone fails to produce complete molecular responders (ie, individuals with a negative RT-PCR for the BCR-ABL transcript), let alone eradicate the Ph-positive stem-cell population.17-19
We have previously reported that although BCR-ABL retains the 3 nuclear localization signals of the ABL moiety, the oncoprotein localizes exclusively to the cell cytoplasm because of a lack of nuclear import. Inactivation of BCR-ABL tyrosine kinase by IM partially restores the import of the oncoprotein that can be trapped in the nucleus by the sequential addition of the nuclear export inhibitor leptomycin B (LMB). In the nucleus, reactivation of BCR-ABL catalytic activity by drug washout triggers an apoptotic response both in immortalized cell lines and in murine bone marrow cells infected with a BCR-ABL retroviral vector.20
In order to assess the efficacy of this approach on primary human CML cells, we isolated myeloid progenitors from the bone marrow of 35 patients affected with CML. As a control, we used CD34-positive cells from 6 healthy volunteers. The cells were treated with IM, LMB, or with the sequential association of both compounds. We found that the latter treatment regimen was the most effective in reducing the proliferative potential of CML myeloid progenitors. Moreover, the combination of IM and LMB induced preferential killing of BCR-ABL-positive cells, partially preserving normal progenitors present in the bone marrow of individuals diagnosed with CML. The efficacy of the 2-drug treatment appeared to be independent of any clinical characteristic of the patients.
Patients, materials, and methods
Patient accrual
Bone marrow specimens were obtained from 35 patients with CML followed by the Division of Clinical and Experimental Hematology within the Department of Biomedical Science. All patients signed an informed consent notice that was approved by the Bioethical Committee of the University of Catania.
Isolation of myeloid precursors
CML-committed hematopoietic cells were isolated from 5 mL of bone marrow by gradient centrifugation on a Ficoll-Hypaque gradient (Amersham Biosciences, Uppsala, Sweden) following the manufacturer's instructions. In the presence of significant erythrocyte contamination, cells were incubated for 10 minutes in ice with an isotonic erythrocyte lysis buffer (0.8% NH4Cl, 10 μM EDTA) in a 1:4 cell-lysis buffer ratio. After vigorous vortexing, cells were centrifuged for 5 minutes at 300g. This procedure was repeated up to 3 times depending on the amount of erythrocytes left after each lysis. Viable cells were then counted by trypan blue exclusion, and placed in RPMI 1640 (Sigma-Aldrich, St Louis, MO) supplemented with 10% fetal bovine serum (FBS; Cambrex, Baltimore, MD) at a density of 1 × 106/mL.
As a control, CD34-positive cells were obtained from the bone marrow of 6 consenting healthy volunteers by immuno-affinity selection using MiniMacs paramagnetic beads (Miltenyi Biotec, Auburn, CA) as previously described.21
Drug treatments and colony-forming assays
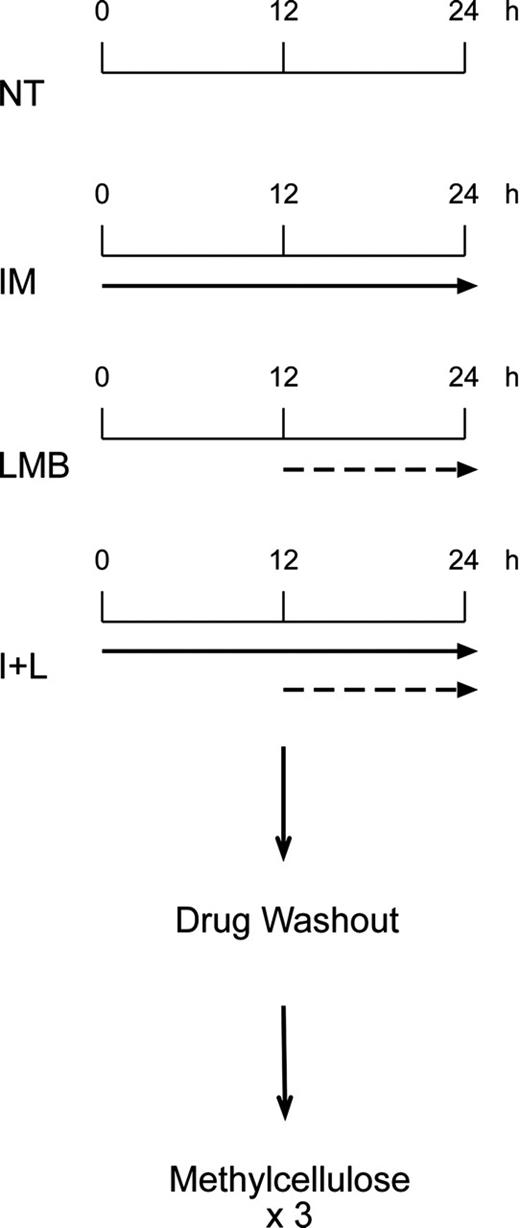
Myeloid precursors were left in plain media, or exposed to 10 μM IM (Novartis Pharma, Basel, Switzerland), 10 nM LMB in 70% methanol (Sigma-Aldrich), or a combination of the 2 drugs as indicated in the scheme depicted in Figure 1. At the end of treatments, cells were rinsed 3 times in phosphate-buffered saline (PBS), diluted 1:10 in methylcellulose media (Methocult GF H4434; Stem Cell Technologies, Vancouver, BC, Canada) and plated in triplicates (1 × 105) in 35-mm cell-culture dishes (Corning, Acton, MA). After a 3- to 4-week incubation at 37°C, erythroid burst-forming unit (BFU-E) and granulocyte-macrophage colony-forming unit (CFU-GM) colonies were counted by 2 different operators unaware of the treatment conditions of each plate. Results were calculated both as total number of colonies per each condition and as the percentage of colony growth after different treatments, with untreated colonies arbitrarily set at 100%.
RNA extraction and RT-PCR reactions
Using a 2.5-μL pipette we plucked 12 individual colonies (4 from each plate forming a triplicate) for each treatment condition. Thus, we collected a total of 48 colonies (12 untreated [NT], 12 IM, 12 LMB, and 12 IMB+LMB) from each patient. Total RNA was extracted from individual colonies by resuspending them in 250 μL Trizol reagent (Invitrogen, Carlsbad, CA) and following the manufacturer's indications. The RNA obtained from this procedure was eluted in 8 μL DEPC (diethyl pyrocarbonate) water.
To detect the presence of the BCR-ABL transcript we used 4 μL total RNA to perform a 1-step RT-PCR (Qiagen, Valencia, CA). Samples that failed to express the BCR-ABL transcript were subjected to a nested PCR using 1 μL of the first amplification reaction. All specimens that scored negative by nested PCR were analyzed for the expression of the ubiquitous gene NCOA4 to verify quality and quantity of the RNA and exclude false-negative results. In this case, the remaining 4 μL total RNA isolated from negative colonies were used to perform a 1-shot RT-PCR for NCOA4. Unfortunately, in 9 of the 35 patients included in the study we failed to detect both BCR-ABL and NCOA4 expression. Hence, qualitative data from these patients were considered uninterpretable and excluded from the study.
Primers used for our reactions were: p190 first BCR e1A, GACTGCAGCTCCAATGAGAAC; p210 first BCR b1A, GAAGTGTTTCAGAAGCTTCTCC; p190-p210 first ABL a3B, GTTTGGGCTTCACACCATTCC; p190 nested BCR e1C, CAGAACTCGCAACAGTCCTTC; p210 nested BCR B2C, CAGATGCTGACCAACTCGTGT; p190-p210 nested ABL a3D, TTCCCCATTGTGATTATAGCCTA; NCOA4 forward, ATTGAAGAAATTGCAGGCTC; and NCOA4 reverse, TGGAGAAGAGGAGCTGTATCT.
Statistical analysis
The distribution of cell counts was assumed to follow a negative binomial (NB) model. This distribution is able to account for overdispersion not explained by the Poisson distribution usually used to model counts. In order to estimate the effect of each of the 2 treatments (IM and LMB), we employed a NB random effect model accounting for the correlation of data collected on the same subjects. The coefficients estimated by the model represent multiplicative factors estimating the effect of each treatment as compared to the untreated group: N predicted = N0 * exp(B1*STI) × exp(B2*LMB) * exp(B3*INTERACTION) where N predicted indicates the number of cell counts predicted by the model; N0, number of cell counts in the untreated group; Exp(B1), effect of IM; Exp(B2), effect of LMB; and Exp(B3), multiplicative effect of IM plus LMB. A coefficient of 0.60, for example, indicates that the average cell count in that specific treatment group was 60% that of the corresponding untreated group. The percentage of cell-count reduction in treated colonies is obtained by subtracting the estimated coefficient from unity (for example, [1-0.60] × 100 = 40% reduction). When the interaction coefficient is equal to 1 this indicates that the combined effect of the 2 treatments is that expected under the multiplicative model. When it is lower than 1, it indicates that the cell-count reduction is higher than that expected by an independent effect of both treatments (positive interaction). Conversely, when the coefficient is higher than 1, it indicates that the cell-count reduction is lower than that expected by an independent effect of both treatments (negative interaction).
To establish whether any of our treatment conditions could discriminate between BCR-ABL-negative and BCR-ABL-positive cells, we analyzed 12 colonies from each experimental condition for the expression of the chimeric oncogene. The proportion of colonies positive for the BCR-ABL gene in the 4 experimental conditions (no treatment, IM, LMB, and both) was analyzed using a logistic regression model stratified per patient.
A first model was fitted evaluating, as the main effects, the role of IM and of LMB alone in reducing the probability of having a positive colony. Again, in order to assess whether the effect of each of the 2 treatments (IM and LMB) was independent of the presence of the other treatment an interaction term was included in the model. The coefficients estimated by the model represent multiplicative factors estimating odds ratios (ORs): an OR is a measure of the probability of being positive for each treated colony compared with the untreated colonies. An OR of 1 means no effect of the treatment. An OR below unity indicates a reduction in the probability of being positive under a specific treatment condition, while an OR above unity means an increase in the probability of being positive under treatment.
All clinical parameters were coded as two-level variables. The same logistic model was run for both levels of each clinical variable in order to investigate whether differences in the effects of the 2 treatments alone or combined could be observed in subgroups with different clinical characteristics. To summarize the results of this analysis we chose a graphical presentation, presenting the ORs together with the 95% confidence intervals (CIs) for each treatment and for their combination.
Treatment schedule followed in the study. CD34-positive cells from healthy individuals and Ficoll-gradient-purified myeloid progenitors from patients with CML were either left untreated (NT) or exposed to 10 μM IM (solid arrow) for 24 hours, 10 nM LMB (dashed arrow) for 12 hours, or a combination of the 2 drugs (I+L). In this case, cells were kept for 24 hours in IM, and LMB was added for the last 12 hours. At the end of each treatment cells were washed free of drugs and plated in triplicates in methylcellulose.
Treatment schedule followed in the study. CD34-positive cells from healthy individuals and Ficoll-gradient-purified myeloid progenitors from patients with CML were either left untreated (NT) or exposed to 10 μM IM (solid arrow) for 24 hours, 10 nM LMB (dashed arrow) for 12 hours, or a combination of the 2 drugs (I+L). In this case, cells were kept for 24 hours in IM, and LMB was added for the last 12 hours. At the end of each treatment cells were washed free of drugs and plated in triplicates in methylcellulose.
Results
Effect of different drug regimens on the colony-forming capacity of healthy myeloid progenitors
We have previously reported that the combined treatment of IM and LMB can efficiently kill murine bone marrow cells expressing the BCR-ABL oncogene or human cell lines derived from individuals diagnosed with CML. In this study we wanted to analyze the effect of the combination of IM and LMB on myeloid progenitors isolated from healthy donors (as a control) or patients with CML.
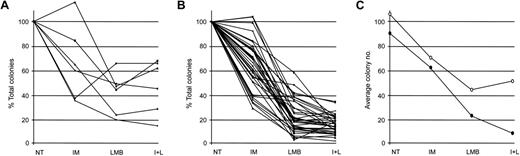
We initially studied the effect of IM and LMB—alone or in combination—on the proliferative potential of normal myeloid progenitors. We collected CD34-positive cells from 6 healthy donors and placed them in drug-free media or in culture media containing 10 μM IM, 10 nM LMB, or a combination of the 2 compounds (Figure 1). At the end of treatment, cells were washed 3 times with PBS and plated in triplicates in methylcellulose. Three to 4 weeks later, each triplicate was evaluated for the total number of colonies (BFU-E and CFU-GM) grown. Numbers obtained from our experiments were analyzed with a random effects negative binomial regression model to evaluate the toxicity of the different drug regimens on the growth of healthy myeloid progenitors. The model included IM alone, LMB alone, and their interaction (Figure 2A). Incubation with IM reduced the number of total colonies by 40% (Table 1), thus generating a coefficient of 0.60 (Table 2). Treatment with LMB caused a 61% decrease in the total number of colonies (Table 1), producing a coefficient of 0.39 (Table 2). Exposure to the combination of IM and LMB produced a 59% decrease in the overall number of colonies (Table 1). This effect was much lower than that expected, assuming independence of the 2 compounds (a cell-count reduction of 77%). Indeed, the interaction term of the 2 drugs was higher than 1 (interaction coefficient, 1.75), suggesting that the addition of both compounds on healthy myeloid progenitors determines a negative interaction (Table 2).
Average number of colonies in healthy controls and in CML patients and reduction in the colony-forming ability of healthy and CML myeloid progenitors after treatment with IM, LMB, or their combination
. | No treatment . | IM . | LMB . | IM + LMB . |
|---|---|---|---|---|
| Healthy controls | ||||
| Colonies, no. | 105 | 73 | 44 | 51 |
| Reduction, % (95% CI) | — | 40 (29-49) | 61 (53-68) | 59 (41-72) |
| Patients with CML | ||||
| Colonies, no. | 91 | 62 | 21 | 13 |
| Reduction, % (95% CI) | — | 37 (33-40) | 79 (78-81) | 88 (82-92) |
. | No treatment . | IM . | LMB . | IM + LMB . |
|---|---|---|---|---|
| Healthy controls | ||||
| Colonies, no. | 105 | 73 | 44 | 51 |
| Reduction, % (95% CI) | — | 40 (29-49) | 61 (53-68) | 59 (41-72) |
| Patients with CML | ||||
| Colonies, no. | 91 | 62 | 21 | 13 |
| Reduction, % (95% CI) | — | 37 (33-40) | 79 (78-81) | 88 (82-92) |
— indicates not applicable.
Coefficients generated by the NB random effect model estimating the effect of different drug regimens on normal and CML progenitors
. | Healthy controls* . | P . | Patients with CML* . | P . |
|---|---|---|---|---|
| IM | 0.60 (0.51-0.71) | < .001 | 0.63 (0.60-0.67) | < .001 |
| LMB | 0.39 (0.32-0.47) | < .001 | 0.21 (0.19-0.22) | < .001 |
| IM+LMB | 1.75 (1.32-2.31) | < .001 | 1.08 (0.95-1.24) | .22 |
. | Healthy controls* . | P . | Patients with CML* . | P . |
|---|---|---|---|---|
| IM | 0.60 (0.51-0.71) | < .001 | 0.63 (0.60-0.67) | < .001 |
| LMB | 0.39 (0.32-0.47) | < .001 | 0.21 (0.19-0.22) | < .001 |
| IM+LMB | 1.75 (1.32-2.31) | < .001 | 1.08 (0.95-1.24) | .22 |
Data expressed as odds ratio (95% confidence interval)
Effect of IM, LMB, or their combination on the proliferative capacity of normal or Ph-positive myeloid progenitors. (A) CD34-positive cells isolated from 6 healthy donors and (B) myeloid progenitors from 35 patients with CML were either left untreated (NT) or exposed to IM, LMB, or a combination of the 2 drugs(I+L) following the schedule indicated in Figure 1. Cells were then plated in methylcellulose media and after 21 to 28 days total number of BFU-E and CFU-GM colonies was determined for each experimental condition. Data are presented as percentage of variations in total colony number with NT cells arbitrarily set at 100%. (C) Average number of colonies derived from healthy (○) or CML (•) cells.
Effect of IM, LMB, or their combination on the proliferative capacity of normal or Ph-positive myeloid progenitors. (A) CD34-positive cells isolated from 6 healthy donors and (B) myeloid progenitors from 35 patients with CML were either left untreated (NT) or exposed to IM, LMB, or a combination of the 2 drugs(I+L) following the schedule indicated in Figure 1. Cells were then plated in methylcellulose media and after 21 to 28 days total number of BFU-E and CFU-GM colonies was determined for each experimental condition. Data are presented as percentage of variations in total colony number with NT cells arbitrarily set at 100%. (C) Average number of colonies derived from healthy (○) or CML (•) cells.
Our data demonstrated that each drug treatment produced a statistically significant decrease in the colony-forming ability of normal myeloid progenitors (Table 2). However, a fair number of normal myeloid cells were able to proliferate after each drug treatment.
Effect of different drug regimens on the colony-forming capacity of CML myeloid progenitors
We next wanted to determine the effect of the different treatments on the proliferative potential of CML cells. Hence, we employed a Ficoll gradient to purify myeloid progenitors from 35 patients with CML with the clinical characteristics reported in Table 4. As we had done for healthy CD34-positive cells, leukemic progenitors were subjected to the treatment regimens described in Figure 1 and then plated in triplicates in methylcellulose to perform colony-forming assays (Figure 2B).
Clinical profile of the patients involved in the study
UPN . | Age, y/sex . | Diagnosis, mo . | Sokal index . | Current therapy . | Previous therapy . | FISH, % Ph . | BCR-ABL variant . | PLT/μL, 1 × 103 . | WBC/μL . | LDH, U/L . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1* | 68/M | 14 | Intermediate | IM | HU+IFN | 26 | e13a2 | 266 | 6 480 | 483 |
| 2 | 32/F | 160 | Low | IM | HU | 100 | e13a2 | 222 | 4 070 | 431 |
| 3 | 69/F | 36 | Intermediate | IM | HU+IFN | 96 | e14a2 | 127 | 3 470 | 202 |
| 4 | 30/F | 0 | Low | None | None | 100 | e14a2 | 508 | 45 340 | 530 |
| 5* | 50/M | 22 | Intermediate | IM | HU+IFN | 2 | e13a2 | 134 | 5 730 | 447 |
| 6* | 51/F | 13 | Intermediate | IM | HU+IFN | 0 | e13a2 | 130 | 4 810 | 324 |
| 7* | 62/F | 13 | Low | IFN | IFN | 26 | e14a2 | 181 | 5 610 | 296 |
| 8 | 52/M | 3 | Intermediate | IM | HU | 100 | e14a2 | 114 | 31 220 | 539 |
| 9 | 53/F | 30 | Intermediate | IM | HU+IFN | 100 | e14a2 | 163 | 4 600 | 228 |
| 10 | 66/F | 28 | Intermediate | IM | HU+IFN | 100 | e13a2 | 190 | 72 830 | 1321 |
| 11 | 52/F | 10 | Low | IFN | HU | 100 | e14a2 | 202 | 3 910 | 484 |
| 12* | 54/M | 25 | Low | IFN | HU | 0 | e13a2 | 204 | 5 870 | 261 |
| 13 | 69/F | 0 | Intermediate | HU | None | 100 | e13a2 | 521 | 50 930 | 815 |
| 14 | 71/F | 0 | Intermediate | None | None | 72 | e14a2 | 160 | 15 220 | 551 |
| 15 | 44/F | 0 | Intermediate | HU | None | 100 | e13a2 | 240 | 8 600 | 468 |
| 16 | 71/F | 0 | Intermediate | HU | None | 100 | e14a2 | 318 | 4 860 | 745 |
| 17 | 73/M | 2 | Low | HU | None | 100 | e14a2 | 121 | 19 650 | 520 |
| 18 | 67/M | 57 | Intermediate | IM | HU+IFN | 100 | e13a2 | 230 | 9 330 | 522 |
| 19* | 49/F | 18 | Low | IFN | HU | 27 | e13a2 | 140 | 4 260 | 107 |
| 20 | 55/M | 0 | Intermediate | HU | None | 100 | e14a2 | 199 | 10 790 | 947 |
| 21 | 53/M | 44 | Intermediate | IM | HU+IFN | 72 | e14a2 | 387 | 15 320 | 476 |
| 22 | 39/M | 47 | Low | IM | HU+IFN | 56 | e13a2 | 404 | 7 080 | 335 |
| 23* | 50/F | 333 | Low | IM | HU+IFN | 100 | e13a2 | 373 | 27 540 | 530 |
| 24 | 53/F | 23 | Intermediate | IM | HU+IFN | 34 | e14a2 | 192 | 6 050 | 221 |
| 25 | 56/M | 0 | Intermediate | IM | HU+IFN | 96 | e14a2 | 229 | 4 320 | 305 |
| 26 | 61/M | 78 | Intermediate | IM | HU+IFN | 96 | e14a2 | 131 | 15 480 | 590 |
| 27 | 65/M | 160 | Low | IM | HU+IFN | 100 | e13a2 | 158 | 4 980 | 547 |
| 28* | 57/F | 19 | High | IM | HU+IFN | 0 | e14a2 | 228 | 7 220 | 477 |
| 29 | 50/M | 0 | Intermediate | HU | None | 100 | e13a2 | 442 | 20 840 | 552 |
| 30 | 56/F | 14 | Intermediate | IFN | HU | 100 | e1a2 | 198 | 17 370 | 713 |
| 31* | 60/F | 20 | Intermedaite | IM | HU+IFN | 0 | e13a2 | 188 | 5 230 | 396 |
| 32 | 63/M | 1 | Intermediate | HU | None | 70 | e14a2 | 630 | 45 830 | 588 |
| 33 | 78/M | 15 | Low | HU | None | 90 | e1a2 | 220 | 18 770 | 852 |
| 34 | 63/M | 0 | Intermediate | HU | None | 100 | e14a2 | 356 | 90 440 | 891 |
| 35 | 36/M | 0 | Intermediate | HU | None | 100 | e13a2 | 400 | 87 440 | 1373 |
UPN . | Age, y/sex . | Diagnosis, mo . | Sokal index . | Current therapy . | Previous therapy . | FISH, % Ph . | BCR-ABL variant . | PLT/μL, 1 × 103 . | WBC/μL . | LDH, U/L . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1* | 68/M | 14 | Intermediate | IM | HU+IFN | 26 | e13a2 | 266 | 6 480 | 483 |
| 2 | 32/F | 160 | Low | IM | HU | 100 | e13a2 | 222 | 4 070 | 431 |
| 3 | 69/F | 36 | Intermediate | IM | HU+IFN | 96 | e14a2 | 127 | 3 470 | 202 |
| 4 | 30/F | 0 | Low | None | None | 100 | e14a2 | 508 | 45 340 | 530 |
| 5* | 50/M | 22 | Intermediate | IM | HU+IFN | 2 | e13a2 | 134 | 5 730 | 447 |
| 6* | 51/F | 13 | Intermediate | IM | HU+IFN | 0 | e13a2 | 130 | 4 810 | 324 |
| 7* | 62/F | 13 | Low | IFN | IFN | 26 | e14a2 | 181 | 5 610 | 296 |
| 8 | 52/M | 3 | Intermediate | IM | HU | 100 | e14a2 | 114 | 31 220 | 539 |
| 9 | 53/F | 30 | Intermediate | IM | HU+IFN | 100 | e14a2 | 163 | 4 600 | 228 |
| 10 | 66/F | 28 | Intermediate | IM | HU+IFN | 100 | e13a2 | 190 | 72 830 | 1321 |
| 11 | 52/F | 10 | Low | IFN | HU | 100 | e14a2 | 202 | 3 910 | 484 |
| 12* | 54/M | 25 | Low | IFN | HU | 0 | e13a2 | 204 | 5 870 | 261 |
| 13 | 69/F | 0 | Intermediate | HU | None | 100 | e13a2 | 521 | 50 930 | 815 |
| 14 | 71/F | 0 | Intermediate | None | None | 72 | e14a2 | 160 | 15 220 | 551 |
| 15 | 44/F | 0 | Intermediate | HU | None | 100 | e13a2 | 240 | 8 600 | 468 |
| 16 | 71/F | 0 | Intermediate | HU | None | 100 | e14a2 | 318 | 4 860 | 745 |
| 17 | 73/M | 2 | Low | HU | None | 100 | e14a2 | 121 | 19 650 | 520 |
| 18 | 67/M | 57 | Intermediate | IM | HU+IFN | 100 | e13a2 | 230 | 9 330 | 522 |
| 19* | 49/F | 18 | Low | IFN | HU | 27 | e13a2 | 140 | 4 260 | 107 |
| 20 | 55/M | 0 | Intermediate | HU | None | 100 | e14a2 | 199 | 10 790 | 947 |
| 21 | 53/M | 44 | Intermediate | IM | HU+IFN | 72 | e14a2 | 387 | 15 320 | 476 |
| 22 | 39/M | 47 | Low | IM | HU+IFN | 56 | e13a2 | 404 | 7 080 | 335 |
| 23* | 50/F | 333 | Low | IM | HU+IFN | 100 | e13a2 | 373 | 27 540 | 530 |
| 24 | 53/F | 23 | Intermediate | IM | HU+IFN | 34 | e14a2 | 192 | 6 050 | 221 |
| 25 | 56/M | 0 | Intermediate | IM | HU+IFN | 96 | e14a2 | 229 | 4 320 | 305 |
| 26 | 61/M | 78 | Intermediate | IM | HU+IFN | 96 | e14a2 | 131 | 15 480 | 590 |
| 27 | 65/M | 160 | Low | IM | HU+IFN | 100 | e13a2 | 158 | 4 980 | 547 |
| 28* | 57/F | 19 | High | IM | HU+IFN | 0 | e14a2 | 228 | 7 220 | 477 |
| 29 | 50/M | 0 | Intermediate | HU | None | 100 | e13a2 | 442 | 20 840 | 552 |
| 30 | 56/F | 14 | Intermediate | IFN | HU | 100 | e1a2 | 198 | 17 370 | 713 |
| 31* | 60/F | 20 | Intermedaite | IM | HU+IFN | 0 | e13a2 | 188 | 5 230 | 396 |
| 32 | 63/M | 1 | Intermediate | HU | None | 70 | e14a2 | 630 | 45 830 | 588 |
| 33 | 78/M | 15 | Low | HU | None | 90 | e1a2 | 220 | 18 770 | 852 |
| 34 | 63/M | 0 | Intermediate | HU | None | 100 | e14a2 | 356 | 90 440 | 891 |
| 35 | 36/M | 0 | Intermediate | HU | None | 100 | e13a2 | 400 | 87 440 | 1373 |
UPN indicates unique patient number; HU, hydroxyurea; and IFN, α-interferon.
Diagnosis refers to time from first diagnosis.
Patients whose colonies were not analyzed for BCR-ABL expression
In our CML patient cohort, the coefficient related to the IM effect was 0.63 (Table 2), indicating that exposure to IM led to a 37% reduction in the total number of colonies (Table 1). Treatment with LMB resulted in a 79% decrease in overall colony number, indicative of a coefficient of 0.21 (Tables 1-2). The combination of IM and LMB produced the highest decrease in colony formation, inducing an 88% reduction (Table 1). Again, to determine if the combination of the 2 drugs generated a multiplicative effect or instead determined negative or positive interactions, we calculated an interaction coefficient. The resulting value of 1.08 suggested that addition of the 2 drugs produces the effect expected assuming independence of their activity (a colony reduction of 87%; Table 2).
The reduction in colony formation caused by LMB alone or by LMB and IM is higher in CML progenitors than in normal myeloid cells
In order to be successful, a pharmacologic treatment requires that the effects observed on pathologic cells be significantly higher than those observed on normal cells. Since the different drug treatments that we used caused a reduction in the proliferative capacity of both healthy and CML myeloid progenitors, we wanted to compare the effect of the different drugs in the 2 groups (ie, healthy individuals and patients with CML). We therefore compared the average number of colonies in the different experimental conditions in a unique model that calculated an interaction term to compare the effect of the different drug treatments on healthy and leukemic myeloid progenitors (Figure 2C, Table 3).
Interaction analysis
. | Odds ratio (CI) . | P . |
|---|---|---|
| IM vs disease group | 1.05 (0.89-1.25) | .55 |
| LMB vs disease group | 0.54 (0.44-0.66) | < .001 |
| IM + LMB vs disease group | 0.62 (0.46-0.84) | .002 |
. | Odds ratio (CI) . | P . |
|---|---|---|
| IM vs disease group | 1.05 (0.89-1.25) | .55 |
| LMB vs disease group | 0.54 (0.44-0.66) | < .001 |
| IM + LMB vs disease group | 0.62 (0.46-0.84) | .002 |
The effect of IM was not statistically different on cells derived from healthy donors or individuals diagnosed with CML (interaction coefficient, 1.05; Table 3). Unexpectedly, LMB caused a reduction in the colony-forming ability of CML progenitors that was significantly higher than that observed in healthy myeloid cells (interaction coefficient, 0.54; Table 3). Likewise, the combination of IM and LMB induced a decline in the number of leukemic colonies that was much higher than the one observed in colonies derived from healthy myeloid progenitors (interaction coefficient, 0.62).
The combination of IM and LMB induces the highest reduction in the number of BCR-ABL-positive colonies
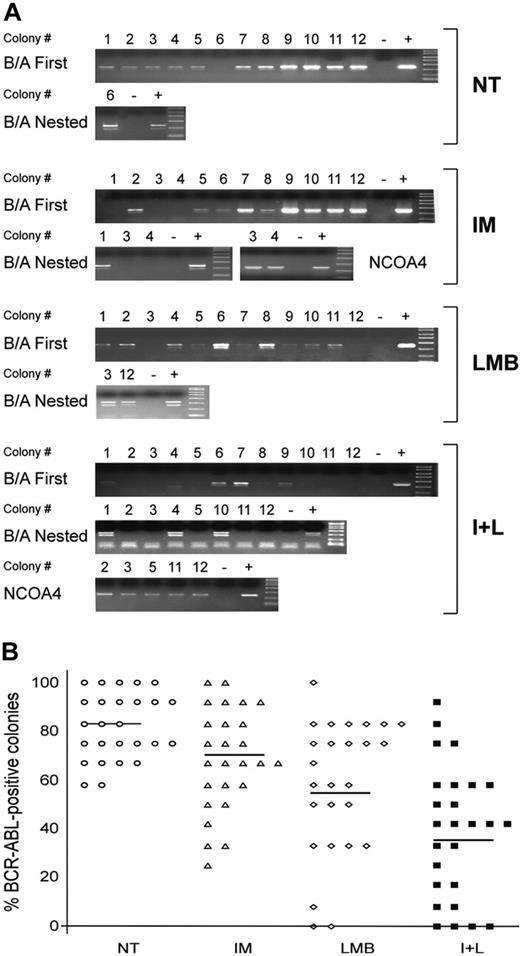
The reduction in colony formation observed after exposure to LMB alone or the combination of IM and LMB suggests that either treatment can significantly reduce the proliferation of CML myeloid progenitors. However, our ultimate goal was to selectively eliminate CML cells while preserving the growth of normal myeloid progenitors. To establish whether any of our treatment conditions could discriminate between BCR-ABL-negative and BCR-ABL-positive cells, we analyzed 12 colonies from each experimental condition for the expression of the chimeric oncogene. RNA extracted from each colony was reverse transcribed and used to perform a first PCR reaction for BCR-ABL (Figure 3A). Negative colonies were subjected to a second (nested) PCR for BCR-ABL. If these reactions scored negative, the remaining RNA was used to perform an RT-PCR for the ubiquitously expressed NCOA4 gene, to confirm that the negative results were due to absence of the BCR-ABL transcript and not to poor quality or quantity of the RNA (Figure 3A). Colonies were obtained from 26 of the 35 patients included in the study, since 8 patients in major or complete cytogenetic response produced an insufficient number of colonies for our assay and we could not isolate good-quality RNA from a ninth patient (Table 4).
Our experiments showed that most colonies derived from CML cells that were left untreated scored positive for BCR-ABL expression (Figure 3B). On average, 82% of untreated CML progenitors generated BCR-ABL-positive BFU-E or CFU-GM colonies. After treatment with IM alone or LMB alone we observed a reduction of this number to 69% and 58%, respectively (Figure 3B). However, the combination of IM and LMB was much more effective in reducing the number of BCR-ABL-positive cells, as only 39% of colonies that survived the combined drug regimen expressed the BCR-ABL transcript (Figure 3B).
To analyze the statistical significance of these reductions we used a logistic regression model stratified per patient. This analysis generated ORs that could be at unity (no effect), above unity (increased probability of a colony to express BCR-ABL), or below unity (decreased probability of a colony to express BCR-ABL). Our data showed that both IM (OR = 0.46) and LMB (OR = 0.27) significantly reduced the probability to be BCR-ABL-positive (Table 5). As we expected, exposure to IM plus LMB led to the highest decrease in the number of BCR-ABL-positive colonies (OR = 0.10; Table 5).
Effect of the different drug regimens on the number of BCR-ABL-positive colonies
Treatment . | Odds ratio (95% CI) . | P . |
|---|---|---|
| IM | 0.46 (0.31-0.69) | < .001 |
| LMB | 0.27 (0.18-0.40) | < .001 |
| IM+LMB | 0.10 (0.07-0.16) | < .001 |
Treatment . | Odds ratio (95% CI) . | P . |
|---|---|---|
| IM | 0.46 (0.31-0.69) | < .001 |
| LMB | 0.27 (0.18-0.40) | < .001 |
| IM+LMB | 0.10 (0.07-0.16) | < .001 |
An OR below unity indicates a reduction in the likelihood that colonies in the indicated treatment group may express BCR-ABL.
Correlation between response to different drug regimens and the clinical characteristics of patients with CML
Once we had established the efficacy of the different drug regimens in reducing the proliferative potential of BCR-ABL-positive myeloid progenitors, we wanted to determine whether we could correlate the molecular response of the patients with their clinical profile. To this end, we selected 11 clinical parameters (Table 4) and used a graphical representation showing the ORs together with the 95% CIs for each drug treatment and for the combination of the 2 compounds. We found that the combination of the 2 compounds produced a significantly higher reduction in the number of BCR-ABL-positive cells independently of any clinical characteristic of the patients (Figure 4). A detailed analysis of the possible correlations between clinical variables and number of BCR-ABL-expressing cells showed that the 2 drug combination showed a slightly higher effect on patients with a short time from diagnosis (< 1 year) compared to those with a longer disease duration (> 1 year). However, a larger number of patients are required to draw any absolute conclusions.
Discussion
Data emerging from short-term follow-up of CML patients treated with IM has consistently shown that 20% to 30% of chronic-phase patients and virtually all individuals in blast crisis develop resistance to the drug.6,22 Moreover, it appears increasingly clear that IM can induce remarkable results in terms of hematologic, cytogenetic, and molecular response, but is unlikely to eradicate BCR-ABL-positive cells.23 Thus, there is an increasing need for pharmacologic compounds that may be used in addition to or in place of IM.24
We have previously found that nuclear sequestration of the BCR-ABL oncoprotein by the combination of IM and LMB can selectively kill both CML cell lines and murine bone marrow cells infected with a BCR-ABL retroviral vector.20 Here we report that the combination of IM and LMB significantly reduces the colony-forming ability of human CML myeloid progenitors (Figure 2). Moreover, the combination of IM and LMB preferentially targets BCR-ABL-expressing cells since it generates the strongest decrease in the number of Ph-positive colonies (Figure 3). Although the number of patients included in this study does not allow definitive conclusions, this reduction appears to be independent of the clinical characteristics of the patients (Figure 4). Thus, this pharmacologic approach should be equally effective on patients with CML who have unfavorable prognostic characteristics (higher age, intermediate or high Sokal index, and 100% Ph-positive metaphases).
What are the possible applications of these results in the clinical setting? Is it possible to hypothesize the use of oral IM and intravenous LMB for the treatment of patients with CML? Our data show that IM, LMB, and their combination decrease the number of both healthy and leukemic myeloid progenitors. However, when both drugs are combined, IM appears to reduce the toxicity of LMB on normal myeloid progenitors (Figure 2A, Table 1). On the contrary, the 2-drug combination displays a multiplicative effect on CML cells, strongly reducing their proliferative potential (Figure 2B). Most of the toxicity observed after treatment of healthy myeloid cells is likely attributable to LMB. This inhibitor of nuclear export has been previously administered intravenously to a limited number of individuals diagnosed with various forms of solid cancer.25 Results were not encouraging because patients experienced gastrointestinal side effects, malaise, and anorexia. No consequences were reported in other tissues or organs. It is possible that the lower dosage (160-fold) and the limited exposure to the drug (12 hours) used in our regimen may significantly reduce the side effects of LMB. However, until this is appropriately verified in a phase 1 clinical trial or until a better nuclear export inhibitor (less toxic and/or specific for the ABL and BCR-ABL proteins) is available, the more likely use for the IM-LMB combination will be in an ex vivo setting for allografting purposes.15,26
Assessment of BCR-ABL expression in leukemic colonies grown in different experimental conditions. In 26 of the 35 patients reported in Table 4 we randomly selected 48 single colonies (12 for each treatment condition). (A) RNA extracted from each colony was used to perform a 1-step RT-PCR for BCR-ABL. Negative RT-PCR reactions were subjected to a nested PCR for BCR-ABL. Colonies that again scored negative were further analyzed for the ubiquitously expressed gene NCOA4 to verify the quality of the RNA obtained. Each PCR reaction included a negative (-) and positive (+) control. One representative patient (UPN 30) is shown here. (B) Average number of BCR-ABL-positive colonies in the 4 experimental conditions (NT is represented by ○; IM, ▵; LMB, ⋄; and I+L, ▪). Data are presented as percentage of variations with 12 positive colonies set at 100%. Horizontal bars indicate the average number of BCR-ABL-positive colonies in each experimental condition.
Assessment of BCR-ABL expression in leukemic colonies grown in different experimental conditions. In 26 of the 35 patients reported in Table 4 we randomly selected 48 single colonies (12 for each treatment condition). (A) RNA extracted from each colony was used to perform a 1-step RT-PCR for BCR-ABL. Negative RT-PCR reactions were subjected to a nested PCR for BCR-ABL. Colonies that again scored negative were further analyzed for the ubiquitously expressed gene NCOA4 to verify the quality of the RNA obtained. Each PCR reaction included a negative (-) and positive (+) control. One representative patient (UPN 30) is shown here. (B) Average number of BCR-ABL-positive colonies in the 4 experimental conditions (NT is represented by ○; IM, ▵; LMB, ⋄; and I+L, ▪). Data are presented as percentage of variations with 12 positive colonies set at 100%. Horizontal bars indicate the average number of BCR-ABL-positive colonies in each experimental condition.
Our data have also shown that CML cells are more sensitive to the nuclear export inhibitor LMB than their normal counterparts. While we do not have an explanation for this observation, several evidences in the literature suggest that this result should have been partially expected. An initial report indicated that LMB greatly reduced the survival of a leukemia cell line.25 More recently, 2 different groups have demonstrated that LMB-mediated activation and nuclear entrapment of p53 induced cell death in cervical and prostate carcinoma cells.27,28 Further evidence suggests that LMB triggers an apoptotic response in a leukemia cell line by down-regulating Mcl-1 and XIAP and stimulating caspase activity.29
A critical point concerns the possible efficacy of the combination of IM and LMB in patients with CML who have acquired resistance to IM. It is obvious that the 2-drug approach will be ineffective in the infrequent cases of BCR-ABL-independent resistance to IM. As for BCR-ABL-dependent IM resistance, nuclear translocation of BCR-ABL requires inactivation of its tyrosine kinase activity. Hence, BCR-ABL mutants that can no longer bind to IM will not migrate to the nucleus. In this scenario, the combination of IM and LMB would lose its efficacy. Indeed, in a murine pro-B cell line expressing the BCR-ABL oncoprotein with point mutations that completely abrogate IM binding (Ba/F3p210Y253F and Ba/F3p210T315I),30 the combination of IM and LMB produced antiproliferative effects that were equivalent to those obtained by each drug alone (Supplemental Figure S1A, available at the Blood website; click on the “Supplemental Figures” link at the top of the online article). However, BCR-ABL mutants that bind IM with lower affinity should remain sensible to this approach, especially considering that for ex vivo treatments it may be possible to increase the dose of IM above the one used in this study. In fact, BaF cells expressing either wild-type BCR-ABL (BaF/BCR-ABL) or a mutant with a moderate (3-fold) increase in IM 50% inhibitory concentration (IC50; Ba/F3p210D276G)31 were still responsive to the combination of IM and LMB (Figure S1A). A further possibility would be to associate LMB with other ABL inhibitors that have been reported to abrogate the catalytic activity of most or all BCR-ABL mutants found in IM-resistant patients with CML.32-35 About one-third of IM-resistant individuals have lost their response to the drug because of amplification or overexpression events occurring at the BCR-ABL locus.22 The ensuing increase in the number of BCR-ABL molecules causes a loss in IM efficacy that can sometimes be countered by an increase in the dose of the drug. Under these circumstances, the combination of IM and LMB should still be effective since we have demonstrated that translocating 30% of total BCR-ABL from the cytoplasm to the nucleus is sufficient to induce cell death. Indeed, when we compared the efficacy of IM and LMB, alone or in combination, on LAMA84r cells that had become resistant to IM because of gene amplification,36 we found that the association of the 2 drugs produced the highest growth inhibition (Supplemental Figure S1B).
Correlation between response to the different drug regimens and the clinical characteristics of the patients. For 26 of the 35 patients reported in Table 4 we calculated odd ratios (ORs) with 95% confidence intervals (error bars) estimating the probability for each treated colony to express BCR-ABL compared with untreated colonies, and correlated the results to 11 clinical characteristics. An OR of 1 means no effect; an OR greater than 1 means an increased probability to express BCR-ABL, while an OR less than 1 indicates a reduced probability to express BCR-ABL. Treatment conditions are indicated on the bottom.
Correlation between response to the different drug regimens and the clinical characteristics of the patients. For 26 of the 35 patients reported in Table 4 we calculated odd ratios (ORs) with 95% confidence intervals (error bars) estimating the probability for each treated colony to express BCR-ABL compared with untreated colonies, and correlated the results to 11 clinical characteristics. An OR of 1 means no effect; an OR greater than 1 means an increased probability to express BCR-ABL, while an OR less than 1 indicates a reduced probability to express BCR-ABL. Treatment conditions are indicated on the bottom.
Another interesting question is how the combination of IM and LMB compares with the numerous pharmacologic associations reported in the literature between IM and other chemotherapeutic agents.37-39 To address this issue we treated 3 human CML lines (K562, KCL22, and LAMA84) and BaF/BCR-ABL cells with the combination of IM and LMB or with associations between IM and cytosine arabinoside (AraC), arsenic trioxide (As2O3), hydroxyurea (HU), or etoposide (VP-16). The combination of IM and LMB obtained the best antiproliferative effect, with the association of IM and As2O3 producing similar results in K562 and LAMA84 cells. All other combinations caused a lower growth inhibition (Supplemental Figure S2).
A further point concerns the possible efficacy of the IM-LMB combination on early quiescent CML progenitors that have been reported to be insensitive to IM.23 While a detailed study of the efficacy of IM and LMB on this cell population was beyond the aims of this study, we hypothesize that our strategy would be ineffective if CD34-positive CML cells were not reached by IM. On the contrary, if in these cells IM successfully inactivates BCR-ABL kinase activity, the ensuing nuclear entrapment of the oncoprotein caused by LMB should induce cell death.
A final issue involves the leukemic cells that in our experimental conditions were not killed by the combination of IM and LMB. We are unsure if these cells would have been eradicated by a different (longer) treatment schedule or have acquired resistance to the 2-drug treatment. The brief exposure to the drugs (total of 24 hours) argues against the possibility that leukemic cells may have become resistant to the pharmacologic treatment. Furthermore, sequencing of the BCR-ABL kinase domain in randomly selected Ph-positive colonies that grew after exposure to the combination of IM and LMB failed to detect any mutations. Preliminary evidence obtained in human CML cell lines points to the cytoplasmic retention of BCR-ABL (even after tyrosine kinase inactivation) as a possible mechanism of resistance to the treatment with IM and LMB (A.A. and P.V., unpublished results, May 2005). However, we found no evidence of such a phenomenon in this study.
In summary, we have found the combination of IM and LMB to effectively reduce the proliferative potential of human CML myeloid progenitors. We have also found that this 2-drug treatment preferentially kills BCR-ABL-expressing cells. Given the emerging problems related to IM monotherapy and the possibility of extending this association strategy to other BCR-ABL kinase inhibitors, it seems reasonable to propose that these results be investigated further in the setting of an appropriate clinical trial.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-05-2123.
Supported by a fellowhip from Fondazione Italiana per la Ricerca sul Cancro (A.A.), a regional grant from Associazione Italiana per la Ricerca sul Cancro (R.G. and A.M.), the Leukemia Research Foundation (P.V.), MIUR (P.V.), and a national grant from Associazione Italiana per la Ricerca sul Cancro (P.V.). P.V. is a fellow of the American-Italian Cancer Foundation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Livia Manzella for a critical revision of the manuscript and Agata Copani for helpful suggestions and insightful criticism.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal