Assembly of a signaling complex around the transmembrane adapter LAT is essential for the transmission of T-cell receptor (TCR)-mediated signaling. However, a LAT-like molecule responsible for the initial activation events in B-cell receptor (BCR) signaling has not yet been identified. Here, we show that LIME is a transmembrane adaptor required for BCR-mediated B-cell activation. LIME was found to be expressed in mouse splenic B cells. Upon BCR cross-linking, LIME was tyrosine phosphorylated by Lyn and associated with Lyn, Grb2, PLC-γ2, and PI3K. Reduction of LIME expression by the introduction of siRNA resulted in the disruption of BCR-mediated activation of MAPK, calcium flux, NF-AT, PI3K, and NF-κB. Taken together, these results establish that LIME is an essential transmembrane adaptor linking BCR ligation to the downstream signaling events that lead to B-cell activation.
Introduction
The binding of a multivalent antigen (Ag) to an Ag-specific B-cell receptor (BCR) is necessary for B-cell activation, survival, proliferation, and differentiation.1 BCR cross-linking leads to translocation of BCR into a lipid raft.2 In the lipid raft, tyrosine residues located within immunoreceptor tyrosine-based activation motifs (ITAMs) of the intracellular domains of Ig-α/Ig-β are phosphorylated by Lyn, a Src family tyrosine kinase. Subsequently, the tyrosine-phosphorylated ITAMs serve as docking sites for the recruitment of the tyrosine kinase Syk. At the same time, the Tec family kinase Btk is activated by BCR engagement.3,4 These tyrosine kinases act in combination to phosphorylate and activate various signaling molecules, such as SLP-65/BLNK/BASH, PI3K, PLC-γ2, and small G proteins. These signaling molecules in turn regulate downstream pathways leading to the activation of transcription factors such as AP-1, nuclear factor of activated T-cell (NF-AT), and nuclear factor of κ binding (NF-κB).5,6
Transmembrane adaptors play important roles in the transmission of immune receptor signals by recruiting intracellular effector proteins from the cytosol to the membrane-proximal region.7,8 In T-cell receptor (TCR)-mediated signaling, the importance of the linker for activation of T cells (LAT) is well characterized. LAT is expressed in T, natural killer, and mast cells and is located in lipid rafts. In T cells, LAT is phosphorylated by ZAP-70 and associates with SLP-76, PLC-γ1, Vav, Cbl, Grb2, and Gads in response to TCR cross-linking.9-11 LAT-deficient Jurkat cells show defects in TCR-induced calcium flux, MAPK, NF-AT, and AP-1 activation.12 In addition, peripheral T cells are absent in mice deficient in LAT due to arrest at the CD4-CD8- double negative (DN) stage of development.13 Together, these reports demonstrate the pivotal role of LAT for T-cell activation.
In BCR-mediated signaling, linker for B-cell activation/non-T-cell activation linker (LAB/NTAL) was first suggested as a functional analog of LAT in B cells.14,15 Upon BCR ligation, LAB becomes phosphorylated by Syk/ZAP-70 and interacts with Grb2. Reduction of LAB expression results in the attenuation of BCR-induced MAPK activation and calcium mobilization. However, LAB either could not rescue normal T-cell activation in LAT knockout mice or provided only minimal rescue.14 Moreover, LAB is not involved in the BCR-induced initial calcium increase but rather regulates the sustained release of intracellular calcium and subsequent entry of extracellular calcium.16 These functions of LAB suggest the presence of a second transmembrane adaptor involved in BCR signaling.14-16
Here, we provide evidence that another transmembrane adaptor, Lck-interacting membrane protein (LIME), plays an important role in linking BCR stimulation to B-cell activation. LIME was previously identified as a transmembrane adaptor protein linking TCR stimulation to downstream signaling pathways via an association with Lck.17,18 LIME localizes to lipid rafts in T cells in response to TCR stimulation. LIME is phosphorylated by Lck and recruits signaling molecules such as Lck, PI3K, Grb2, Gads, and SHP-2.17,18 Overexpression of LIME induces extracellular signal-regulated kinase (ERK), c-jun N-terminal kinase (JNK), and IL-2 promoter activity.17 Therefore, LIME has been suggested to be a positive regulator of TCR-mediated T-cell activation.
Here, we found that LIME is expressed in primary B cells. Based on this finding, we studied the role of LIME in BCR signaling events and found that LIME was phosphorylated by Lyn upon BCR cross-linking. Upon BCR stimulation, LIME also interacted with signaling molecules such as Grb2, PLC-γ2, Lyn, and PI3K p85. Moreover, the reduction of LIME expression by siRNA abrogated the BCR-induced activation of MAPK, calcium flux, NF-AT, PI3K, and NF-κB. These results indicate that LIME serves as the transmembrane adapter linking BCR-induced membrane-proximal signaling to B-cell activation.
Materials and methods
Cells and antibodies
Mouse B-cell line A20 (American Type Culture Collection, Manassas, VA) was maintained in RPMI 1640 medium with 10% FBS, 50 μM β-mercapto-ethanol, 1 mM sodium pyruvate, and penicillin/streptomycin at 37°C in 5% CO2. 293T cells were maintained in Dulbecco modified Eagle medium with 10% FBS and penicillin/streptomycin at 37°C in 5% CO2. Thy1.2+ T cells and B220+ B cells were purified from splenocytes of C57BL/6 mice using anti-Thy1.2 or anti-B220 magnetic beads with the SuperMACS magnetic cell separation system following the supplier's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Polyclonal mouse and rabbit anti-LIME antisera were raised against GST-fused LIME (amino acids 159 to 269) as described previously.17 For BCR activation, an F(ab′)2 goat anti-mouse IgG or F(ab′)2 goat anti-mouse IgM (Jackson ImmunoResearch Labs, West Grove, PA) was used. The other antibodies (Abs) used in this study were obtained from commercial sources and include anti-Lyn (sc-15), anti-PLC-γ2 (Q-20), anti-His, and anti-p65 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-ERK and anti-ERK (Cell Signaling Technology, Beverly, MA), antiflag (Sigma-Aldrich, St Louis, MO), antiphosphotyrosine (4G10), antiflotillin-2 (BD, San Jose, CA), anti-Grb2 and anti-p85 (Upstate Biotechnology, Lake Placid, NY), and Alexa 568-conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR).
Construction of plasmids
The expression plasmids encoding C-terminal flag-tagged mouse LIME and tyrosine point mutants were described previously.17 The plasmid encoding C-terminal flag-tagged mouse LAB was generated by polymerase chain reaction (PCR) amplification with specific primers using cDNA from A20 cells and subsequent ligation of the PCR product into pcDNA3.1 (+) (Invitrogen, Carlsbad, CA). Human Lyn and Syk cDNAs were kind gifts from Dr S. G. Lee (Yonsei University, Seoul, Korea) and were used for the construction of expression vectors in pcDNA3.1 (-) myc/his (Invitrogen). The PLC-γ2 expression vector was a gift from Dr Y. S. Bae (Ewha Womens University, Seoul, Korea).
Construction of siRNA-expressing plasmids
The pSUPER plasmid was used to synthesize small interfering RNAs (siRNAs). Three 19-mer sequences corresponding to nucleotides (nts) 380 to 398 (CTCTGCCTTCCCACCACGGCAG), 582 to 600 (GGCTAAGGTGATCCCAGCT), and 678 to 695 (CCTGCAGGATGGGAGAACAAG) in the coding sequence of murine Lime were selected as targets for the construction of siRNA-expressing constructs in pSUPER plasmids, which were produced according to the method of Brummelkamp et al.19 To examine the effects of the siRNAs, the expression plasmids encoding LIME-flag and each siRNA were cotransfected into 293 cells. Subsequent analysis of LIME levels by immunoblotting with an antiflag Ab revealed that the most effective construct was the one encoding the sequence of nts 678 to 695. Therefore, this construct was used for the rest of the experiments. Using the same methods, the target sequences for Lab and Lat were determined to be the sequences corresponding to nts 247 to 265 (GAACAAGATGCTGTTCTCC) for Lab and nts 363 to 381 (GAATGTGGATGCAGATGAG) for Lat. For the experiments to determine the localization of NF-κB, a pLL3.7 lentivirus vector, kindly provided by Dr Luk Van Parijs (Massachusetts Institute of Technology, Cambridge),20 was employed for the siRNA expression. This vector coexpresses enhanced green fluorescent protein (EGFP), permitting transfected cells to be identified with fluorescent microscopy. pLL3.7 encoding LIME siRNA was also constructed by targeting the sequence corresponding to nts 678 to 695 (CCTGCAGGATGGGAGAACAAG) of murine Lime.
Transient transfection and B-cell activation
293T cells were transfected using the calcium phosphate precipitation method. For transient transfection, 1.5 × 107 A20 cells were mixed with 20 μg target DNA and electroporated with 280 V, 25 mA pulses. After 2 days, the cells were harvested, suspended in RPMI at 1 × 107/mL, and incubated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 30 minutes on ice. The cells were then stimulated by switching to 37°C and incubating for the indicated time. For the inhibition of the kinase activity of Lyn and Syk, cells were pretreated with 100 μM PP1 (Sigma-Aldrich) and 50 μM piceatannol (Calbiochem, San Diego, CA), respectively, for 30 minutes at 37°C. Subsequently, cells were stimulated with 20 μg/mL F(ab′)2 goat anti-mouse IgG.
Immunoprecipitation and Western blotting
For immunoprecipitation, 2 × 107 A20 cells were lysed with RIPA lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Brij 35, 0.5% Na-deoxycholate, 1 mM Na3VO4, 5 mM NaF, 25 μg/mL aprotinin, 1 mM PMSF, 25 μg/mL leupeptin, 1 μg/mL pepstatin). After incubation on ice for 1 hour, the lysates were centrifuged at 15 000g for 20 minutes at 4°C. The supernatants were harvested and used for immunoprecipitation. Subsequent procedures for immunoprecipitation and Western analysis were described previously.17
Immunofluorescent microscopy
To visualize NF-κB p65, cells were incubated for 20 minutes at 37°C on poly-l-lysine-coated slides (Sigma-Aldrich) and fixed by incubation with 4% paraformaldehyde for 10 minutes. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 15 minutes and incubated with primary Ab for 30 minutes at 37°C. Subsequently, the cells were washed and incubated with Alexa 568-conjugated anti-rabbit IgG for 30 minutes at 37°C. After the cells were washed, they were mounted with VECTA-SHIELD mounting medium (Vector Laboratories, Burlingame, CA). Images were obtained using an LSM 510 confocal microscope equipped with an Axiovert 100 ×/1.4 oil-immersion objective lens (Zeiss, Oberkochen, Germany). Captured images were processed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
Calcium mobilization
A20 cells (2 × 106) were washed, suspended in RPMI, and loaded with the acetoxymethyl ester of fura-2 (fura-2/AM; Molecular Probes) for 30 minutes at 37°C. After 2 washes with PBS, the cytosolic-free calcium concentration was monitored with an RF-5301PC Spectrofluorophotometer (Shimadzu, Kyoto, Japan).
Luciferase assay
Cells were transfected by electroporation with an NF-AT-luciferase reporter construct along with either pSUPER or LIME siRNA. After 48 hours, the cells were harvested, plated in 48-well plates at 3 × 105 cells per well, and incubated with medium alone, 10 μg/mL F(ab′)2 goat anti-mouse IgG, or 50 ng/mL PMA (Calbiochem) and 100 ng/mL ionomycin (AG. Scientific, San Diego, CA). After 6 hours, the cells were lysed and the luciferase activity was measured with an LB953 luminometer (Berthold, Bad Wildbad, Germany). To control for transfection efficiency, the luciferase activities of the medium- and BCR-stimulated samples were normalized to those of PMA/ionomycin-treated samples. The experiments were performed twice in triplicate.
RT-PCR
Total RNA (1 μg) was prepared from siRNA-expressing A20 cells using Trizol (Gibco BRL, Carlsbad, CA) and converted into cDNA with ImProm-II (Promega, Madison, WI). For PCR amplification, the following primers were used: LAB sense 5′-ttccagaagccctcagaaga-3′, LAB antisense 5′-gcacccatagtcctcttgga-3′, GAPDH sense 5′-ttagcacccctggccaagg-3′, GAPDH antisense 5′-cttactccttggaggcatg-3′. Cycling condition was 1 cycle at 95°C for 5 minutes, 27 cycles at 95°C for 30 seconds, 56°C for 30 seconds, 72°C for 1 minute, and 1 cycle corresponding to 10 minutes at 72°C.
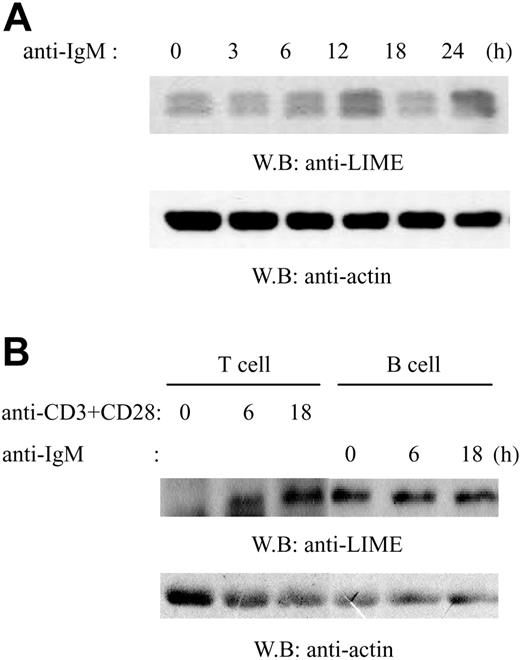
LIME expression in primary mouse splenic B cells. (A) Splenic B cells were isolated from C57BL/6 mice using anti-B220 (CD45R) microbeads. Cells were activated by F(ab′)2 goat anti-mouse IgM for 0, 3, 6, 12, 18, or 24 hours and then lysed. Lysates were analyzed by immunoblotting with mouse polyclonal anti-LIME or antiactin Abs. (B) Splenic T cells were isolated from C57BL/6 mice using anti-Thy1.2 microbeads. T cells were activated by anti-CD3 and CD28 Ab. B cells were purified and activated as described in panel A. Lysates were analyzed by immunoblotting with rabbit polyclonal anti-LIME or antiactin Abs.
LIME expression in primary mouse splenic B cells. (A) Splenic B cells were isolated from C57BL/6 mice using anti-B220 (CD45R) microbeads. Cells were activated by F(ab′)2 goat anti-mouse IgM for 0, 3, 6, 12, 18, or 24 hours and then lysed. Lysates were analyzed by immunoblotting with mouse polyclonal anti-LIME or antiactin Abs. (B) Splenic T cells were isolated from C57BL/6 mice using anti-Thy1.2 microbeads. T cells were activated by anti-CD3 and CD28 Ab. B cells were purified and activated as described in panel A. Lysates were analyzed by immunoblotting with rabbit polyclonal anti-LIME or antiactin Abs.
Results
LIME is expressed in B cells
Previously, we identified LIME as a transmembrane adaptor that mediates TCR activation.17 In addition to its expression in T cells, LIME protein was also detected in most hematopoietic cell lines tested, including B-cell lines.17 Therefore, we examined whether LIME is expressed in primary B cells. Mouse splenic B cells were isolated using anti-B220 magnetic beads and were activated by F(ab′)2 goat anti-mouse IgM for 0, 3, 6, 12, 18, or 24 hours. Lysates were analyzed by immunoblotting with anti-LIME Ab or antiactin Ab (Figure 1A). LIME was detected as doublet bands of 28 and 30 kDa and was expressed constitutively regardless of BCR stimulation. The doublet bands sometimes appeared depending on the electroporesis condition. Although we have no evidence, they may represent differential phosphorylation status of LIME, because LIME has 5 potential phosphorylation sites. Next, to compare the expression levels of LIME, mouse splenic T and B cells were isolated using anti-Thy1.2 or anti-B220 magnetic beads, respectively. A combination of anti-CD3 and anti-CD28 Ab was used to activate T cells while B cells were activated by anti-mouse IgM. After activation for 0, 6, or 18 hours, lysates were analyzed by immunoblotting with anti-LIME Ab or antiactin Ab (Figure 1B). As reported previously,17 LIME expression was up-regulated upon TCR stimulation, and the level of LIME in B cells was comparable to the induced level in T cells. LIME, therefore, likely has important functions in B cells.
Upon BCR cross-linking, LIME is tyrosine-phosphorylated by the tyrosine kinase of the Src family
Next, we tested whether LIME is phosphorylated upon BCR cross-linking. At the same time, we were interested in determining the kinase responsible for the phosphorylation of LIME. A20 cells were transiently transfected with the expression construct encoding LIME-flag. Cells were pretreated either with the Src family PTK inhibitor PP1 or the Syk inhibitor piceatannol for 30 minutes and were subsequently stimulated with F(ab′)2 goat anti-mouse IgG for 0 or 1 minutes. Cell lysates were subjected to immunoprecipitation with an antiflag Ab, and the precipitates were analyzed by immunoblotting with an antiflag or antiphosphotyrosine Ab. As shown in Figure 2A, LIME was phosphorylated upon BCR cross-linking, and treatment with PP1 but not with piceatannol significantly suppressed BCR-dependent phosphorylation of LIME. To control for the effectiveness of the inhibitors, the same cell lysates were analyzed by immunoblotting with antiphosphotyrosine Ab. Both PP1 and piceatannol effectively reduced the overall tyrosine phosphorylation of cellular proteins (Figure 2A, third panel). These data suggest that LIME is phosphorylated upon BCR stimulation by a tyrosine kinase of the Src family.
As another approach to determine the kinase responsible for the phosphorylation of LIME, flag-tagged LIME was transiently coexpressed with Lyn or Syk in 293T cells. LIME was strongly phosphorylated when it was coexpressed with Lyn but not with Syk (Figure 2B), showing that LIME is a substrate of Lyn. Taken together, these studies suggest that Lyn is responsible for the phosphorylation of LIME upon BCR-cross linking. However, we do not exclude the possibility that other Src family kinases, such as Blk and Fyn, are also involved in the phosphorylation of LIME.
Upon BCR cross-linking, LIME is phosphorylated by Lyn tyrosine kinase. (A) A20 cells were transfected with constructs encoding LIME-flag. Subsequently, cells were pretreated with either DMSO, PP1 (a Src family tyrosine kinase inhibitor), or piceatannol (a Syk inhibitor) and stimulated with F(ab′)2 goat anti-mouse IgG for 1 minute. LIME was immunoprecipitated from cell lysates using an antiflag Ab, and the precipitates were analyzed by immunoblotting with an antiphosphotyrosine (4G10) or antiflag Ab. As a control, the same cell lysates were analyzed by immunoblotting with an antiphosphotyrosine Ab (third panel). (B) LIME-flag was coexpressed with Lyn or Syk in 293T cells. Lysates were subjected to immunoprecipitation with an antiflag Ab, and the precipitates were analyzed by Western blotting with antiphosphotyrosine (4G10) and antiflag Abs.
Upon BCR cross-linking, LIME is phosphorylated by Lyn tyrosine kinase. (A) A20 cells were transfected with constructs encoding LIME-flag. Subsequently, cells were pretreated with either DMSO, PP1 (a Src family tyrosine kinase inhibitor), or piceatannol (a Syk inhibitor) and stimulated with F(ab′)2 goat anti-mouse IgG for 1 minute. LIME was immunoprecipitated from cell lysates using an antiflag Ab, and the precipitates were analyzed by immunoblotting with an antiphosphotyrosine (4G10) or antiflag Ab. As a control, the same cell lysates were analyzed by immunoblotting with an antiphosphotyrosine Ab (third panel). (B) LIME-flag was coexpressed with Lyn or Syk in 293T cells. Lysates were subjected to immunoprecipitation with an antiflag Ab, and the precipitates were analyzed by Western blotting with antiphosphotyrosine (4G10) and antiflag Abs.
LIME associates with Lyn, Grb2, and PLC-γ2 upon BCR stimulation
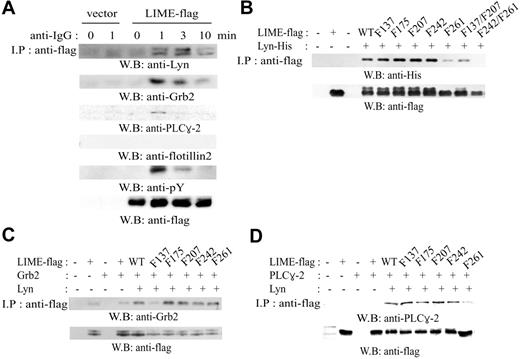
We next examined whether LIME interacts with proximal signaling molecules such as Lyn, Grb2, and PLC-γ2. A20 cells expressing LIME-flag were activated for 0, 1, 3, or 10 minutes with F(ab′)2 goat anti-mouse IgG, and the cell lysates were analyzed by immunoprecipitation with an antiflag Ab and subsequent Western blotting with anti-Lyn, -Grb2, -PLC-γ2, and -flotillin-2 Abs (Figure 3A). Flotillin-2 Ab was employed as a negative control because flotillin-2 is a raft protein21 that is not expected to associate with LIME. The association of LIME with Lyn, Grb2, and PLC-γ2 was induced upon BCR stimulation with the maximal binding at 1 to 3 minutes after BCR cross-linking, and the extent of binding was proportional to the phosphorylation of LIME. Flotillin-2 was not detected in LIME immunoprecipitates, although the protein was detected in the total lysates (data not shown). These data show that LIME associates with Lyn, Grb2, and PLC-γ2 upon BCR stimulation.
The LIME motifs responsible for binding to Lyn were mapped by coexpressing tyrosine site mutants of LIME with Lyn in 293T cells and analyzing binding ability (Figure 3B). All of the single tyrosine mutants of LIME coprecipitated with Lyn, although Y261F mutant showed reduced binding. The double tyrosine mutant of LIME, Y242/261F, failed to bind Lyn. Following the same approach, we mapped the domain of LIME responsible for binding to Grb2. As shown in Figure 3C, the association between LIME and Grb2 was dependent on the presence of Lyn, and Y137F mutant showed significantly reduced binding, suggesting that tyrosine 137 is responsible for binding to Grb2. The sequence of the motif, Y137SNV, resembles the consensus binding site of the Grb2 SH2 domain.22,23 We also mapped the binding motif for PLC-γ2 in 293T cells (Figure 3D). Again, the association between LIME and PLC-γ2 was dependent on the presence of Lyn, and the Y261F mutant showed almost completely abolished binding, suggesting that tyrosine 261 is involved in association with PLC-γ2. However, the sequence of this motif, Y261ESI, varies significantly from that of the consensus binding motif for the PLC-γ2 SH2 domains,22,23 and we therefore do not exclude the possibility of indirect association.
LIME associates with Lyn, Grb2, and PLC-γ2. (A) Expression plasmids encoding LIME-flag were transfected into A20 cells. Subsequently, cells were activated by cross-linking with F(ab′)2 goat anti-mouse IgG for the indicated times. Cell lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using anti-Lyn, anti-Grb2, anti-PLC-γ2, antiphosphotyrosine (4G10), antiflotillin-2, and antiflag Abs. In antiphosphotyrosine blot (fifth panel), a 32-kDa band corresponding to the size of LIME was detected upon BCR stimulation. (B) Mapping of the binding sites for association with Lyn. His-tagged Lyn was coexpressed with flag-tagged tyrosine mutants of LIME (tyrosine→phenylalanine) in 293T cells. Lysates were analyzed by immunoprecipitation using an antiflag Ab followed by immunoblotting with an anti-His, antiflag, or antiphosphotyrosine (4G10) Ab. (C) Mapping of the binding sites for association with Grb2. Flag-tagged tyrosine mutants of LIME were coexpressed with Grb2 and Lyn in 293T cells. Cell lysates were processed as described in panel B. (D) Mapping of the binding sites for association with PLC-γ2. Flag-tagged tyrosine mutants of LIME were coexpressed with PLC-γ2 and Lyn in 293T cells. Cell lysates were processed as described in panel B.
LIME associates with Lyn, Grb2, and PLC-γ2. (A) Expression plasmids encoding LIME-flag were transfected into A20 cells. Subsequently, cells were activated by cross-linking with F(ab′)2 goat anti-mouse IgG for the indicated times. Cell lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using anti-Lyn, anti-Grb2, anti-PLC-γ2, antiphosphotyrosine (4G10), antiflotillin-2, and antiflag Abs. In antiphosphotyrosine blot (fifth panel), a 32-kDa band corresponding to the size of LIME was detected upon BCR stimulation. (B) Mapping of the binding sites for association with Lyn. His-tagged Lyn was coexpressed with flag-tagged tyrosine mutants of LIME (tyrosine→phenylalanine) in 293T cells. Lysates were analyzed by immunoprecipitation using an antiflag Ab followed by immunoblotting with an anti-His, antiflag, or antiphosphotyrosine (4G10) Ab. (C) Mapping of the binding sites for association with Grb2. Flag-tagged tyrosine mutants of LIME were coexpressed with Grb2 and Lyn in 293T cells. Cell lysates were processed as described in panel B. (D) Mapping of the binding sites for association with PLC-γ2. Flag-tagged tyrosine mutants of LIME were coexpressed with PLC-γ2 and Lyn in 293T cells. Cell lysates were processed as described in panel B.
LIME mediates BCR-dependent MAP kinase activation, calcium mobilization, and NF-AT activation
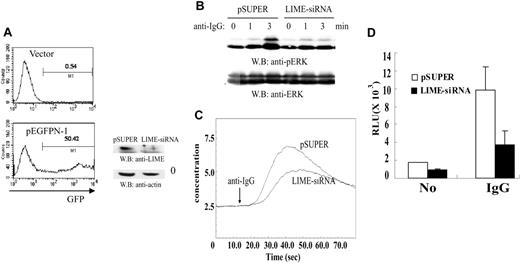
Having found that LIME recruits Grb2 and PLC-γ2 in a BCR-dependent manner, we were interested in testing whether LIME is involved in BCR-dependent downstream events such as MAPK activation and calcium mobilization. pSUPER or a pSUPER construct expressing LIME siRNA along with pEGFPN-1 plasmids encoding EGFP were introduced into A20 cells by electroporation. We routinely obtained the transfection efficiency of more than 50% as measured by the fluorescence-activated cell sorter (FACS) analysis of GFP-expressing cells. (Figure 4A, left panels). Analysis of LIME levels by Western blotting showed that LIME levels were effectively reduced by the siRNA (Figure 4A, right panel). To determine whether BCR-mediated MAPK activation was also affected by LIME siRNA, A20 cells were stimulated with F(ab′)2 goat anti-mouse IgG for 0, 1, or 3 minutes, and the cell lysates were subjected to immunoblotting with an anti-phospho-ERK Ab. The phosphorylation of ERK observed at 3 minutes after BCR cross-linking was abolished in cells expressing LIME siRNA. Analysis of ERK as a control showed that the lysates were loaded equally (Figure 4B).
Next, to examine the role of LIME in calcium mobilization, cells expressing LIME siRNA were loaded with fura-2 and stimulated with F(ab′)2 goat anti-mouse IgG. Intracellular calcium concentrations were then measured. As shown in Figure 4C, the reduction of LIME expression by siRNA resulted in a significant decrease in calcium flux. To examine whether LIME is involved in the activation of NF-AT, A20 cells were transfected with an NF-AT-Luc reporter construct along with pSUPER expressing LIME siRNA. In cells expressing LIME siRNA, reporter activity mediated by BCR and NF-AT was suppressed to approximately 35% of that of pSUPER-transfected cells (Figure 4D).
LIME, but not LAB, is required for BCR-induced PI3K activation
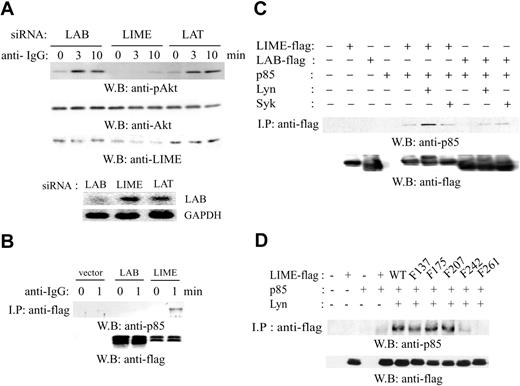
To test the effect of LIME and LAB siRNA expression on BCR-dependent activation of PI3K, we transfected A20 cells with pSUPER plasmids expressing LAB siRNA or LIME siRNA and stimulated with F(ab′)2 goat anti-mouse IgG Ab for 0, 3, or 10 minutes. pSUPER plasmids expressing LAT siRNA were used as a negative control, because LAT is not expressed in mature B cells. Subsequently, lysates were subjected to immunoblotting with anti-phospho-Akt, anti-Akt, or anti-LIME Abs. In cells expressing LIME siRNA, but not in those expressing LAB or LAT siRNA, Akt phosphorylation 3 or 10 minutes after BCR cross-linking was abolished (Figure 5A, top three panels). As a control, the level of endogenous Lab was examined by reverse transcriptase (RT)-PCR (Figure 5A, bottom two panels).
LIME is required for BCR-mediated activation of p42/44 MAPK, calcium flux, and NF-AT. (A) A20 cells were transfected with the control vector or plasmids expressing LIME siRNA along with pEGFPN-1. Forty-eight hours after electroporation, transfection efficiency was analyzed by FACS (left panels). The reduction of LIME expression upon expression of siRNA was assessed by Western blotting with an anti-LIME Ab (right panel). (B) LIME mediates BCR-dependent MAPK activation. A20 cells were transfected with pSUPER or pSUPER encoding siRNA directed against murine LIME. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 1 and 3 minutes at 37°C. Lysates were analyzed by Western blotting with an anti-phospho-ERK or anti-ERK Ab. (C) LIME is required for the BCR-mediated Ca2+ response. A20 cells were transfected with pSUPER vector or pSUPER expressing an siRNA against LIME. After 48 hours, transfected cells were loaded with fura-2/AM for 30 minutes at 37°C. Subsequently, cells were stimulated with 20 μg/mL F(ab′)2 goat anti-mouse IgG, and the intracellular calcium level was measured. (D) LIME mediates BCR-dependent NF-AT activation. The plasmids expressing siRNA against LIME were cotransfected into A20 cells with an NF-AT-luciferase reporter construct. After 48 hours, the cells were incubated for 6 hours with medium alone, F(ab′)2 goat anti-mouse IgG, or PMA/ionomycin, and then harvested. Subsequently, the cells were lysed and the luciferase activity was assayed. To control for the transfection efficiency, the luciferase activities of medium or F(ab′)2 goat anti-mouse IgG-treated samples were normalized to those of PMA/ionomycin-treated samples. The experiments were performed twice in triplicate. Error bars indicate SD.
LIME is required for BCR-mediated activation of p42/44 MAPK, calcium flux, and NF-AT. (A) A20 cells were transfected with the control vector or plasmids expressing LIME siRNA along with pEGFPN-1. Forty-eight hours after electroporation, transfection efficiency was analyzed by FACS (left panels). The reduction of LIME expression upon expression of siRNA was assessed by Western blotting with an anti-LIME Ab (right panel). (B) LIME mediates BCR-dependent MAPK activation. A20 cells were transfected with pSUPER or pSUPER encoding siRNA directed against murine LIME. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 1 and 3 minutes at 37°C. Lysates were analyzed by Western blotting with an anti-phospho-ERK or anti-ERK Ab. (C) LIME is required for the BCR-mediated Ca2+ response. A20 cells were transfected with pSUPER vector or pSUPER expressing an siRNA against LIME. After 48 hours, transfected cells were loaded with fura-2/AM for 30 minutes at 37°C. Subsequently, cells were stimulated with 20 μg/mL F(ab′)2 goat anti-mouse IgG, and the intracellular calcium level was measured. (D) LIME mediates BCR-dependent NF-AT activation. The plasmids expressing siRNA against LIME were cotransfected into A20 cells with an NF-AT-luciferase reporter construct. After 48 hours, the cells were incubated for 6 hours with medium alone, F(ab′)2 goat anti-mouse IgG, or PMA/ionomycin, and then harvested. Subsequently, the cells were lysed and the luciferase activity was assayed. To control for the transfection efficiency, the luciferase activities of medium or F(ab′)2 goat anti-mouse IgG-treated samples were normalized to those of PMA/ionomycin-treated samples. The experiments were performed twice in triplicate. Error bars indicate SD.
The observed involvement of LIME in BCR-dependent PI3K activation led us to examine the interaction between LIME and the PI3K p85 subunit. After transfection of the plasmid-encoding vector, LIME-flag or LAB-flag, A20 cells were activated with F(ab′)2 goat anti-mouse IgG for 1 minute. Subsequently, cell lysates were subjected to immunoprecipitation with an antiflag Ab, and the immunoprecipitates were examined for the presence of PI3K p85. LIME but not LAB associated with p85 upon BCR cross-linking (Figure 5B). In addition, we examined whether the interaction between LIME and p85 is dependent on the phosphorylation of LIME. 293T cells were transfected with the plasmids encoding p85 along with the plasmid encoding LIME-flag or LAB-flag. As shown in Figure 5C, the interaction of LIME with p85 was observed only when Lyn was coexpressed. LAB interacted with p85 very weakly regardless of the coexpression of Lyn or Syk. In addition, the motif of LIME responsible for binding to p85 was mapped by coexpressing tyrosine mutants of LIME with Lyn in 293T cells (Figure 5D). The association between LIME and p85 was dependent on the presence of Lyn, and both the Y242F and Y261F mutant showed almost completely or completely reduced binding. These results suggest that tyrosines 242 and 261 are responsible for the association with p85. However, the sequences of these motifs, Y242EAI and Y261ESI, are quite different from that of the consensus binding motif for the SH2 domain of p85,22,23 and we therefore do not exclude the possibility of indirect interaction.
LIME is required for BCR-dependent Akt activation. (A) A20 cells were transfected with plasmids expressing siRNAs directed against murine LAB, LIME, or LAT. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 3 or 10 minutes at 37°C. Lysates were analyzed by Western blotting with anti-phospho-Akt, anti-Akt, or anti-LIME Abs. The level of Lab mRNA was assessed by RT-PCR. (B) p85 associates with LIME. A20 cells were transfected with the plasmids expressing LIME-flag or LAB-flag and stimulated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 0 or 1 minutes at 37°C. The lysates were immunoprecipitated using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab. (C) p85 was coexpressed with LIME-flag or LAB-flag in 293T cells in combination with Lyn or Syk. Lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab. (D) Mapping of the motifs responsible for association with p85. Flag-tagged tyrosine mutants of LIME were coexpressed with p85 and Lyn in 293T cells. Lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab.
LIME is required for BCR-dependent Akt activation. (A) A20 cells were transfected with plasmids expressing siRNAs directed against murine LAB, LIME, or LAT. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 3 or 10 minutes at 37°C. Lysates were analyzed by Western blotting with anti-phospho-Akt, anti-Akt, or anti-LIME Abs. The level of Lab mRNA was assessed by RT-PCR. (B) p85 associates with LIME. A20 cells were transfected with the plasmids expressing LIME-flag or LAB-flag and stimulated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 0 or 1 minutes at 37°C. The lysates were immunoprecipitated using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab. (C) p85 was coexpressed with LIME-flag or LAB-flag in 293T cells in combination with Lyn or Syk. Lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab. (D) Mapping of the motifs responsible for association with p85. Flag-tagged tyrosine mutants of LIME were coexpressed with p85 and Lyn in 293T cells. Lysates were subjected to immunoprecipitation using an antiflag Ab, and the precipitates were analyzed by Western blotting using an anti-p85 or antiflag Ab.
Taken together, these results show that LIME, but not LAB, mediates BCR-induced PI3K activation via interaction with the p85 subunit.
LIME is required for BCR-dependent NF-κB activation
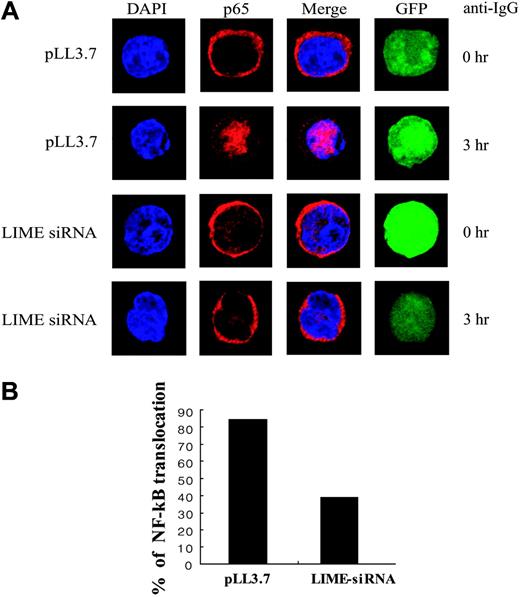
Transcriptional activation by NF-κB is an important event in B-cell activation. NF-κB activation is regulated by Iκ-B, which retains NF-κB dimers within the cytosol in an inactive state. Upon BCR-ligation, Iκ-Bα becomes degraded following phosphorylation by IKK, thereby releasing NF-κB, which moves to the nucleus. We therefore tested whether LIME is involved in NF-κB activation using nuclear translocation as an indicator of NF-κB activation. We used a pLL3.7 lentivirus vector instead of pSUPER to introduce LIME siRNA expression; the pLL3.7 vector coexpresses EGFP, permitting transfected cells to be identified. A20 cells transfected with pLL3.7 or pLL3.7 expressing LIME siRNA were activated with F(ab′)2 goat anti-mouse IgG for 3 hours. Cells were stained with the anti-RelA (p65) Ab and Alexa 568-conjugated anti-rabbit IgG. In control cells transfected with pLL3.7 vector, NF-κB localized in the cytoplasm before stimulation and translocated to the nucleus upon BCR cross-linking. However, in cells expressing LIME siRNA, BCR-dependent NF-κB translocalization was blocked (Figure 6A). For each experiment, we analyzed approximately 30 GFP-positive cells and counted the number of cells showing p65 nuclear translocation. The number of cells with nuclear translocation of NF-κB was reduced by approximately 70% in cells expressing LIME siRNA compared with pLL3.7-transfected cells (Figure 6B). The observed suppression of the activation of MAPK, calcium flux, NF-AT, PI3K, and NF-κB by LIME siRNA indicates that LIME is essential for BCR-mediated B-cell activation.
LIME is required for BCR-dependent NF-κB translocation. (A) LIME mediates BCR-dependent NF-κB translocation to the nucleus. A20 cells were transfected with pLL3.7 plasmids or pLL3.7 expressing siRNA against LIME. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 3 hours at 37°C. The cells were fixed and permeabilized with 0.1% Triton X-100, after which the localization of NF-κB was detected by an anti-p65 Ab and Alexa 568-conjugated anti-rabbit IgG. (B) Graph shows the number of cells with nuclear translocation of NF-κB.
LIME is required for BCR-dependent NF-κB translocation. (A) LIME mediates BCR-dependent NF-κB translocation to the nucleus. A20 cells were transfected with pLL3.7 plasmids or pLL3.7 expressing siRNA against LIME. After 48 hours, transfected cells were activated with 20 μg/mL F(ab′)2 goat anti-mouse IgG for 3 hours at 37°C. The cells were fixed and permeabilized with 0.1% Triton X-100, after which the localization of NF-κB was detected by an anti-p65 Ab and Alexa 568-conjugated anti-rabbit IgG. (B) Graph shows the number of cells with nuclear translocation of NF-κB.
Discussion
Previously, LIME was identified as a regulator of TCR-mediated signaling. In this report, we show that LIME also functions as a mediator of BCR-dependent B-cell activation. LIME was phosphorylated by the Src family kinase Lyn but not Syk upon BCR cross-linking. LIME interacted with Lyn and recruited Grb2, PLC-γ2, and the p85 subunit of PI3K in response to BCR cross-linking. Decreased LIME expression led to abrogation of BCR-dependent ERK activation, calcium flux, and subsequent activation of NF-AT, PI3K, and NF-κB. These data strongly support that LIME acts as a transmembrane adaptor mediating BCR-induced B-cell activation.
Recently, 2 groups identified LAB as a transmembrane adaptor that is expressed in B cells.14,15 Although both LIME and LAB are rapidly phosphorylated upon BCR cross-linking, these 2 molecules have different properties during B-cell activation signaling. First, while LIME is phosphorylated by the Src family kinase Lyn, LAB is phosphorylated by Syk, suggesting that these 2 proteins regulate distinct signal pathways. Second, although an RNA interference study supported a positive regulator role for LIME and LAB in the initial BCR-dependent calcium flux, PLC-γ2 associates with LIME but not LAB in B cells.14 Because the generation of the initial calcium flux from an intracellular source is regulated by the molecular complex of SLP-65/BLNK/BASH and PLC-γ2,24 it is tempting to speculate that LIME rather than LAB may mediate the BCR signaling-dependent initial calcium flux. In BCR signaling, BLNK has been proposed as a central adaptor protein linking the proximal receptor-mediated signal and the downstream activation pathways.25 Following BCR activation, BLNK is rapidly phosphorylated by Syk and translocates to the membrane. Subsequently, BLNK associates with several intracellular signaling molecules, including Grb2, PLC-γ2, Vav, Btk, and Nck, and regulates BCR-mediated MAPK activation via the Grb2-Sos pathway, Ca2+ flux via PLC-γ2, and NF-κB activation via the PLC-γ2-PKC pathway.26-28 However, despite the importance of BLNK in BCR signaling, it has not been clearly defined how BLNK is recruited to the membrane during BCR engagement. It has been reported that BLNK might be recruited to Ig-α by direct interaction, but this has not yet been clarified in vivo.29 By analogy to TCR signaling, it seems likely that a transmembrane adaptor molecule is responsible for the recruitment of BLNK to the membrane upon BCR signaling. Therefore, we tested the possible association of LIME with BLNK upon BCR stimulation in A20 cells but failed to detect BLNK in LIME immunoprecipitates (data not shown). Although the association was not detected, we do not exclude the possibility that LIME is involved in the recruitment of BLNK via interactions with PLC-γ2 or Grb2, because BLNK is known to associate with PLC-γ224 and Grb226 upon BCR stimulation.
LIME may be involved in the recruitment of PH domain-containing molecules, such as Btk and Bam32, to the membrane through activation of PI3K activity. Btk, a Tec family kinase, has a PH domain that binds PtdInsP3.30 Btk is activated when tyrosine phosphorylated by Lyn and plays a critical role in BCR signaling. Activated Btk associates with the BLNK/PLC-γ2 complex and activates PLC-γ2 through phosphorylation.30,31 Therefore, LIME may contribute to the activation of PLC-γ2 and the subsequent calcium flux through 2 pathways: via activation of PI3K and subsequent activation of Btk and via recruitment of PLC-γ2 to the membrane by association. Bam32 is an adaptor protein with a PH domain and an SH2 domain.32 Bam32 mediates activation of JNK and ERK and is essential for BCR-induced B-cell proliferation.33 In addition, Bam32 contributes to BCR-dependent activation of NF-κB activity.32,33 Because Bam32 is recruited to the plasma membrane via its PH domain, LIME may contribute to this event by activating PI3K activity, which elevates the PtdInsP3 level. Thereby, LIME may regulate Bam32-mediated events such as JNK activation and NF-κB activation.
Taken together, we have shown that LIME links the BCR to downstream events leading to B-cell activation. LIME is therefore a transmembrane adaptor protein essential for BCR signaling.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-05-1859.
Supported by Nano/Bio Science & Technology Program grant 2004-00640 from the Korean Ministry of Science and Technology and Korea Science Foundation through the Center for Cell Signaling Research at Ewha Womans University.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We appreciate Dr Jaesang Kim for critical reading of the manuscript. We thank Dr Agami for pSUPER vector, Dr Luk Van Parijs for pLL3.7 vector, Dr Yunsoo Bae for PLC-γ2 expression vector, and Dr S. G. Lee for human Lyn, Syk expression vector.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal