Abstract
Studies in unrelated donor (UD) hematopoietic stem cell transplantations (HSCT) show an effect of the matching status of HLA-DPB1 on complications. We analyzed 423 UD-HSCT pairs. Most protocols included T-cell depletion (TCD). All pairs had high-resolution tissue typing performed for 6 HLA loci. Two hundred eighty-two pairs were matched at 10 of 10 alleles (29% were DPB1 matched). In 141 HLA-mismatched pairs, 28% were matched for DPB1. In the 10 of 10 matched pairs (n = 282), the 3-year probability of relapse was 61%. This was significantly higher in DPB1-matched pairs (74%) as compared with DPB1-mismatched pairs (56%) (log rank, P = .001). This finding persisted in multivariate analysis. In the group overall (n = 423), relapse was also significantly increased if DPB1 was matched (log rank; P < .001). These results were similar in chronic myeloid leukemia (CML; P < .001) and acute lymphoblastic leukemia (ALL; P = .013). In ALL, DPB1-matched pairs had a significantly worse overall survival (log rank; P = .025). Thus, in recipients of TCD UD-HSCT, a match for DPB1 is associated with a significantly increased risk of disease relapse, irrespective of the matching status for the other HLA molecules. It is possible that this effect is especially apparent following TCD transplantations and invites speculation about the function of DPB1 within the immune system.
Introduction
The study and understanding of HLA molecules have greatly increased the safety and applicability of hematopoietic stem cell transplantations (HSCTs) over the years, and there is good evidence to support the fact that high-resolution tissue typing and matching for the HLA-A, -B, -C, and -DRB1 loci results in an improved outcome.1-3 Nevertheless there remain unanswered questions. The observation that complications (especially graft-versus-host disease [GvHD]) continue to occur even in the setting of a matched sibling transplantation or a 10 of 10 allele-matched unrelated donor transplantation, led to the search for additional molecules able to provoke immunogenicity. HLA-DPB1 is one of these.
HLA-DPB1 is in the MHC class II region lying centromeric to the other class II loci. It is highly polymorphic (120 alleles have been described to date4 ). Increased recombination events are found in the region between the HLA-DP loci and the other class II loci, explaining the relative lack of linkage disequilibrium (LD) between HLA-DP and the rest of the extended MHC haplotype.5 For this reason it is difficult to find a donor matched for DPB1 in addition to the other classic HLA molecules. In sibling donors the rate of incompatibility has been estimated to be as high as 10.9%, and complications as a result of mismatching for this molecule have been shown.6,7 This is even more evident in unrelated donors, where studies report a mismatch rate of up to 89%.8-10
Broadly speaking, HLA-DPB1 resembles the other HLA class II molecules in terms of structure and function. As with other class II molecules, it presents peptide in a MHC-restricted fashion to CD4+ T cells. DPB1 has been shown to present a range of peptides, including those from infectious agents,11,12 foreign bodies,13 and tumors,14 as well as allogeneic15 or autologous16,17 peptides, including those derived from other HLA molecules.12,18 DPB1-specific responses have been reported in a number of settings, including in rejection of transplanted tissue,19,20 susceptibility to chronic beryllium disease,13 and acute GvHD.15 DPB1 serves as a marker for susceptibility for certain diseases.9,12,13 DPB1 is thought to be present on the same cell types as other class II molecules, but with a reduced cell-surface expression, and has been shown to be expressed on leukemic cells in levels similar to (acute myeloid leukemia [AML], chronic lymphocytic leukemia [CLL]) or greater than (B-cell acute lymphoblastic leukemia [B-ALL]) DQB1, although in much lower numbers than in class I and DRB1.21
Studies of an effect because of this molecule lagged behind those of other HLA loci. However, there is now a bulk of evidence supporting a role for HLA-DPB1-matching status on the outcome in unrelated donor (UD)-HSCT.8,9,22 We have previously published results23 showing a significant increase in disease relapse and decrease in acute GvHD (aGvHD) in patients matched for DPB1 with their unrelated donor. The aim of the current study was to assess whether these results were borne out in a much larger study cohort, to look at similar outcomes in specific disease subgroups, and, in addition, to ascertain whether DPB1 could be considered as an individual risk factor predictive of transplantation complications. Our results have enabled us to provide advice to transplantation physicians regarding the choice of the best unrelated donor, with respect to DPB1-matching status.
Patients, materials, and methods
Study population
The study group comprised 423 transplant recipients who had received an HSC transplant from an UD through the Anthony Nolan Trust (ANT). Both recipient and donor were required to have 4-digit allele typing results at 6 HLA loci (HLA-A, -B, -C, -DRB1, -DQB1, -DPB1). Because the study was registry based, the blood samples and clinical data were provided by the individual transplantation centers (TCs). Clinical data were collected on proformas which were sent to data managers at the individual TC.
The transplantations took place between September 1996 and January 2003. Abroad range of diseases were indications for transplantation, and the patient and donor pretransplantation factors can be seen in Tables 1 and 2. The majority of the transplantation protocols included T-cell depletion (86%), in most cases using alemtuzumab in vivo (92%); however, in the remaining 8% of recipients antithymocyte globulin (ATG) was used. A roughly equal number of patients received transplants for each of the diagnoses: AML, ALL, chronic myeloid leukemia (CML), and other malignant disease (Table 3). A small number received transplants for nonmalignant disease (Table 3). Early disease stage was defined as first complete remission (CR1) in acute leukemia and first chronic phase (1CP) in CML. Untreated patients (eg, with MDS) were included in this definition. All patients with disease beyond this were grouped as late-stage disease. The median age of the patients was 30 years with a broad range, including infants. The donors had a median age of 35 years, although once again with a relatively broad range. Two thirds of the female donors were multiparous.
Patient- and donor-specific demographics
Characteristic . | Value . |
|---|---|
| Recipient age, y, median (range) | 30 (<1-65) |
| Recipient sex | |
| Male, no. (%) | 258 (61) |
| Donor sex | |
| Male, no. (%) | 315 (74) |
| Recipient/donor sex, no. (%) | |
| Sex matched | 260 (61) |
| Male/female | 53 (13) |
| Female/male | 110 (26) |
| Parity of donor, no. (%) | |
| Multiparous | 67 (62) |
| Nulliparous | 36 (33) |
| Missing | 5 (5) |
| Donor age, y, median (range) | 35 (21-56) |
| Recipient CMV serostatus, no. (%) | |
| Negative | 287 (68) |
| Positive | 121 (29) |
| Missing | 15 (3) |
| Donor CMV serostatus, no. (%) | |
| Negative | 325 (77) |
| Positive | 98 (23) |
| Recipient/donor CMV status, no. (%) | |
| Negative/negative | 241 (57) |
| Negative/positive | 46 (11) |
| Positive/negative | 72 (17) |
| Positive/positive | 49 (12) |
| Missing | 15 (4) |
Characteristic . | Value . |
|---|---|
| Recipient age, y, median (range) | 30 (<1-65) |
| Recipient sex | |
| Male, no. (%) | 258 (61) |
| Donor sex | |
| Male, no. (%) | 315 (74) |
| Recipient/donor sex, no. (%) | |
| Sex matched | 260 (61) |
| Male/female | 53 (13) |
| Female/male | 110 (26) |
| Parity of donor, no. (%) | |
| Multiparous | 67 (62) |
| Nulliparous | 36 (33) |
| Missing | 5 (5) |
| Donor age, y, median (range) | 35 (21-56) |
| Recipient CMV serostatus, no. (%) | |
| Negative | 287 (68) |
| Positive | 121 (29) |
| Missing | 15 (3) |
| Donor CMV serostatus, no. (%) | |
| Negative | 325 (77) |
| Positive | 98 (23) |
| Recipient/donor CMV status, no. (%) | |
| Negative/negative | 241 (57) |
| Negative/positive | 46 (11) |
| Positive/negative | 72 (17) |
| Positive/positive | 49 (12) |
| Missing | 15 (4) |
Disease- and transplantation-specific demographics
Characteristic . | No. (%) . |
|---|---|
| Disease | |
| Chronic myeloid leukemia | 93 (22) |
| Acute myeloid leukemia | 102 (24) |
| Acute lymphoblastic leukemia | 103 (24) |
| Other malignant | 101 (24) |
| Nonmalignant | 24 (6) |
| Disease stage (excluding nonmalignant) | |
| Early | 217 (54) |
| Late | 176 (44) |
| Missing | 6 (2) |
| Conditioning regimen | |
| Myeloablative conditioning | 292 (69) |
| Reduced intensity conditioning | 101 (24) |
| No conditioning | 2 (< 1) |
| Missing | 28 (7) |
| Stem cell source | |
| Bone marrow | 348 (82) |
| PBSC | 75 (18) |
| T-cell deletion | |
| Yes | 363 (86) |
| No | 29 (7) |
| Missing | 31 (7) |
| Posttransplantation immunosuppression | |
| Cyclosporin A and methotrexate | 200 (47) |
| Cyclosporin A alone | 138 (33) |
| Other | 12 (3) |
| None | 31 (7) |
| Missing | 42 (10) |
Characteristic . | No. (%) . |
|---|---|
| Disease | |
| Chronic myeloid leukemia | 93 (22) |
| Acute myeloid leukemia | 102 (24) |
| Acute lymphoblastic leukemia | 103 (24) |
| Other malignant | 101 (24) |
| Nonmalignant | 24 (6) |
| Disease stage (excluding nonmalignant) | |
| Early | 217 (54) |
| Late | 176 (44) |
| Missing | 6 (2) |
| Conditioning regimen | |
| Myeloablative conditioning | 292 (69) |
| Reduced intensity conditioning | 101 (24) |
| No conditioning | 2 (< 1) |
| Missing | 28 (7) |
| Stem cell source | |
| Bone marrow | 348 (82) |
| PBSC | 75 (18) |
| T-cell deletion | |
| Yes | 363 (86) |
| No | 29 (7) |
| Missing | 31 (7) |
| Posttransplantation immunosuppression | |
| Cyclosporin A and methotrexate | 200 (47) |
| Cyclosporin A alone | 138 (33) |
| Other | 12 (3) |
| None | 31 (7) |
| Missing | 42 (10) |
PBSC indicates peripheral blood stem cells.
Specific diagnoses for other malignant disease and for nonmalignant disease
Specific diagnosis . | No. cases . |
|---|---|
| Other malignant disease | |
| CLL | 6 |
| Hodgkin disease | 9 |
| CMML | 2 |
| JMML | 2 |
| MDS | 36 |
| Myeloma | 13 |
| PCL | 1 |
| WM | 1 |
| Myelofibrosis | 2 |
| NHL | 29 |
| Nonmalignant disease | |
| Aplastic anemia | 4 |
| Hurler syndrome | 7 |
| SCID | 5 |
| Schwachmann-Diamond | 2 |
| Wiskott Aldrich | 1 |
| Osteopetrosis | 1 |
| CD40L deficiency | 1 |
| Omenn syndrome | 1 |
| ALD | 1 |
| Cartilage hair hypoplasia | 1 |
Specific diagnosis . | No. cases . |
|---|---|
| Other malignant disease | |
| CLL | 6 |
| Hodgkin disease | 9 |
| CMML | 2 |
| JMML | 2 |
| MDS | 36 |
| Myeloma | 13 |
| PCL | 1 |
| WM | 1 |
| Myelofibrosis | 2 |
| NHL | 29 |
| Nonmalignant disease | |
| Aplastic anemia | 4 |
| Hurler syndrome | 7 |
| SCID | 5 |
| Schwachmann-Diamond | 2 |
| Wiskott Aldrich | 1 |
| Osteopetrosis | 1 |
| CD40L deficiency | 1 |
| Omenn syndrome | 1 |
| ALD | 1 |
| Cartilage hair hypoplasia | 1 |
CMML indicates chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; MDS, myelodysplastic syndrome; PCL, plasma cell leukemia; WM, Waldenström macroglobulinemia; NHL, non-Hodgkin lymphoma; SCID, severe combined immunodeficiency; ALD, adrenoleukodystrophy.
Transplantation pairs could score 0, 1, or 2 for DPB1 allele incompatibilities. In those with CML 38% of pairs were DPB1 compatible, compared with 30% in AML, 27% in ALL, and 18% in the malignant other subgroup (38% in the nonmalignant group) (chi-squared test, P = .031). Because the majority of these transplantations did not have DPB1 typing performed before transplantation (ie, were not selected on this criteria), this is an unexpected finding.
Ethical permission
The study received appropriate ethical permission and all patients and donors signed informed consent. Approval was obtained from the Anthony Nolan Trust's institutional review board for these studies.
Statistical analysis
Statistical analyses of clinical outcome were done using SPSS (version 10.0) software (SPSS, Chicago, IL). The association between variables was assessed using the chi-squared test. Time-dependent variables (overall survival, chronic graft-versus-host disease, and disease relapse rates) were analyzed using Kaplan-Meier methods24 and were compared using the log-rank statistic. Probabilities are quoted at 3 years unless otherwise stated. For time-dependent variables, multivariate analysis was performed by using Cox regression analysis. Acute graft-versus-host disease was analyzed as a binary variable by using logistic regression models.
Results
HLA matching status
Of the 423 pairs, 282 pairs (67%) were matched for 5 classic HLA loci (HLA-A, -B, -C, -DRB1, -DQB1; ie, 10 of 10 matched) and 141 were mismatched. Of the 141 pairs, 111 were class I mismatched only, 19 were class II (DRB1, DQB1) mismatched only, and 11 were mismatched at class I and class II.
HLA-DPB1 matching status
Of the 423 pairs, 71 (17%) shared complete DPB1 identity, 214 (51%) had a mismatch at 1 allele, and 138 (33%) were mismatched at 2 alleles. When DPB1 compatibility in a GvH direction was considered; 121 (29%) pairs were completely DPB1 compatible, 174 (41%) had an incompatibility at 1 allele, and 128 (30%) had an incompatibility at 2 alleles. The reason for the discrepancy between DPB1 identity and compatibility is the high percentage of DPB1 homozygous individuals seen.
Of the 282 pairs, when DPB1 compatibility in a graft-versus-host direction was considered, 82 (29%) pairs were completely DPB1 compatible, 118 (42%) had an incompatibility at 1 allele, and 82 (29%) had an incompatibility at 2 alleles.
Results in 282 pairs matched for HLA-A, -B, -C, -DRB1, -DQB1
aGvHD. The overall incidence of aGvHD in this cohort was 48% (120 of 251). This was reported to be grade I in 53 cases (44%), grade II in 57 cases (48%), grade III in 5 cases (4%), and grade IV in 5 cases (4%). Thus, overall the incidence of clinically significant aGvHD (grade III-IV) was 4% (10 of 251). There was a trend toward increased incidence of aGvHD in those pairs who were incompatible for DPB1 (93 of 180, 52%) compared with pairs with DPB1 compatibility (27 of 71, 38%) (chi-squared test, P = .051). This was not significantly different when comparing the compatible group with the 1 allele incompatible group (56 of 103, 54%) and the 2 allele incompatible group (37 of 77, 48%) (chi-squared test, P = .105). The severity of aGvHD was not significantly affected by DPB1 compatibility. When other factors known to affect aGvHD were included in a multivariate logistic regression model (recipient age, donor age, conditioning), HLA-DPB1 mismatching remained significantly associated with aGvHD (odds ratio [OR], 1.89; 95% confidence interval [CI],1.04-3.43; P = .035). There was no effect of DPB1 compatibility on the incidence of cGvHD or transplantation-related mortality (TRM).
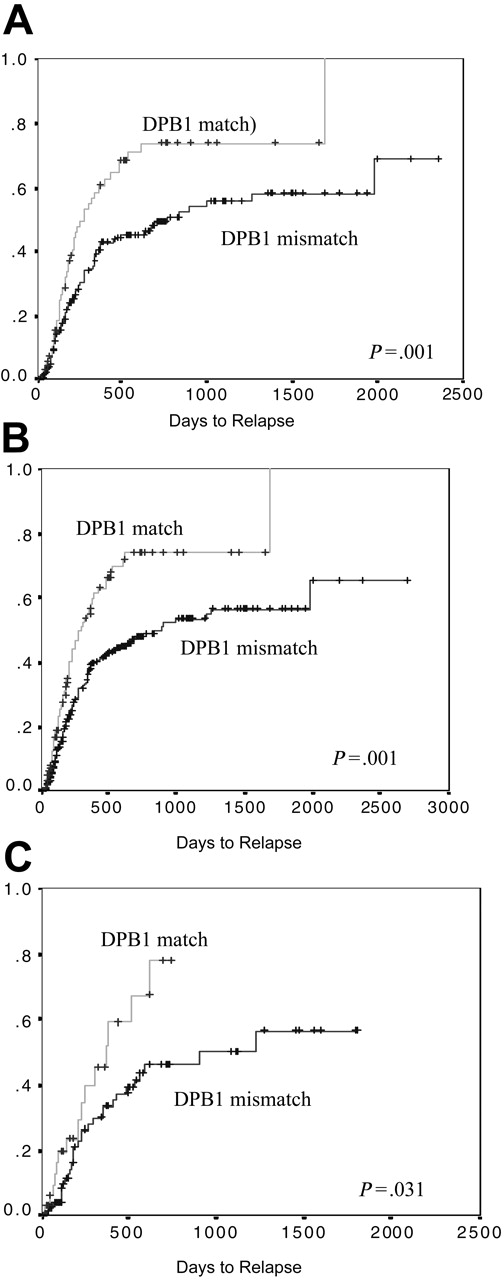
Relapse. The 3-year probability of relapse was 61% (median, 461 days). There was a highly significantly increase in disease relapse in those transplantation pairs with DPB1 compatibility (74%) as compared with those pairs with either 1 or 2 DPB1 incompatibilities (56%) (log rank, P = .001) (Figure 1A). There was no difference in this effect as the number of DPB1 incompatibilities increased (ie, 1 or 2 incompatibilities were equally protective, 56% versus 55%, respectively, log rank P = .959). There was a significant difference in the probability of relapse between the disease groups (log rank, P = .014). At 3 years this was 82% in CML, 65% in AML, 51% in other malignant diseases, and 43% in ALL. Other factors that resulted in a higher rate of relapse were older patient age (P = .002) and patients who received transplants in relapse (P < .001), and the use of a male donor (P = .085) showed a trend toward significance, irrespective of the sex of the patient (Table 4). Factors that were not significantly associated with disease relapse included type of conditioning, GvHD prophylaxis, stem cell source, donor age, and class I or II (DRB1, DQB1) matching status. In a Cox proportional hazard regression model, including the significant factors, mismatching for DPB1 remained significantly protective against disease relapse (OR, 0.56; 95% CI, 0.38-0.83; P = .004) (Table 4).
Factors associated with disease relapse (univariate and multivariate analysis)
. | Univariate . | Multivariate . | . | . | ||
|---|---|---|---|---|---|---|
| Variable . | P . | Odds ratio . | 95% CI . | P . | ||
| Patient older than 30 y | .002 | 1.62 | 1.05-2.50 | .027 | ||
| Transplantation in relapse | < .001 | 4.52 | 2.38-8.59 | < .001 | ||
| Female donor | .085 | 0.76 | 0.44-1.31 | NS | ||
| Other malignant (constant) | .014 | – | – | – | ||
| CML | – | 2.19 | 1.26-3.80 | .005 | ||
| AML | – | 1.88 | 1.05-3.37 | .033 | ||
| ALL | – | 1.56 | 0.80-3.03 | NS | ||
| DPB1 mismatch | .001 | .563 | 0.38-0.83 | .004 | ||
. | Univariate . | Multivariate . | . | . | ||
|---|---|---|---|---|---|---|
| Variable . | P . | Odds ratio . | 95% CI . | P . | ||
| Patient older than 30 y | .002 | 1.62 | 1.05-2.50 | .027 | ||
| Transplantation in relapse | < .001 | 4.52 | 2.38-8.59 | < .001 | ||
| Female donor | .085 | 0.76 | 0.44-1.31 | NS | ||
| Other malignant (constant) | .014 | – | – | – | ||
| CML | – | 2.19 | 1.26-3.80 | .005 | ||
| AML | – | 1.88 | 1.05-3.37 | .033 | ||
| ALL | – | 1.56 | 0.80-3.03 | NS | ||
| DPB1 mismatch | .001 | .563 | 0.38-0.83 | .004 | ||
The presence of aGvHD was significantly associated with a decreased relapse rate (the probability of relapse with aGvHD was 51% compared with 73% without aGvHD; log rank, P = .041). This stratified according to grade: those with grade I to II had a relapse rate of 52% compared with 38% in those with grade III to IV. The presence of chronic GvHD (cGvHD) was also significantly associated with a decreased relapse rate (53% with cGvHD relapsed compared with 65% with no cGvHD; log rank, P = .025). Those with extensive cGvHD were less likely to experience disease relapse (41%) than were those with limited cGvHD (54%). There was no difference in those with de novo cGvHD or with disease occurring after aGvHD.
The effect of DPB1 matching on disease relapse. Matched pairs are significantly more at risk of relapse than are mismatched pairs. This can be seen (A) in those pairs matched for HLA-A, -B, -C, -DRB1, and -DQB1; (B) in the group overall, irrespective of matching status for HLA-A, -B, -C, -DRB1, and -DQB1; and (C) in only those pairs mismatched for one or more of HLA-A, -B, -C, -DRB1, and -DQB1.
The effect of DPB1 matching on disease relapse. Matched pairs are significantly more at risk of relapse than are mismatched pairs. This can be seen (A) in those pairs matched for HLA-A, -B, -C, -DRB1, and -DQB1; (B) in the group overall, irrespective of matching status for HLA-A, -B, -C, -DRB1, and -DQB1; and (C) in only those pairs mismatched for one or more of HLA-A, -B, -C, -DRB1, and -DQB1.
Overall survival. Despite the effect of DPB1 on aGvHD and relapse, there was no significant difference in overall survival (OS) between those pairs who were compatible and those who were incompatible for HLA-DPB1 (log rank, P = .803). There was a survival advantage seen in those with grade I to II aGvHD compared with no aGvHD or grade III to IV disease (log rank, P = .046). A Kaplan-Meier analysis to assess the effect of cGvHD on OS revealed a significant survival advantage in those with limited cGvHD (83%) disease compared with those with extensive disease (38%) or no cGvHD (49%) (log rank, P < .001).
Results in group overall including 423 pairs
As mentioned, there was no association between compatibility for DPB1 and matching for the other HLA types. Given this finding, we considered that DPB1 could be analyzed as an individual risk factor for complications following transplantation, irrespective of the matching status at the other HLA loci.
Acute GvHD was significantly more likely to occur in DPB1 mismatched pairs, in the group overall (423 pairs) (150 of 267, 56% compared with 44 of 105, 42%; chi-squared test, P = .013). The incidence of severe aGvHD was not affected. There was a trend toward an increase in cGvHD in the DPB1-mismatched pairs (P = .061), but no effect on TRM.
As in the pairs who were 10 of 10 HLA matched, we found that there was a highly significant increased risk of relapse in the group overall (423 pairs) if DPB1 was matched (log rank, P < .001) (Figure 1B). This was similar if either 1 or 2 alleles were mismatched. This persisted, although was less significant, when only the 141 pairs with one or more mismatches for 10 of 10 alleles were considered (P = .031) (Figure 1C). Acute GvHD was protective against disease relapse, which stratified dependent on the grade of aGvHD (in those with no aGvHD there was a probability of relapse of 69% compared with 51% with grade I-II and 29% with grade III-IV, P = .015). Interestingly, the presence of cGvHD was not associated with relapse rate (log rank, P = .156).
There was no significant difference in OS in the group overall whether they were matched or mismatched for DPB1 (log rank, P = .385). aGvHD did have a significant effect on survival. The best OS was seen in those with grade I disease (58%), next grade 0 (46%) and grade II (46%), with the worst overall survival in those with grade III (27%) or grade IV (17%) disease (P = .026). Likewise, those with cGvHD had an OS of 65% compared with those without, 48% (P < .001), with limited cGvHD offering a highly significant survival benefit compared with either no cGvHD or extensive disease (78% versus 48% versus 41%, P < .001).
Individual disease groups
In view of the obvious difference between the different disease groups, in particular in the rate of disease relapse, and strategies to treat this complication, the analysis was performed in each of the main disease groups (CML, ALL, AML) individually
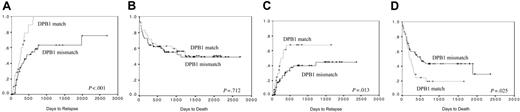
CML. Ninety-three patients with CML were included in this group. The incidence of aGvHD was 59%, in 5 cases greater than grade II (6% overall). There was no significant effect of DPB1 matching on this complication (chi-squared test, P = .446). In contrast there was a significant effect because of DPB1 matching on disease relapse. The 1-year incidence of relapse in the DPB1-mismatched pairs was 42%, whereas in the matched pairs this was 69% (log rank, P < .001) (Figure 2A). In multivariate analysis, including factors associated with relapse in univariate analysis (patient age and DPB1-matching status), a mismatch for DPB1 was the only factor significantly protective against disease relapse (OR, 0.44; 95% CI, 0.23-0.83; P = .012). There was no significant effect of aGvHD on disease relapse. Of those who experienced relapse, 40 (83%) received donor lymphocyte infusion (DLI), of whom 30 (75%) remain alive. The 5-year probability of OS was 50%. There was no significant difference in OS dependent on matching status for DPB1 (log rank, P = .712) (Figure 2B).
ALL. One hundred three patients with ALL were included in this group. The incidence of aGvHD was 53%, in 3 cases this was greater than grade II (3% overall). There was a trend toward a significant effect of DPB1 matching on this complication, in matched pairs the incidence was 38%, whereas in mismatched pairs this was 59% (chi-squared test, P = .067). In addition, there was a significant effect due to DPB1 matching on disease relapse. The 2 year incidence of relapse in the DPB1-mismatched pairs was 40%, whereas in the matched pairs this was 78% (log rank, P = .013) (Figure 2C). In multivariate analysis, including factors associated with relapse in univariate analysis (disease stage, patient age, and DPB1-matching status), a mismatch for DPB1 remained significantly protective against disease relapse (OR, 0.31; 95% CI, 0.15-0.64, P = .002) (Table 5). The presence of aGvHD was protective against relapse (P = 0.045). Of those who experienced relapse, 7 (19%) received DLI, one of whom remains alive.
Factors associated with disease relapse and OS in ALL (univariate and multivariate analysis)
. | Relapse . | . | . | . | Overall survival . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Univariate . | Multivariate . | . | . | Univariate . | Multivariate . | . | . | ||||||
| Variable . | P . | Odds ratio . | 95% CI . | P . | P . | Odds ratio . | 95% CI . | P . | ||||||
| Patient older than 30 y | .086 | 1.2 | 0.53-2.96 | NS | .028 | 1.5 | 0.74-3.04 | NS | ||||||
| Transplantation in relapse | < .001 | 8.2 | 2.51-27.3 | .001 | .016 | 2.8 | 1.10-7.59 | .030 | ||||||
| Female patient | .047 | 0.49 | 0.21-1.12 | NS | NS | – | – | – | ||||||
| Patient CMV positive | NS | – | – | – | .068 | 1.2 | 0.60-2.57 | NS | ||||||
| Donor older than 30 y | NS | – | – | – | .072 | 1.5 | 0.75-3.25 | NS | ||||||
| DPB1 mismatch | .013 | 0.31 | 0.15-0.64 | .002 | .025 | 0.58 | 0.31-1.07 | .084 | ||||||
. | Relapse . | . | . | . | Overall survival . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Univariate . | Multivariate . | . | . | Univariate . | Multivariate . | . | . | ||||||
| Variable . | P . | Odds ratio . | 95% CI . | P . | P . | Odds ratio . | 95% CI . | P . | ||||||
| Patient older than 30 y | .086 | 1.2 | 0.53-2.96 | NS | .028 | 1.5 | 0.74-3.04 | NS | ||||||
| Transplantation in relapse | < .001 | 8.2 | 2.51-27.3 | .001 | .016 | 2.8 | 1.10-7.59 | .030 | ||||||
| Female patient | .047 | 0.49 | 0.21-1.12 | NS | NS | – | – | – | ||||||
| Patient CMV positive | NS | – | – | – | .068 | 1.2 | 0.60-2.57 | NS | ||||||
| Donor older than 30 y | NS | – | – | – | .072 | 1.5 | 0.75-3.25 | NS | ||||||
| DPB1 mismatch | .013 | 0.31 | 0.15-0.64 | .002 | .025 | 0.58 | 0.31-1.07 | .084 | ||||||
DPB1 matching is associated with disease relapse and overall survival. A significantly increased risk of relapse is seen in CML (A) and ALL (C). There is a survival benefit seen in ALL (D) but not CML (B).
DPB1 matching is associated with disease relapse and overall survival. A significantly increased risk of relapse is seen in CML (A) and ALL (C). There is a survival benefit seen in ALL (D) but not CML (B).
The 5-year probability of OS was 37%. This was significantly different depending on matching status for DPB1: the 2-year probability in the DPB1-matched group was 20% compared with 43% in the DPB1-mismatched group (log rank, P = .025) (Figure 2D). In multivariate analysis, including factors associated with OS in univariate analysis (disease stage, patient age, DPB1-matching status, patient CMV status, donor age), a mismatch for DPB1 showed a trend toward a protective effect on OS (OR, 0.58; 95% CI, 0.31-1.07; P = .084) (Table 5). The only other factor retaining significance in this model was transplantation performed in relapse.
AML. One hundred two patients with AML were included in this group. The incidence of aGvHD was 50%, in 5 cases greater than grade II (5% overall). There was no significant effect of DPB1 matching on this complication (chi-squared test, P = .252). There was a suggestion of a trend toward an effect resulting from DPB1 matching on disease relapse. The 2-year incidence of relapse in the DPB1-mismatched pairs was 49%, whereas in the matched pairs this was 66% (log rank, P = .106). There was no significant effect of aGvHD on disease relapse. Of those who experienced relapse, 21 (47%) received DLI, 9 (43%) of whom remain alive. The 5-year probability of OS was 36%. There was no significant difference in OS dependent on matching status for DPB1 (log rank, P = .502).
Discussion
We are able to show, in this study, that the presence of DPB1 allele incompatibility resulted in significant differences in the incidence of aGvHD and disease relapse. Most previous studies examining DPB1 effects, including our own, have been performed in pairs who are completely matched for the other 5 classic HLA loci, 10 of 10 (HLA-A, -B, -C, -DRB1, -DQB1), with an aim to exclude any additive or confounding effects that result from other HLA mismatches. However, it is also known that, although strong linkage disequilibrium exists between the other 5 HLA loci, this does not extend to DP. We were able to show that in these unrelated donor transplantations, the chance of matching for DPB1 was the same whether the other HLA loci were matched or not. Therefore, we considered it possible to analyze DPB1 matching as an individual risk factor for transplantation complications. Consequently, our findings held true not only to those pairs matched for 10 of 10 HLA molecules, but also to the group overall irrespective of matching status at the other loci. In addition, there was a significant increase in disease relapse in patients with CML and ALL, and a decrease in overall survival in ALL, in patients receiving DPB1-matched grafts.
A number of other studies have reported an effect of DPB1 matching on aGvHD. In a study of 205 patients with CML,9 those with 2 allele disparities had an increase in both the incidence overall and the severity of aGvHD (the incidence of severe aGvHD was 32%). These data are in agreement with another recent study.22 A different group has shown an increase in severe aGvHD in DPB1-incompatible pairs, but this was not dependent on the number of incompatibilities present.8
In the current study, although there was a difference in the incidence of aGvHD overall, there was no effect on the incidence of severe aGvHD and no threshold effect was seen; that is, the presence of an incompatibility scoring 2 did not result in a higher incidence of aGvHD than did an incompatibility scoring one. Although about half of the patients developed aGvHD, there was a very low incidence of clinically severe (grade III-IV) disease. This is likely to be explained by the use of T-cell depletion (TCD) (usually alemtuzumab in vivo), as the chief method of GvHD prophylaxis in this study. This represents a major difference between this study and most other studies reported in the literature.
A novel finding in this transplantation cohort is the highly significant effect of DPB1 compatibility on disease relapse. This effect is not dependent on the number of incompatibilities present (ie, in keeping with the lack of threshold effect seen in aGvHD). In addition, it remains highly significant in multivariate analysis, in fact, the only factor to have a greater effect than this is patients who have received a transplant in disease relapse. In this study the probability of relapse was high (61% at 3 years) related in part to the use of TCD, but also because of the large number of high-risk patients receiving transplants.
It is likely that a degree of genetic disparity between recipient and donor is necessary to contribute to a graft-versus-leukemia (GvL) effect. This is supported by the observation that relapse rates may be greater when using sibling than unrelated donors.25 In the current study, no major HLA allodisparity remains to contribute to a GvL response in those pairs who are matched for DPB1 in addition to the other 10 HLA alleles, thus accounting for the increased risk of relapse.
Both GvHD and GvL may be mediated by T cells. In the ALL cohort, the significant effect of DPB1 on both aGvHD and relapse, as well as the protective effect of aGvHD on relapse (a situation which is well recognised26,27 ), argue strongly in favor of a common T-cell mechanism mediating both outcomes. HLA-DPB1-specific T cells have been demonstrated to be involved in acute GvHD,15 as well as directed against leukemic blasts.21
The association between GvHD and relapse in the myeloid patients (AML, CML) was less marked. It has been reported that a GvL effect may operate in the absence of GvHD,28 in particular in a T-cell-depleted setting26 and also that T cells mediating GvL may be different from those mediating GvHD.29 Alternately, other mechanisms may be implicated.30-32
None of the other HLA loci (either individually or in any combination (eg, class II alone or class I alone) were found to have an effect on disease relapse (data not shown). A potential explanation in this cohort, for the lack of effect because of other class II loci on this complication, is the small number of mismatches for DRB1 and DQB1 which were present (consequent on the selection procedures in the time period for unrelated donors). However, we may also speculate that DPB1 may differ in function to some extent from the other HLA molecules. The uncharacteristic overrepresentation of a single DP molecule (encoded by DPB1*0401), and the fact that the polymorphism at this locus appears to be subject to directional selection (rather than balancing selection as at the other HLA loci), may suggest a different evolutional history for this molecule,33,34 as well as a different function within the immune system.35 Clearly, more experimental work is required to investigate further the role of DPB1 in diverse transplantation settings, especially with a view to uncovering a GvL effect and the mediators thereof.
Some studies have not shown an effect of DPB1 matching on OS,9 whereas others have.8,22,36 The methodologies used in those studies, and the patient groups studied, are not coherent, making comparison difficult. In addition, many studies have not considered the outcome in patients by disease type. In the current study, there was a lack of a significant effect of DPB1-matching status on OS in the group overall, and in the 10 of 10 matched cohorts. However, there was a significantly worse outcome in patients with ALL receiving DPB1-matched grafts.
We may speculate about the reasons for this. The increase in aGvHD with DPB1 mismatches, and consequently cGvHD, is associated with a survival advantage in its mild form compared with more severe forms of this complication. We were unable to show a correlation between the severity of GvHD and DPB1 matching, which supports the contention that both forms may be predicted for, but influenced by additional factors. In addition, there is an interplay between GvHD and disease relapse, which may be more prevalent in some diseases than others. In patients who relapse certain methods may be used to return them to a CR or PR. This is particularly the case in CML, in which DLI is an efficient treatment modality, and further studies are warranted to assess the effect of DPB1 matching on the response and complications following DLI. In diseases such as ALL, however, where salvage after relapse is usually unsuccessful,37 DPB1 matching is associated with a significantly worse outcome.
Thus, knowing the matching status for DPB1 prior to transplantation is not only important in anticipating complications for the patient but also in counseling the unrelated donor appropriately. A graft compatible for DPB1 is more likely to result in disease relapse in this T-cell-depleted setting. Strikingly, patients with CML appeared almost inevitably to relapse if DPB1 is compatible. Equally striking is the finding that patients with ALL receiving a DPB1-matched graft have a significantly worse OS. These findings should be confirmed by other groups, and it is hoped that future studies will help to elucidate the role of DPB1.
Prepublished online as Blood First Edition Paper, October 18, 2005; DOI 10.1182/blood-2005-08-3121.
B.E.S., S.G.E.M., and J.A.M. contributed to the design of the research; N.H.R. contributed significantly to the number of patients recruited; B.E.S. and N.P.M. contributed to the data collection and generation; and B.E.S. analyzed the data.
All authors contributed to the writing of the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the United Kingdom transplantation teams who have provided us with the data and samples required to perform this study. We also thank Hazael Maldonado for help with the analysis of allele frequencies and Glenys Thomson for her input and ideas regarding the evolutionary history of DP.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal