The poliovirus receptor CD155 and its family member CD112 (nectin-2) are the ligands for the activating cell-surface receptor DNAM-1 on CD8+ T cells and natural killer (NK) cells. Here, we demonstrate that, whereas the RMA tumor grew in syngeneic mice, DNAM-1 ligand-transduced RMA was rejected, in which CD8+ T cells and NK cells played an essential role. Importantly, CD8+ memory cytotoxic T cells to parental RMA were generated in these mice. We found that DNAM-1 was also expressed on CD8α+, rather than CD8α-, dendritic cells (DCs). Cross-linking DNAM-1 induced maturation of CD8α+ DCs. Antigen presentation by these stimulated DCs drove Th1 cells. Moreover, the rejection of DNAM-1 ligand-transduced RMA was canceled in CD4+ T-cell–depleted and major histocompatibility complex class II–deficient mice. Taken together, these results suggest that DNAM-1 ligands stimulate innate immunity by CD8α+ DCs as well as NK cells, which efficiently prime cell-mediated tumor-specific immunity.
Introduction
Immune surveillance is mediated by both cellular and humoral immunities. Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells are major players in cell-mediated immunity against tumors or virus-infected cells. CTLs and NK cells often share adhesion or costimulatory molecules, including CD2 and leukocyte function–associated antigen-1 (LFA-1; CD11a/CD18), CD27, 2B4, CD28, inducible costimulator (ICOS), NKG2D, and others.1,2 These cell-surface molecules not only mediate intercellular binding between these cytotoxic lymphocytes and target cells, but they also participate in signal transduction for cytotoxicity and cytokine secretion.1,3
DNAM-1 (CD226) is an adhesion molecule that is a member of the immunoglobulin superfamily containing 2 immunoglobulin-like domains of the V-set.4-6 DNAM-1 is an approximate 65-kDa glycoprotein expressed on the majority of NK cells, T cells, monocytes, and platelets and a subset of B lymphocytes.4,7,8 The monoclonal antibody (mAb) against DNAM-1 inhibited antigenspecific CTL-mediated and NK-cell–mediated cytolysis of some, but not all, tumor targets in vitro, suggesting that DNAM-1 is involved in cytotoxicity against certain tumor cells expressing the DNAM-1 ligand. Recently, we and others identified the poliovirus receptor (PVR) CD155 and its family member CD112 (PVRrelated family 2 [PRR-2], also called as nectin-2), as the ligands for human DNAM-1.9,10 Ectopic expression of human CD155 or CD112 rendered tumor cell lines more susceptible to interleukin 2 (IL-2)–activated T and NK-cell–mediated cytotoxicity in vitro. Although the human DNAM-1 ligands CD112 and CD155 are broadly distributed on epithelial and endothelial cells in many tissues,11,12 they are overexpressed on certain human tumors, including colorectal carcinomas,13 myeloid leukemias,14 and neuroblastomas,15 suggesting that the expression of DNAM-1 ligands is inducible by “stress” such as transformation and infection. However, it has still been an open question how the interaction of DNAM-1 with its ligands is involved in immune responses in vivo.
We recently identified the murine homologues of DNAM-1 and its ligands.16 In the present study, we investigated an in vivo role of DNAM-1 and DNAM-1 ligands on tumor immunosurveillance.
Materials and methods
Mice, antibodies, and cells
C57BL/6 (B6) mice were purchased from Charles River (Yokohama, Japan) and were bred and housed under specific pathogen-free conditions. OVA peptide (ISQAVHAAHAEINEAGR; OVA + 323 - 339)–specific CD4 TCR transgenic mice (OT-II) and major histocompatibility complex (MHC) class II–deficient mice were kindly provided by William R. Heath (WEHI, Parkville, Australia) and Diane Mathis (Harvard University, Boston, MA), respectively. For depletion of CD8+ and CD4+ T cells and NK cells, anti–mouse CD8α (19/178; a gift from Eiichi Nakayama, Okayama University, Okayama, Japan), anti–mouse CD4 (GK1.5), and anti-NK1.1 (PK136; obtained from American Type Culture Collection, Manassas, VA) were used. The other mAbs used in this study were purchased from BD Biosciences (San Jose, CA). RMA cells stably expressing mouse CD112α and CD155 (RMA-CD112 and RMA-CD155, respectively) were established, as described previously.16 Control RMA cells were also established by transducing pMX vector alone and this transduction was confirmed by genomic polymerase chain reaction (PCR) for the vector backbone sequence, using the primer combination of 5′-ggtggaccatcctctagact-3′ and 5′-ccctttttctggagactaaat-3′. Antimouse DNAM-1 (TX42) mAb was generated in our laboratory by standard methods.16 TX42 mAb was a blocking antibody that inhibited binding of DNAM-1-Fc chimeric protein with RMA-CD155 or RMA-CD112, as determined by flow cytometry. In vivo injection (200 μg intraperitoneally on day 1 and day 3) of TX42 mAb did not affect the populations of T cells, B cells, and NK cells in spleen at day 7, as determined by flow cytometry, whereas anti-CD8α (19/178) and anti-NK1.1 (PK136) almost completely depleted CD8+ T cells and NK cells, respectively, by the same procedure. TX42 mAb was free from endotoxin, as examined using the limulus amebocyte lysate (LAL) test (Sigma, St Louis, MO).
Tumor growth assay and survival of mice
Groups of 5 or 10 C57BL/6 mice were pretreated by intraperitoneal injection of 200 μg mAb for depletion of CD8+ T, CD4+ T, or NK cells at days -1, 1, 8, 15, and 22, where day 0 is the day of primary or secondary tumor inoculation. Depletion of each cell type was confirmed by analyzing splenocytes from a group of mice treated with each mAb. For blocking of interaction of DNAM-1 with its ligands in vivo, mice were pretreated by intraperitoneal injection of 200 μg anti–DNAM-1 at days -1, 1, 6, and 10. Control mice received the equivalent amounts of normal mouse IgG. At day 0, groups of 5 or 10 mice per experiment were given subcutaneous injections in the back with 1 × 105 RMA cells that were transduced with Cd112d, CD155, or mock-control vector. Mice were examined twice a week for tumor size with a caliper square. For the rechallenge experiments, mice that had completely rejected the initial tumor cells were given injections with RMA tumor cells 70 days after the first tumor inoculation.
ELISPOT assay
Enzyme-linked immunospot (ELISPOT) assay was performed with a mouse IFN-γ–specific ELISPOT kit (eBioscience, San Diego, CA) according to the manufacturer's instructions. Splenocytes from naive C57BL/6 mice were used as antigen-presenting cells (APCs) after pulse with 1 μM gagL peptide (CCLCLTVFL; gagL75-83), which is a leader sequence of Rauscher murine leukemia virus, or a control OVA peptide (SIINFEKL; OVA257-264) for 30 minutes at 37°C, and then plated at 5 × 106 cells/well. CD8+ T cells from splenocytes of mice inoculated with RMA-CD155 or RMA-mock were purified by positive selection using magnetically activated cell sorting (MACS; Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions, and plated across a range of cell concentrations to achieve 10 to 100 spots/well for accurate counting, and cultured for 24 hours with APCs prepulsed with peptides. The colored spots were counted using a stereomicroscope. Frequencies of antigen-specific IFN-γ–secreting cells were calculated based on the numbers of CD8+ cells and the number of spots per well after subtraction of background, which was obtained by incubating CD8+ T cells stimulated with the control OVA peptides. Experiments were performed in triplicate.
In vitro stimulation of DCs
CD11c+ dendritic cells (DCs) were purified from spleen cells of C57BL/6 mice by positive selection using MACS (Miltenyi Biotec). Cells were incubated with anti-CD16/32 mAb (2.4G2) to block nonspecific binding to Fcγ receptors by antibodies and then stimulated with plate-coated control immunoglobulin or anti–DNAM-1 (TX42) for 4 days. Cells were then stained with phycoerythrin (PE)–conjugated anti-CD11c mAb, allophycocyanin (APC)–conjugated anti-CD8 mAb, and either fluorescein isothiocyanate (FITC)–conjugated anti-CD80, anti-CD86, anti–MHC class II (I-Ab) mAbs or control IgG and analyzed by flow cytometry. For analysis of function of DNAM-1–expressing DCs, CD11c+CD8α+ or CD11c+CD8α- DCs were purified from splenocytes of C57BL/6 mice by positive selection by using MACS and then by flow cytometry. Each DC population was stimulated with plate-coated anti–DNAM-1, anti-CD40, or control immunoglobulin for 4 days. Cells were pulsed with the OVA peptide and then cocultured with CD4+ T cells from splenocytes of OT-II mice for 7 days. Cells were then stimulated with anti-CD3 and anti-CD28 for 48 hours, and IFN-γ concentrations in the culture supernatants were determined with a mouse IFN-γ–specific enzyme-linked immunosorbent assay (ELISA) kit (Biosource International, Camarillo, CA).
Immunohistochemistry
Immunostaining was performed with some modifications as previously described.17 Cryostat sections (6 μm) were processed for immunohistochemical procedures based on the Labeled Polymer method (Dako Japan, Kyoto, Japan). Briefly, sections were treated with 3% hydrogen peroxidase in absolute methanol for 30 minutes to quench endogenous peroxidase activity. After antigen retrieval with Target Retrieval Solution (Dako Japan), sections were preincubated in 0.01 M phosphate-buffered saline (PBS) containing 5% normal goat serum (Jackson ImmunoResearch, West Grove, PA) at room temperature (RT) for 1 hour, and then incubated in anti–PRR-2 mAb (1:20 dilution) containing 0.5% bovine serum albumin (BSA) at RT for 2 hours. After washes with 0.1M PBS, the sections were incubated in goat anti–mouse immunoglobulin conjugated to peroxidase-labeled dextran polymer (EnVision+, Dako Japan) at RT for 30 minutes. After washes with 0.1M PBS, the horseradish peroxidase reaction was developed in 0.1M Tris-buffered saline, pH 7.4, containing 0.05% 3,3′diaminobenzidine tetrahydrochloride (Sigma), 0.02% nickel sulfate, and 0.01% hydrogen peroxide. Methyl green was used for counterstaining. The following controls were performed: (1) incubation with isotype control antibody instead of the primary antibody and (2) incubation without the primary antibody or without primary and secondary antibodies. All controls revealed no signals. Images of tumor sections were obtained via analysis using a Zeiss Axiovert 135 microscope equipped with Plan-Neofluar 5 ×/0.15 and 10 ×/0.30 objective lenses (Carl Zeiss, Oberkochen, Germany) and using a Keyence VB-7010 camera (Keyence Osaka, Japan). Image processing was performed using VH analyzer VH-H1A5 (Keyence) and Adobe Photoshop software (Adobe Systems, San Jose, CA).
Quantitative RT-PCR
Total RNA was extracted from gastric cancer tissues and the adjacent normal tissues in 3 patients with poorly differentiated gastric cancer with ISOGEN reagent (Nippongene, Toyama, Japan). Reverse transcription was performed with 5 μg total RNA using Thermoscript reverse transcription–polymerase chain reaction (RT-PCR) system and 50 ng random hexamers (Invitrogen, Carlsbad, CA) in a final volume of 20 μL. Real-time PCR analysis of CD112 was carried out on an ABI7700 sequence detector (Applied Biosystems, Foster City, CA). The primers for Cd112 were as follows: upper primer, 5′-atgtggccgccttccaccctaa-3′; lower primer, 5′-cagactgtccactgcgtggatg-3′. The 18S rRNA level was measured as an internal control (upper primer, 5′-ccctgccctttgtacacacc-3′; lower primer, 5′-cgatccgagggcctcacta-3′). The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Standard curves were generated using serial dilutions of cDNA from HOS cells (human osteosarcoma cell line), covering 4 orders of magnitude and yielding correlation coefficients of at least 0.98 in all experiments. Each standard and sample value was determined in triplicate in 3 independent experiments. In each experiment, CD112 expression under a given experimental condition was calculated relative to the control condition.
Results
Expression of the DNAM-1 ligand on human poorly differentiated gastric cancer
Recent evidence indicates that the DNAM-1 ligands CD155 and CD112 are overexpressed on certain human tumors, including colorectal carcinomas, neuroblastomas, and myeloid leukemias.13-15 We also investigated the expression of the DNAM-1 ligand CD112 on specimens from 5 patients with gastric cancer, including 2 well-differentiated and 3 poorly differentiated histologic types, and 2 patients with colon cancer. Immunohistochemical analyses demonstrated that CD112 was overexpressed preferentially in the poorly but not the well-differentiated histologic type of gastric cancer (Figure 1A). We could not clearly detect CD112 in the untransformed normal tissue area bordering transformed cancer tissue in each specimen. To further assess whether CD112 is specifically expressed in poorly differentiated cancer tissue but not in normal gastric tissue, we divided the gastric tissues resected under surgery into transformed and normal areas of slices and compared mRNA expression of CD112 in each patient by quantitative RT-PCR analysis. As shown in Figure 1B, the relative amount of CD112 mRNA expressions are about 3 times higher in the poorly differentiated gastric cancer compared with that in normal gastric tissue in all 3 patients. We also observed the overexpression of CD112 on colon cancer as well (Figure 1A). These observations, along with recent reports,13-15 suggest that DNAM-1 ligands can be targets for tumor immunosurveillance in vivo.
RMA tumor expressing DNAM-1 ligands is rejected in mice
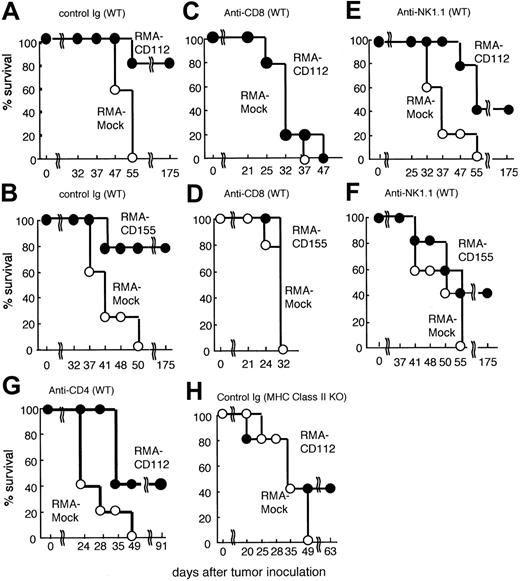
To investigate the in vivo consequences of expression of DNAM-1 ligands on tumor cells, B6 mice were inoculated subcutaneously with a murine T-cell lymphoma RMA transfected with control vector (RMA-mock) or Cd112 (RMA-CD112). Whereas mice that received injections of 1 × 105 RMA-mock all developed tumors and died by 55 days after tumor inoculation, most mice rejected the same number of RMA-CD112 (Figure 2A). These recipient mice of RMA-CD112 survived significantly longer than those given RMA-mock (Figure 2A). Similarly, most mice rejected 1 × 105 RMA transfected with Cd155 (RMA-CD115) and survived more than 175 days (Figure 2B). Because all these RMA transfectants showed similar growth kinetics in vitro, as determined by bromodeoxyuridine (BrdU)–incorporated proliferation assay (data not shown), these results indicate that ectopic expression of DNAM-1 ligands can result in rejection of tumors in syngeneic mice.
Up-regulation of CD112 expression in gastric and colon cancers. (A) Specimens from human gastric and colon cancer tissues were stained with anti-CD112 mAb (TX31). Positive signals were observed in the poorly differentiated (black arrow), but not well-differentiated (white arrow), transformed epithelial cells in gastric cancer (left). The transformed epithelial cells also show the highly positive signal (black arrow) in colon cancer (right). (B) mRNA was obtained from gastric cancer tissues from 3 patients with poorly differentiated histologic type. In each patient, mRNA was also obtained from untransformed normal tissue area. RT-PCR was performed with Cd112-specific primers and the amount of cDNA was measured by quantitative PCR analysis. CD112 mRNA expression was normalized to the internal standard 18S ribosomal RNA.
Up-regulation of CD112 expression in gastric and colon cancers. (A) Specimens from human gastric and colon cancer tissues were stained with anti-CD112 mAb (TX31). Positive signals were observed in the poorly differentiated (black arrow), but not well-differentiated (white arrow), transformed epithelial cells in gastric cancer (left). The transformed epithelial cells also show the highly positive signal (black arrow) in colon cancer (right). (B) mRNA was obtained from gastric cancer tissues from 3 patients with poorly differentiated histologic type. In each patient, mRNA was also obtained from untransformed normal tissue area. RT-PCR was performed with Cd112-specific primers and the amount of cDNA was measured by quantitative PCR analysis. CD112 mRNA expression was normalized to the internal standard 18S ribosomal RNA.
Rejection of DNAM-1 ligand-expressing RMA tumor cells. Groups of 5 C57BL/6 mice were pretreated with control immunoglobulin or mAbs indicated at days -1, 1, 8, 15, and 22. The mice were inoculated subcutaneously in the back at day 0 with 1 × 105 mock-transduced RMA tumor cells (○) or Cd112- or Cd155-transduced RMA tumor cells (•). Survival data for 5 mice per group are shown.
Rejection of DNAM-1 ligand-expressing RMA tumor cells. Groups of 5 C57BL/6 mice were pretreated with control immunoglobulin or mAbs indicated at days -1, 1, 8, 15, and 22. The mice were inoculated subcutaneously in the back at day 0 with 1 × 105 mock-transduced RMA tumor cells (○) or Cd112- or Cd155-transduced RMA tumor cells (•). Survival data for 5 mice per group are shown.
CD8+ T cells are responsible for the rejection of DNAM-1 ligand-transduced RMA cells
DNAM-1 is strongly expressed on the majority of CD8+ T cells and mediates a costimulatory signal in antigen-specific CD8+ T cells in vitro.4,16 We therefore investigated whether CD8+ T cells were responsible for the rejection of RMA-expressing DNAM-1 ligands. Mice were treated in vivo with depleting mAbs specific for CD8 or with a control antibody and then inoculated with RMA transfectants. Depletion of CD8+ T cells completely canceled the tumor rejection of both RMA-CD155 and RMA-CD112, and these mice became moribund at the same time as recipients of RMA-mock (Figure 2C-D). These results indicated that CD8+ T cells were essential for the rejection of DNAM-1 ligand-transduced RMA tumors.
Colonna and colleagues have recently identified the T cell-activated increased late expression (Tactile; also called as CD96), which is expressed on activated T cells,18 as another receptor ligand for human CD155.19 However, it is unclear whether the mouse homologue of CD96 is also a receptor for mouse CD155 or CD112, although it has not yet been identified. To confirm that DNAM-1, but not putative murine CD96, is responsible for the rejection of DNAM-1 ligand-expressing RMA tumors, we generated an anti–DNAM-1 mAb (TX42) that was able to block the ligand binding in vitro (Figure 3A). Mice were pretreated in vivo with TX42 mAb, which did not deplete T cells and NK cells in vivo (Figure 3B), and then inoculated subcutaneously with RMA-CD155. Eight days after tumor inoculation when tumors had not yet grown enough to be visible on the skin, the skin region was resected and subjected to histologic analysis. Whereas RMA-mock substantially proliferated in the skin region, most RMA-CD155 appeared to be dead (Figure 3C). However, pretreatment with anti–DNAM-1 mAb restored RMA-CD155 growth (Figure 3C), and the survival of these mice was significantly shorter than that of mice pretreated with control immunoglobulin (Figure 3D). Similar results were observed in mice inoculated with RMA-CD112 (data not shown). Because TX42 mAb did not affect the lymphocytes population in vivo, these results suggested an essential role of DNAM-1 on CD8+ T cells in the tumor rejection.
DNAM-1 is involved in rejection of DNAM-1 ligand-expressing RMA tumor cells. (A) CD155-RMA and CD112-RMA were stained with DNAM-1-Fc chimeric protein, which had been incubated with an excess of TX42 mAb (bold line) or control IgG (thin line), and then analyzed by flow cytometry. Staining with control human IgG1 (dotted line) and blocking of the DNAM-1-Fc staining with an excess of TX42 mAb or control IgG was performed to show the inhibitory function of TX42 mAb for DNAM-1 ligand recognition. (B) Splenocytes from mice were stained with FITC-conjugated anti-CD3 and PE-conjugated DX5 and analyzed 7 days after injection with TX42 mAb (200 μg intraperitoneally on day 1 and day 3) by flow cytometry. (C) Groups of 5 C57BL /6 mice were pretreated with control immunoglobulin or anti–DNAM-1 mAb at day -1. The mice were inoculated subcutaneously in the back at day 0 with 1 × 105 mock-transduced RMA tumor cells or Cd155-transduced RMA tumor cells. The skin regions were subjected to histologic analysis with hematoxylin-eosin under light microscopy at day 8. Data are representative of 3 mice in each group. (D) Survival data for 5 mice per group are shown.
DNAM-1 is involved in rejection of DNAM-1 ligand-expressing RMA tumor cells. (A) CD155-RMA and CD112-RMA were stained with DNAM-1-Fc chimeric protein, which had been incubated with an excess of TX42 mAb (bold line) or control IgG (thin line), and then analyzed by flow cytometry. Staining with control human IgG1 (dotted line) and blocking of the DNAM-1-Fc staining with an excess of TX42 mAb or control IgG was performed to show the inhibitory function of TX42 mAb for DNAM-1 ligand recognition. (B) Splenocytes from mice were stained with FITC-conjugated anti-CD3 and PE-conjugated DX5 and analyzed 7 days after injection with TX42 mAb (200 μg intraperitoneally on day 1 and day 3) by flow cytometry. (C) Groups of 5 C57BL /6 mice were pretreated with control immunoglobulin or anti–DNAM-1 mAb at day -1. The mice were inoculated subcutaneously in the back at day 0 with 1 × 105 mock-transduced RMA tumor cells or Cd155-transduced RMA tumor cells. The skin regions were subjected to histologic analysis with hematoxylin-eosin under light microscopy at day 8. Data are representative of 3 mice in each group. (D) Survival data for 5 mice per group are shown.
Induction of memory CD8+ T cells in mice inoculated with DNAM-1 ligand-expressing RMA
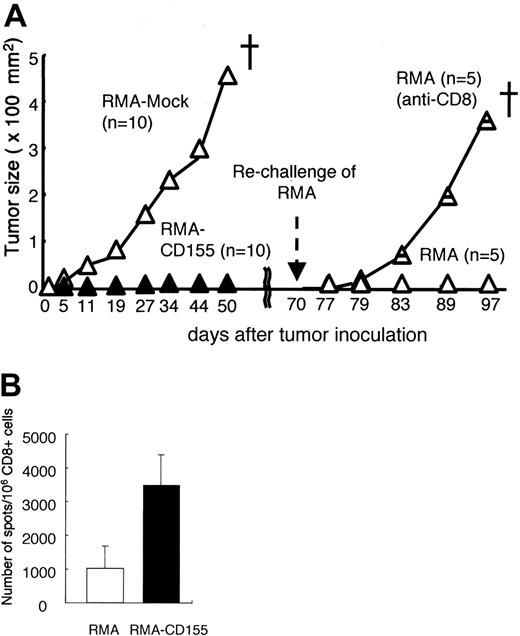
To investigate whether DNAM-1 ligand expression on RMA induces T-cell–mediated immunologic memory, mice that had previously rejected the Cd155-tranfected RMA tumor cells were rechallenged with the parental RMA tumors 70 days after the first inoculation (Figure 4). Whereas RMA-mock grew and killed all naive mice after the first inoculation, they were rejected in all the mice that had once rejected RMA-CD155 (Figure 4). However, depletion of CD8+ T cells with in vivo injection of anti-CD8 mAb before the rechallenge canceled the rejection of RMA (Figure 4). These results suggest that CD8+ memory T cells specific for an RMA tumor–associated antigen were generated in mice inoculated with RMA-CD155.
RMA is a Rauscher murine leukemia virus–transformed cell line bearing H-2Db, which associates with a peptide derived from the gag leader (gagL) sequence of the virus recognized by CTLs as an epitope.20,21 To investigate whether CD8+ T cells generated in mice inoculated with RMA-CD155 were specific to the gagL peptide, we isolated CD8+ T cells from spleen in mice after inoculation with RMA-mock or RMA-CD155, stimulated the CD8+ T cells in vitro with the gagL peptide, and determined the frequency of the peptide-specific IFN-γ–producing CD8+ T cells by ELISPOT assay. Although we could not detect CD8+ T cells that responded to the gagL peptide in mice immunized with RMA-mock or RMA-CD155 at the priming phase around days 3 to 5 after the tumor challenge by this assay (data not shown), mice inoculated with RMA-CD155 generated CD8+ T cells responsive to the gagL peptide significantly more than those inoculated with RMA-mock at days 14 to 18 when RMA-mock tumor become visible (Figure 4B). These results suggested that the gagL peptide–specific CD8+ T cells might play an important role for efficient tumor rejection of RMA-CD155 and memory CTL induction.
NK cells are also involved in tumor rejection
DNAM-1 is expressed on NK cells and involved in NK-cell–mediated cytotoxicity and cytokine secretion in vitro.4,9 We therefore examined whether NK cells are also involved in the rejection of RMA-CD155 and RMA-CD112 that express MHC class I (H-2b). As demonstrated in Figure 2E-F, depletion of NK cells with anti-NK1.1 mAb partially canceled the tumor rejection of RMA-CD112 and RMA-CD155, indicating that NK cells were, in part, involved in the tumor rejection. These results suggested that NK cells might be directly activated by DNAM-1 ligand–expressing RMA.
Memory CD8+ T-cell induction specific to RMA by the DNAM-1 ligands. (A) Groups of 10 C57BL /6 mice were primarily inoculated with mock-transduced or Cd155-transduced RMA tumor cells. Mice that had rejected 1 × 105Cd155-transduced RMA tumor cells were divided into 2 groups and pretreated (intraperitoneally) with either anti-CD8 mAb or control immunoglobulin, and rechallenged subcutaneously in the back with 1 × 105 parental RMA tumor cells at 70 days after the primary tumor inoculation. Tumor size in each mouse was measured twice a week and the mean values were plotted. (B) CD8+ T cells were separated from spleen in mice 14 to 18 days after inoculation with RMA-mock or RMA-CD155 and stimulated in vitro with gagL peptide in the presence of splenocytes that were used as APCs. The frequency of the peptide-specific CD8+ T cells, defined as IFN-γ–producing cells, was determined by ELISPOT assay, as described in “Materials and methods.” Error bars indicate 1 SD.
Memory CD8+ T-cell induction specific to RMA by the DNAM-1 ligands. (A) Groups of 10 C57BL /6 mice were primarily inoculated with mock-transduced or Cd155-transduced RMA tumor cells. Mice that had rejected 1 × 105Cd155-transduced RMA tumor cells were divided into 2 groups and pretreated (intraperitoneally) with either anti-CD8 mAb or control immunoglobulin, and rechallenged subcutaneously in the back with 1 × 105 parental RMA tumor cells at 70 days after the primary tumor inoculation. Tumor size in each mouse was measured twice a week and the mean values were plotted. (B) CD8+ T cells were separated from spleen in mice 14 to 18 days after inoculation with RMA-mock or RMA-CD155 and stimulated in vitro with gagL peptide in the presence of splenocytes that were used as APCs. The frequency of the peptide-specific CD8+ T cells, defined as IFN-γ–producing cells, was determined by ELISPOT assay, as described in “Materials and methods.” Error bars indicate 1 SD.
DNAM-1–mediated signal induces maturation of CD8α+ DCs
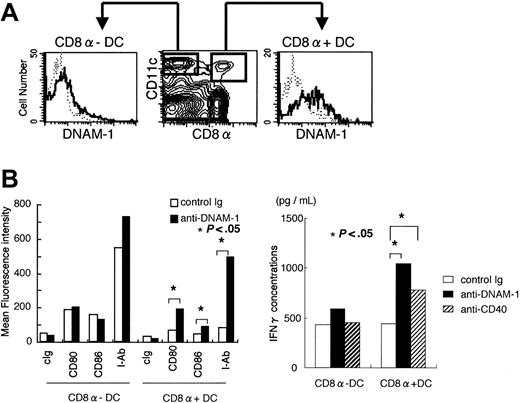
In a detailed study of mouse DNAM-1 expression on immune cells using our newly generated anti–DNAM-1 mAb, we found that a subset of CD11c+ cells also expressed DNAM-1. Further examination demonstrated that DNAM-1 was preferentially expressed on CD8α+, rather than CD8α-, CD11c+ DCs in spleen (Figure 5A), suggesting that DNAM-1 ligands may stimulate also this subset of DCs. To examine this possibility, CD11c+ cells were purified from spleen by MACS, stimulated with plate-coated anti–DNAM-1 mAb, and then stained with anti-CD11c, anti-CD8α, and either anti-CD80, anti-CD86, or anti–MHC class II mAbs. As demonstrated in Figure 5B, cross-linking DNAM-1 with anti–DNAM-1 mAb induced up-regulation of CD80, CD86, and MHC class II expressions in CD8α+, but not CD8α-, CD11c+ cells, indicating that DNAM-1–mediated signal induces maturation of CD8α+ DCs.
CD4+ T cells are also involved in tumor rejection
CD8α+ DCs are a functionally specialized subpopulation (for a review, see Shortman and Liu22 ), which produces much higher level of IL-12 p70 than CD8α- DC population and is involved in Th1 differentiation from naive helper T cells.23,24 Therefore, we assumed that antigen presentation by MHC class II molecules of these stimulated CD8α+ DCs drives differentiation of antigen-specific helper T cells into Th1 cells. To test this hypothesis, CD4+ T cells purified from OVA-specific TCR transgenic mice were cocultured with OVA-peptide-prepulsed CD8α+ or CD8α- DCs that had been stimulated with anti–DNAM-1 mAb. We observed that IFN-γ secretion was significantly increased in cultures of CD4+ T cells with CD8α+, but not CD8α-, DCs stimulated with anti–DNAM-1 mAb (Figure 5C), indicating that DNAM-1 on CD8α+ DCs is involved in Th1 polarization. These results suggested that not only CD8+ T cells but also CD4+ T cells might play an important role in tumor rejection of RMA-CD112 or RMA-CD155. In fact, depletion of CD4+ T cells with anti-CD4 mAb partially canceled the rejection of RMA-CD112, leading to shorter survival of mice without CD4+ T cells than those with CD4+ T cells (Figure 2A,G). Similarly, the survival of mice deficient in MHC class II molecules was significantly shorter than that of wild-type mice after inoculation with RMA-CD112 (Figure 2A,F).
DNAM-1–mediated signal induces maturation and activation of CD8α DCs. (A) Spleen cells from C57BL /6 mice were stained with PE-conjugated anti-CD11c, FITC-conjugated anti-CD8, and biotin-conjugated anti–DNAM-1, followed by APC-conjugated streptavidin. DNAM-1 expressions on CD8α-CD11c+ and CD8α+CD11c+ cells were analyzed by flow cytometry. (B) CD11c+ DCs were separated from splenocytes of C57BL /6 mice by positive selection using MACS. Purified DCs were stimulated for 4 days with plate-coated anti–DNAM-1 mAb or control IgG, and stained with anti-CD11c, anti-CD8, and either control immunoglobulin, anti-CD80, anti-CD86, or anti–MHC class II (I-Ab). Mean fluorescence intensity of CD80, CD86, and I-Ab expressions on CD8+CD11c+ or CD8-CD11c+ DCs were determined by flow cytometry. Data are representative of several independent experiments. (C) CD11c+CD8α+ or CD11c+CD8α- DCs were stimulated with plate-coated mAbs indicated for 4 days, pulsed with the OVA peptide, and then cocultured with CD4+ T cells from splenocytes of OT-II mice for 7 days. CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for 48 hours and IFN-γ concentrations in the culture supernatant were determined by ELISA. Data are representative of several independent experiments.
DNAM-1–mediated signal induces maturation and activation of CD8α DCs. (A) Spleen cells from C57BL /6 mice were stained with PE-conjugated anti-CD11c, FITC-conjugated anti-CD8, and biotin-conjugated anti–DNAM-1, followed by APC-conjugated streptavidin. DNAM-1 expressions on CD8α-CD11c+ and CD8α+CD11c+ cells were analyzed by flow cytometry. (B) CD11c+ DCs were separated from splenocytes of C57BL /6 mice by positive selection using MACS. Purified DCs were stimulated for 4 days with plate-coated anti–DNAM-1 mAb or control IgG, and stained with anti-CD11c, anti-CD8, and either control immunoglobulin, anti-CD80, anti-CD86, or anti–MHC class II (I-Ab). Mean fluorescence intensity of CD80, CD86, and I-Ab expressions on CD8+CD11c+ or CD8-CD11c+ DCs were determined by flow cytometry. Data are representative of several independent experiments. (C) CD11c+CD8α+ or CD11c+CD8α- DCs were stimulated with plate-coated mAbs indicated for 4 days, pulsed with the OVA peptide, and then cocultured with CD4+ T cells from splenocytes of OT-II mice for 7 days. CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for 48 hours and IFN-γ concentrations in the culture supernatant were determined by ELISA. Data are representative of several independent experiments.
Discussion
The DNAM-1 ligands are constitutively expressed at low levels on epithelial and endothelial cells. In the present study, we have shown that the DNAM-1 ligand CD112 is overexpressed on human gastric and colon cancers. There is also evidence that the expression of DNAM-1 ligands is up-regulated on human tumors, including colorectal carcinomas, neuroblastomas, and myeloid leukemias.13-15 These observations suggest that the expression of DNAM-1 ligands may be inducible by “stress” such as transformation, leading us to examine whether these “stress-inducible self-ligands” may potentially stimulate innate or adaptive tumor immunity. Here, we have demonstrated that ectopic expression of DNAM-1 ligands permitted rejection of RMA tumors inoculated in mice. CD8+ T cells played an essential role for the rejection of the DNAM-1 ligand-expressing RMA tumors.
Several lines of evidence have demonstrated that innate immune surveillance is important for efficient priming of adaptive antitumor immunity.25 Several cell-surface receptors, including CD27 and NKG2D, expressed on NK cells have been reported to induce the primary and secondary tumor rejections by NK cells and CD8+ memory T cells, respectively,26,27 suggesting a link between innate and adaptive immunities. Although both human and mouse DNAM-1 are expressed on NK cells and involved in NK cell-mediated cytotoxicity and IFN-γ secretion in vitro,4,9 it remained undetermined whether DNAM-1 ligands on tumors that bear MHC class I molecules could stimulate NK cells in vivo despite inhibitory signals in NK cells mediated by MHC class I receptors. In the present study, we have demonstrated that, although RMA tumor expresses MHC class I (H-2b), NK cells seemed to be partially responsible for rejection of RMA expressing the DNAM-1 ligands. It was likely that NK cells were directly activated by DNAM-1 ligand-expressing RMA tumors and produced cytokines, such as IFN-γ or IFN-α (or both), an efficient priming of adaptive immunity.
One of the most notable findings in the present study was that CD8α +, but not CD8α-, DCs substantially expressed DNAM-1 and cross-linking DNAM-1 with anti–DNAM-1 mAb induced maturation of CD8α+ DCs in vitro. Importantly, CD8α+ DCs are a functionally specialized subpopulation (for a review, see Shortman and Liu22 ). They produce much higher levels of IL-12 p70 than CD8α- DC populations,23,24 resulting in differentiation of Th1 cells that facilitate cell-mediated immunity. In fact, our study showed that antigen presentation by CD8α+ DCs stimulated with anti–DNAM-1 drove Th1 cells. It was of note, however, that depletion of CD4+ T cells or deficiency of MHC class II molecules did not completely cancel the tumor rejection of RMA-CD112, indicating that CD4+ T cells are not necessarily required for the tumor rejection. Recent evidence has demonstrated that CD8α+ DCs selectively endocytose dying cells in vivo,28 and can cross-present exogenous antigens on MHC class I molecules,29,30 resulting in priming CTLs.31 Therefore, it was also likely that the CD8α+ DCs carried out the uptake of tumor debris destroyed by NK cells and directly stimulated CD8+ T cells resulted in cross-presentation of a tumor-associated antigen.
We have demonstrated that CD8+ T cells specific to the Rauscher murine leukemia virus–derived gagL peptide antigen were generated in mice immunized with DNAM-1 ligand–transduced RMA significantly more than those immunized with mock-transduced RMA. Moreover, cross-linking DNAM-1 induced maturation of CD8α+ DCs. Antigen presentation by these stimulated CDs drove Th1 cells, and CD4+ helper T cells were involved in tumor rejection. Taken together, these results suggested that DNAM-1 ligand expression on RMA may play an important role for priming of the gagL peptide–specific CD8+ CTLs. However, we still cannot reach this conclusion by these data at present because we could not directly detect CD8+ T cells that responded to the peptide in the early days of priming around days 3 to 5 after tumor challenge by the ELISPOT assay, and therefore could not exclude the possibility that proliferation of the gagL peptide–specific CD8+ T cells was affected in mice inoculated with RMA-mock by an immunosuppressive effect of growing RMA tumor. Nonetheless, the present studies suggested that interaction of DNAM-1 and DNAM-1 ligands played an important role in tumor rejection mediated by the RMA tumor–associated antigen-specific CD8+ T cells. Although we observed that most tumor cell lines, except RMA, expressed the DNAM-1 ligands, whether these tumors are rejected in vivo may depend on expression of tumor antigen specifically recognized by CD8+ T cells.
In summary, the present study proposes a model that DNAM-1 ligands, whose expression is inducible on tumors, potentially trigger innate immunity mediated by NK cells and CD8α+ DCs, both of which further stimulate each other by cytokines produced by themselves such as IFN-α, IFN-γ, IL-12, IL-15, and IL-18.32 CD8α+ DCs then capture tumor debris destroyed by NK cells and present a tumor-associated antigen on both MHC class I and class II molecules, resulting in priming antigen–specific CD8+ and CD4+ T cells. Further studies are required to clarify molecular and cellular mechanisms of DNAM-1–mediated tumor immunity.
Prepublished online as Blood First Edition Paper, October 25, 2005; DOI 10.1182/blood-2005-04-1684.
Supported in part by the grants provided by the Ministry of Education, Science and Culture of Japan (S.T.-H., K.S., and A.S.), Special Coordination Funds of the Science and Technology Agency of the Japanese Government (A.S.), the Uehara Memorial Foundation (A.S.), and the Yasuda Memorial Foundation (A.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank L. Lanier and Mitsuyasu Kato for helpful discussions. We also thank Ayumi Hara, Yuko Onoda, and Hua Zhang for technical assistance and Yurika Soeda for secretarial assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal